Figure 4.
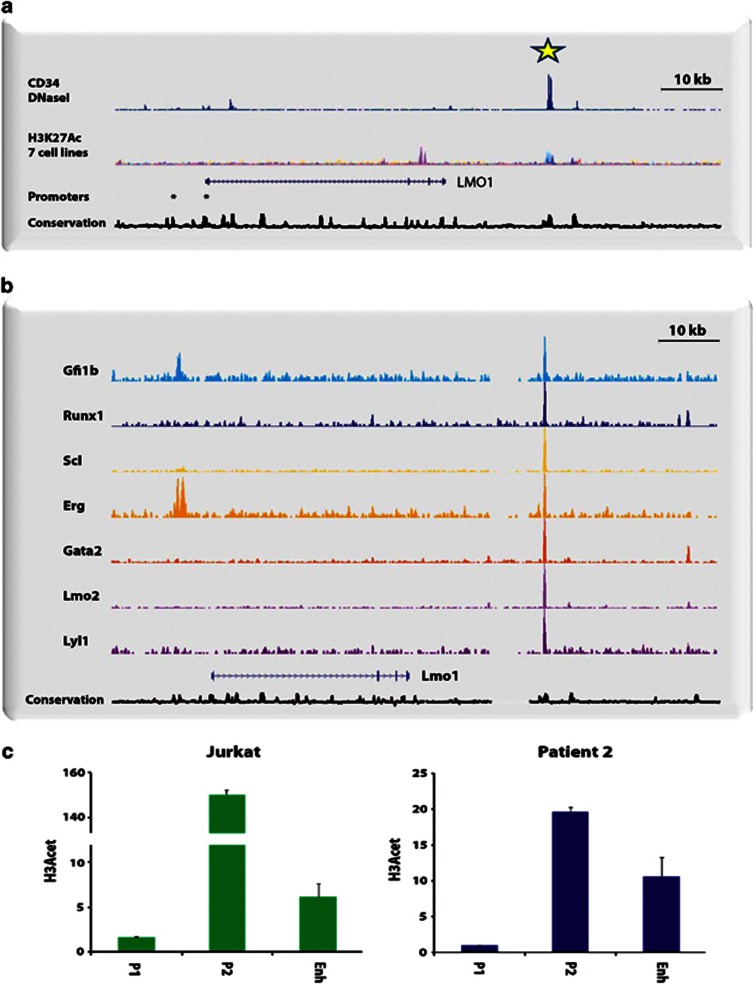
A conserved non-coding region 3' of LMO1 displays active chromatin marks and transcription factor binding in haematopoietic cells: (a) A highly conserved region ∼20 kb 3' to the final exon of the human LMO1 gene (+57 kb from the ATG immediately adjacent to the second exon) was identified in publicly available datasets from the ENCODE project, Roadmap Epigenomics project and UCSC goldenpath. This +57 region displayed marked DNaseI hypersensitivity, had histone H3 acetylation peaks in human umbilical vein endothelial cells (HUVEC) as well as the myeloid leukaemia cell line K562, and was bound by multiple transcription factors in a range of cell types. (b) Binding by multiple transcription factors including Gfi1b, Runx1, Gata2, Erg, Scl/Tal1, Lmo2 and Lyl1 to the mouse equivalent of the human LMO1 +57 region in the HPC7 murine blood progenitor cell line. (c) Real-time PCR analysis of H3 Acetylation ChIP material demonstrated minimal enrichment at promoter 1, but significant enrichment at both promoter 2 and the +57 region, in a nontranslocated LMO1-expressing T-ALL (Jurkat) cell line and a non-translocated T-ALL patient sample.