Abstract
Background
Members of the anaerobic thermophilic bacterial genus Caldicellulosiruptor are emerging candidates for consolidated bioprocessing (CBP) because they are capable of efficiently growing on biomass without conventional pretreatment. C. bescii produces primarily lactate, acetate and hydrogen as fermentation products, and while some Caldicellulosiruptor strains produce small amounts of ethanol C. bescii does not, making it an attractive background to examine the effects of metabolic engineering. The recent development of methods for genetic manipulation has set the stage for rational engineering of this genus for improved biofuel production. Here, we report the first targeted gene deletion, the gene encoding lactate dehydrogenase (ldh), for metabolic engineering of a member of this genus.
Results
A deletion of the C. bescii L-lactate dehydrogenase gene (ldh) was constructed on a non-replicating plasmid and introduced into the C. bescii chromosome by marker replacement. The resulting strain failed to produce detectable levels of lactate from cellobiose and maltose, instead increasing production of acetate and H2 by 21-34% relative to the wild type and ΔpyrFA parent strains. The same phenotype was observed on a real-world substrate – switchgrass (Panicum virgatum). Furthermore, the ldh deletion strain grew to a higher maximum optical density than the wild type on maltose and cellobiose, consistent with the prediction that the mutant would gain additional ATP with increased acetate production.
Conclusions
Deletion of ldh in C. bescii is the first use of recently developed genetic methods for metabolic engineering of these bacteria. This deletion resulted in a redirection of electron flow from production of lactate to acetate and hydrogen. New capabilities in metabolic engineering combined with intrinsic utilization of lignocellulosic materials position these organisms to provide a new paradigm for consolidated bioprocessing of fuels and other products from biomass.
Keywords: ldh, Metabolic engineering, Switchgrass, Biohydrogen, Caldicellulosiruptor
Background
Fuel production from plant biomass offers the opportunity to generate energy from a sustainable feedstock, reduce dependence on petroleum, and reduce the negative environmental impact of increased CO2 emissions. The major obstacle in the use of lignocellulosic feedstocks is the recalcitrance of the biomass itself. Plants have evolved to resist deconstruction by microbes, and plant cell wall components such as cellulose, hemicellulose, and lignin play a major role in recalcitrance [1-3]. Industrial conversion of plant biomass to fuels currently relies on thermal and chemical treatment of biomass to remove hemicellulose and lignin, followed by enzymatic hydrolysis to solubilize the plant cell walls to generate a fermentable substrate for fuel-producing organisms [4-6]. However, these methods add cost, produce hydrolysates that are toxic to microorganisms [7] and are destructive to the sugars in the biomass [8]. An alternative approach is to use consolidated bioprocessing (CBP), in which the fermentative organism is also responsible for production of the biomass-solubilizing enzymes [9]. Members of the genus Caldicellulosiruptor are able to ferment all primary C5 and C6 sugars from plant biomass and are the most thermophilic cellulolytic bacteria known, with growth temperature optima between 78°C ~ 80°C [10]. They can also grow on and degrade biomass containing high lignin content as well as highly crystalline cellulose without conventional pretreatment [11-13], raising the possibility of further economic improvement of biofuel production from plant biomass by reducing or eliminating the pretreatment step.
While Caldicellulosiruptor species are attractive platforms for fuel and chemical production from plant biomass, the dearth of genetic tools for this genus has prevented rational strain development. Recent advances have enabled genetic transformation of Caldicellulosiruptor bescii[14], opening the possibility of metabolic engineering for improved biofuel production in this genus.
During growth on glucose, C. bescii is predicted to utilize the Embden-Meyerhof glycolytic pathway to produce a combination of lactate and acetate + H2 + CO2 (see Figure 1 for a simplified pathway). As these are the only major fermentative products, pyruvate serves as the major metabolic branch point during fermentation, with carbon either being routed to lactate or acetyl-CoA and electrons being routed to lactate or H2. Thus, production of acetate is obligately coupled to H2 production to allow reoxidation of NADH and ferredoxin. While enteric bacteria such as Enterobacter aerogenes, Enterobacter cloacae and Escherichia coli produce 1 ~ 2 moles of H2 per mole of glucose [15,16], and Clostridium spp. can produce similar amounts [17-19], some hyperthermophiles such as Thermococcales spp., Pyrococcus furiosus, Thermotogales spp., and Caldicellulosiruptor spp. produce about 3 ~ 4 moles of H2 per mole of glucose [20-25]. Enterics typically utilize a formate-H2 lyase and Clostridium spp. use a ferredoxin-dependent hydrogenase to avoid the thermodynamically unfavorable formation of H2 from NADH, instead using other pathways such as ethanol or butanol production to reoxidize NADH. Thermotoga (and likely all the above-mentioned hyperthermophiles), on the other hand, use an electron bifurcating hydrogenase [26] to simultaneously oxidize NADH and ferredoxin to produce H2, using the thermodynamically favorable oxidation of ferredoxin to drive the unfavorable oxidation of NADH. This bifurcating hydrogenase allows a theoretical maximum yield of 4 moles of H2 per mole of glucose, whereas wild type E. coli has a theoretical maximum of 2 moles of H2 per mole of glucose. C. bescii encodes a putative bifurcating hydrogenase (Cbes1295-1299). This, combined with the native ability of C. bescii to catabolize plant biomass and the newly developed genetic transformation system, makes C. bescii a compelling platform to engineer high yield H2 production directly from plant biomass without conventional pretreatment.
Figure 1.
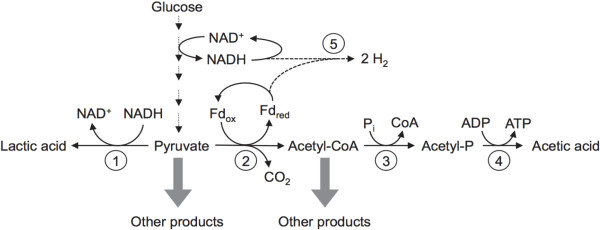
A simplified version of predicted metabolic pathways for fermentation, glycolysis and electron transfer in Caldicellulosiruptor bescii. (1) L-Lactate dehydrogenase; (2) Pyruvate-ferredoxin oxidoreductase; (3) Phosphotransacetylase; (4) Acetate kinase; (5) Bifurcating (reduced ferredoxin: NADH-dependent) hydrogenase. The thick grey arrows represent potential heterologous pathways that do not exist in Caldicellulosiruptor species but could be used for renewable fuel and chemical production.
We hypothesized that by developing the necessary tools to delete genes from the C. bescii chromosome [14,27,28], we would enable metabolic engineering to increase H2 production. Here, we demonstrate the utility of gene deletion in the pyruvate metabolic pathway for rational strain engineering of C. bescii while simultaneously creating a platform for further strain modification for advanced production of fuels and chemicals from renewable plant feedstocks.
Results
Deletion of lactate dehydrogenase (ldh) from the C. bescii chromosome
We recently reported a method for DNA transformation and marker replacement in Caldicellulosiruptor bescii based on uracil prototrophic selection [14,27,28]. C. bescii strain JWCB005 (ΔpyrFA, ura-/5-FOAR) contains a deletion of the pyrFA locus making the strain a uracil auxotroph resistant to 5-fluoroorotic acid (5-FOA) [27], allowing the use of pyrF as both a selectable and counter-selectable marker (Figure 2A). A deletion of the L-lactate dehydrogenase gene (Cbes1918) was constructed by fusing the 5′ and 3′ flanking regions of the ldh gene and cloning the fused product into a non-replicating plasmid vector, resulting in plasmid pDCW121. This vector also contains the wild type pyrF allele under the transcriptional control of a ribosomal protein gene promoter (Cbes2105, 30S ribosomal protein S30EA), allowing both positive (uracil prototrophy) and negative (5-FOA sensitivity) selection. Plasmid pDCW121 was transformed into C. bescii JWCB005 selecting uracil prototrophy resulting from plasmid recombination into the targeted region, followed by counter-selecting 5-FOA resistance (resulting from plasmid excision). The resulting strain, JWCB017, contained a deletion of the ldh wild type gene in the chromosome. To confirm the ldh deletion in JWCB017, the region of the ldh locus was amplified by PCR using primers outside of the plasmid regions of homology used to construct the deletion (Figure 2B). The wild type and the ΔpyrFA strain (JWCB005) gave the same expected 3.0 kb bands, while PCR from JWCB017 resulted in the smaller 2.0 kb band, as predicted. The PCR product was also sequenced to verify that the deletion in the chromosome was the same as that constructed on the plasmid.
Figure 2.
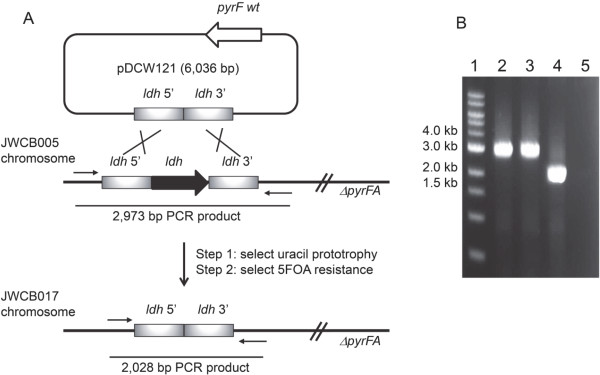
Deletion of the ldh gene in C. bescii. (A) A deletion cassette for the ldh gene was constructed in a non-replicating plasmid that contained a wild type copy of the pyrF gene, resulting in plasmid pDCW121. The cassette contained ldh 5′ and 3’ flanking DNA fragments. The plasmid was transformed into JWCB005, and uracil prototrophs were selected (resulting from plasmid insertion). Counter-selection with 5-FOA selected for strains that underwent a second recombination event, resulting in deletion of the marker and ldh to produce strain JWCB017. (B) Agarose gel showing PCR products amplified from the ldh locus in the wild type (lane 2), JWCB005 (ΔpyrFA parent strain, lane 3) and JWCB017 (ΔpyrFA Δldh, lane 4). Lane 1: 1 kb DNA ladder; Lane 5: no template PCR control. Expected bands: wild type ldh locus – 3 kb; ldh deletion – 2.0 kb.
Deletion of ldh eliminates lactate production and increases acetate and H2 production
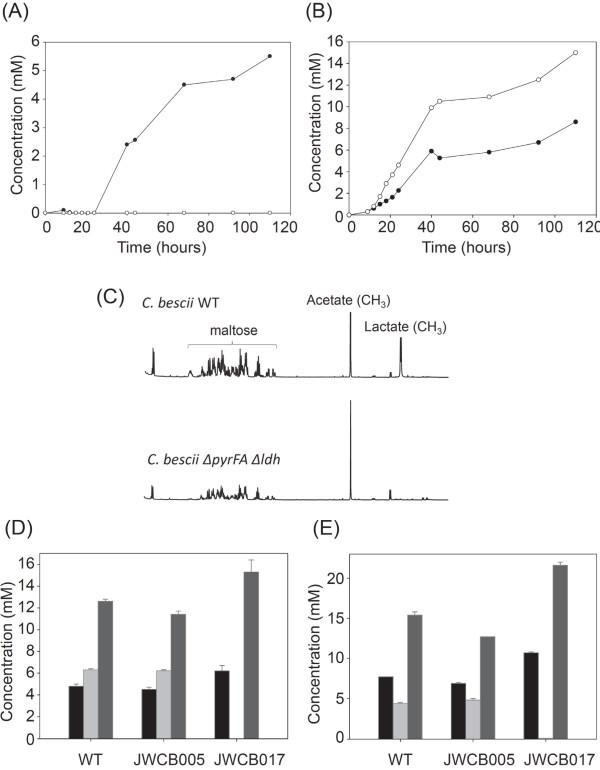
Cbes1918 is the only predicted lactate dehydrogenase gene encoded in the C. bescii genome. To confirm that this gene is solely responsible for the production of lactate in C. bescii, wild type, JWCB005 and JWCB017 were grown on 0.5% maltose, and fermentation products were analyzed by high-performance liquid chromatography (HPLC) (Figure 3AB) and nuclear magnetic resonance (NMR) analysis (Figure 3C). No lactate was detected in the mutant by either method, as compared to approximately 5.0 mM lactate from the wild-type and parental strains.
Figure 3.
Fermentation products by C. bescii mutant strains. (A) Lactic acid and (B) acetic acid production were measured during growth on 0.5% maltose by the parent strain (JWCB005 ΔpyrFA; filled circles) and JWCB017 (ΔpyrFA Δldh; open circles). (C) Production of lactic and acetic acids by wild type and JWCB017 were further measured by NMR analysis after 48 hours incubation. (D) End products of C. bescii wild-type and mutant strains were measured by HPLC on cellobiose after 30 hours incubation, and (E) switchgrass after 120 hours incubation. Acetate, Black; Lactate, Light gray; Hydrogen, Dark gray.
To compare the production of lactate, acetate and hydrogen, C. bescii wild-type and mutant strains were grown in LOD medium [29] with soluble cellodextrans (cellobiose) or plant biomass (switchgrass) as carbon source. When grown on 0.5% cellobiose for 30 hours, JWCB017 showed 29% and 21% more acetate production and 37% and 34% more hydrogen production than wild type and parent strains, respectively (Figure 3D). Cells grown for 120 hours on LOD medium supplemented with 0.5% switchgrass as the sole carbon source showed a similar profile to that on cellobiose, with the Δldh strain producing 38% and 40% more acetate and 55% and 70% more hydrogen than wild-type and parent strains (Figure 3E).
Growth yield increases upon deletion of ldh
Growth of JWCB017 was compared to the wild type and parental strains in defined media [29] supplemented with either 0.5% maltose or 0.5% cellobiose. While growth of the ΔpyrFA parent strain on both maltose (Figure 4A) and cellobiose (Figure 4B) was indistinguishable from the wild type [29], the JWCB017 mutant strain reached a 34-53% higher final optical density than the wild type and parent. Interestingly, while the growth rate was comparable, the exponential growth phase of JWCB017 was extended resulting in higher cell densities.
Figure 4.
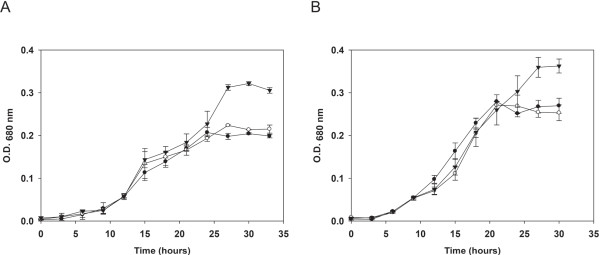
Growth (O.D.680 nm) comparison of wild-type and mutant strains. (A) 0.5% of maltose and (B) 0.5% cellobiose as the carbon source; filled circles, Wild-type; open circles, ΔpyrFA (JWCB005); filled triangles, ΔpyrFA Δldh (JWCB017). Error bars based on three biologically independent experiments.
Discussion
We have built upon recent advances in the genetic manipulation of Caldicellulosiruptor[14,27,28] to delete the gene encoding lactate dehydrogenase. While the wild type strain produced roughly equimolar amounts of acetate and lactate, the JWCB017 mutant strain no longer produced lactate, instead rerouting carbon and electron flux to acetate and H2, respectively. JWCB001 (~1.8 mol/mol of glucose) and JWCB005 (~1.7 mol/mol of glucose) appeared a bit lower in hydrogen yield than reported values for C. saccharolyticus (~2.5 mol/mol of glucose) [22]. However, in this report, C. saccharolyticus was grown in culture media with added yeast extract, which improves yields [22]. JWCB017 (~3.4 mol/mol of glucose) was more than that reported for C. saccharolyticus. Yield and titer of acetate and H2 were increased in the C. bescii ldh deletion strain from both model soluble substrates and real-world plant biomass. A similar approach has been applied to other thermophilic biomass-degrading bacteria, including xylanolytic Thermoanaerobacterium saccharolyticum and cellulolytic Clostridium thermocellum[30-32], with the goal of increasing ethanol production.
Members of the genus Caldicellulosiuptor offer special advantages for biomass conversion to products of interest in that they are hyperthermophiles with optimal growth temperatures between 70°C ~ 80°C and they are capable of using biomass without conventional pretreatment.
Interestingly, deletion of ldh resulted in a higher cell yield and longer exponential growth phase relative to the wild type. The increase in cell density is likely caused by an increase in acetate production, which should increase ATP production per glucose via acetate kinase providing more energy for biosynthesis and growth. The evolutionary pressures that selected for maintenance of ldh are not clear, though it may be related to the partial pressure of H2 found in the environment. Further, the molecular mechanism by which C. bescii switches from production of acetate + H2 to lactate is unknown. C. thermocellum encodes a lactate dehydrogenase that is allosterically activated by fructose-1,6-bisphosphate [33], such that lactate is only produced when the rate of substrate uptake exceeds glycolytic flux. It would be interesting to examine whether C. bescii utilizes a similar mechanism for flux control at the pyruvate node of glycolysis. Independent of mechanism, the fact that C. bescii JWCB017 grows to a higher density without an obvious effect on growth rate suggests that further engineered strains may be able to compete well with the wild type strain and thrive in an industrial setting.
Recent progress in genetic tool development opened the possibility of more advanced metabolic engineering strategies to increase the utility of C. bescii for industrial applications. In addition to the construction of gene deletions, this will enable gene insertion into the chromosome (so called gene knock-ins), simplifying the process of heterologous gene expression by eliminating the need for plasmid maintenance and increasing the number of genes that can be stably expressed. Thus, we have created a new platform for rational strain design for lignocellulosic bioconversion, enabling future efforts to increase the titer of H2, express heterologous pathways for production of liquid fuels and chemicals, increase robustness, and improve upon the native ability of Caldicellulosiruptor species to deconstruct and convert biomass without conventional pretreatment.
Characterization of the JWCB017 mutant also sheds light on the basic physiology of C. bescii, which will inform future metabolic modeling and engineering efforts. For instance, this strain has now been engineered to produce only acetate and H2 from sugars, providing further evidence that C. bescii uses a bifurcating hydrogenase to funnel all the electrons to H2. Examination of the C. bescii genome sequence reveals that glycolysis likely yields NADH from glyceraldehyde-3-phosphate oxidation, based on the presence of glyceraldehyde-3-phosphate dehydrogenase (Cbes1406) and lack of glyceraldehyde-3-phosphate: ferredoxin oxidoreductase. Conversion of pyruvate to acetyl-CoA, on the other hand, presumably reduces ferredoxin, based on the presence of pyruvate: ferredoxin oxidoreductase (Cbes0874-0877) and the lack of pyruvate dehydrogenase and pyruvate-formate lyase. Because H2 evolution from NADH is thermodynamically unfavorable except in extremely low H2 partial pressures [34], this implies that the favorable production of H2 from ferredoxin drives the unfavorable NADH-dependent H2 production. Further genetic modification will increase our understanding of metabolic flux in C. bescii, allowing better metabolic models and further informing metabolic engineering efforts.
Conclusions
Here we show the first application of recently developed genetic methods for metabolic engineering of a member of the genus Caldicellulosiruptor. The method for creating a deletion of the ldh gene in the C. bescii chromosome was efficient enough to allow targeted marker replacement using non-replicating plasmids. The resulting mutant grew to a higher cell density and produced more hydrogen than the wild-type strain. Using the tools developed here, C. bescii JWCB017 will serve as a platform for additional rational strain engineering for production of fuels and chemicals from lignocellulosic feedstocks.
Methods
Strains, growth conditions and molecular techniques
A spontaneous mutant containing a deletion within the pyrFA locus of C. bescii, JWCB005 [27,28], was used in this study to select transformants. C. bescii strains were grown in modified DSMZ516 medium or LOD (low osmolality defined growth medium) [29] containing 0.5% maltose as carbon source, final pH 7.0. Liquid cultures were grown from a 0.5% inoculum or a single colony and incubated at 75°C in anaerobic culture bottles degassed with five cycles of vacuum and argon. A solid medium was prepared by mixing an equal volume of liquid medium at a 2× concentration with the same volume of (wt/vol) agar, 3.6% (Difco, Sparks, MD) that had been previously autoclaved. Both solutions were maintained at 70°C and poured into petri dishes immediately after mixing. A series of dilutions of this culture were mixed with 4 ml of soft top agar (1.5% of agar) and poured across the top of the solid agar medium. The plates were degassed with five cycles of vacuum and argon and incubated at 75°C for 4 days in anaerobic jars. E. coli DH5α was used to prepare plasmid DNA. Cells were grown in LB broth supplemented with apramycin (50 μg/ml). Plasmid DNA was isolated using a Qiagen Mini-prep Kit (QIAGEN inc., Valenica, CA). A complete list of strains, plasmids, and primers used in this study is shown in Tables 1 and 2.
Table 1.
Plasmids and C. bescii strains (JWCB) used in this study
Table 2.
Oligonucleotides used in this study
| Primer | Sequence 5’- 3’ |
|---|---|
| DC081 |
TCCAATGATCGAAGTTAGGCTGGT |
| DC348 |
GAATTCTCTGACGCTCAGTGGAACGAA |
| DC349 |
GAAAACAAATGGGCTTGGGAGGATAGGAGGCTGT |
| DC350 |
TGGGCTTGGGAGGATAGGAGGCTGTCTAAAAACAA |
| DC351 |
TGCCAAGATATGAAATGAGAACT |
| DC356 | CGTCTCATCTGTGCATATGGACAGTTATAATCCCAAAAGGAGGATTGGATCC |
Construction of pDCW121
To construct a plasmid for deletion of the ldh gene (Cbes1918), three cloning steps including overlapping polymerase chain reactions were used. All PCR amplifications were performed using Pfu Turbo DNA polymerase (Agilent Tech., Santa Clara, CA). A 1,009 bp fragment containing a KpnI site upstream of the ldh gene was amplified using primers DC348 and DC349. A 1,011 bp fragment containing an EcoRI site downstream of ldh, was amplified using primers DC350 and DC351. The two fragments were joined by overlapping PCR using primers DC348 and DC351 to generate a 2,020 bp product that was cloned into pDCW88 [28] using the Kpnl and EcoRI sites. The resulting plasmid, pDCW121, was transformed into E. coli DH5α by an electrotransformation via a single electric pulse (1.8 kV, 25 μF and 200 Ω) in a pre-chilled 1 mm cuvette using a Bio-Rad gene Pulser (Bio-Rad, Hercules, CA). Transformants were selected on LB solid medium containing apramycin (50 μg/ml final).
Competent cells, transformation and mutant selection in C. bescii
To prepare competent cells, a 50 ml culture of JWCB005 was grown in LOD minimal medium at 75°C for 18 hours (to mid exponential phase) and 25 ml of the culture was used to inoculate a 500 ml culture of LOD (low osmolarity defined growth medium) supplemented with 40 μM uracil and a mixture of 19 amino acids (5% inoculum, v/v) [29]. The 500 ml culture was incubated at 75°C for 5 hours and cooled to room temperature for 1 hr. Cells were harvested by centrifugation (6000 × g, 20 min) at 25°C and washed three times with 50 ml of pre-chilled 10% sucrose. After the third wash, the cell pellet was resuspended in 50 μl of pre-chilled 10% sucrose in a microcentrifuge tube and stored at −80°C until needed. Before transformation, plasmids from E. coli cells were methylated in vitro with C. bescii methyltransferase (M.CbeI, [14]) and methylated plasmid DNAs (0.5-1.0 μg) were added to the competent cells, gently mixed and incubated for 10 minutes in ice. Electrotransformation of the cell/DNA mixture was performed via single electric pulse (1.8 kV, 25 μF and 350 Ω) in a pre-chilled 1 mm cuvette using a Bio-Rad gene Pulser (Bio-Rad, Hercules, CA). After pulsing, cells were inoculated into 10 ml of LOC medium (low osmolarity complex growth medium, [29]) and incubated for 4 hours at 75°C. 100 μl of the culture was transferred into 20 ml of defined medium without uracil. After 18 hours incubation at 75°C, cells were harvested by centrifugation (at 6000 × g for 20 min) and resuspended in 1 ml of 1× basal salts. 100 microliters of the cell suspension was plated onto solid defined media with 40 μM uracil and 8 mM 5-FOA (5- fluoroorotic acid monohydrate).
Analytical techniques for determining fermentation end products
Batch fermentations were conducted in stoppered 125 ml serum bottles containing 50 ml LOD medium with 5 g/l maltose, cellobiose or switchgrass. Cultures of JWCB005 and JWCB017 were supplemented with 40 μM uracil. Triplicate bottles were inoculated with a fresh 2% (v/v) inoculum and incubated at 75°C without shaking. Total cell dry weight (CDW) was determined by concentrating 25 ml of each culture on dried, preweighed 47 mm Supor membrane filters (0.45, Pall Corp., Ann Arbor, MI) and washed with 10 ml of ddH2O. Cell retentates were dried for 16 hours at 85°C and weighed on an analytical balance. Culture supernatants were analyzed via HPLC using a Waters Breeze 2 system (Waters Chromatography, Milford, MA) operated under isocratic conditions at 0.6 ml/min with 5 mM H2SO4 as a mobile phase. Analytes were separated on an Aminex HPX-87H column (Bio-Rad Laboratories, Hercules, CA) at 60°C and monitored via refractive index (RI) using a Waters 2414 RI detector. Total peak areas were integrated using Waters Breeze 2 software and compared against peak areas and retention times of known standards for each analyte of interest. H2 was measured using an Agilent Technologies 6850 Series II Gas Chromatograph equipped with a thermal conductivity detector at 190°C with a N2 reference flow and a HP-PLOT U Column (30 m * 0.32 mm). To measure organic acid production, Nuclear magnetic resonance (NMR) analysis was performed. One-dimensional 1H-NMR spectra were recorded at 298 K with a Varian Inova-NMR operating at 600 MHz for 1H and equipped with a 5-mm NMR cold probe. Samples (500 μL) of cell free culture media were mixed with 150 μL of D2O as internal lock and immediately analyzed. 128 scans were recorded for each sample using a pre-saturation method to suppress the water resonance. The amounts of the most abundant components in the samples were calculated by integration of the proton signals in the spectra. The data were normalized to the amount of acetic acid in each sample.
Biomass preparation
Air-dried switchgrass (Panicum virgatum, Alamo variety) was reduced to 60 mesh using a Wiley Mini-Mill (Thomas Scientific, Swedesboro, NJ, USA). The ground switchgrass was subjected to a hot water treatment similar to that described by Yang et. al. [12] however the biomass was boiled in distilled H2O (2% w/v) for 1 hour rather than treating overnight at 75°C. The switchgrass was then washed and dried overnight at 50°C before dispensing into serum bottles as previously described [12].
Abbreviations
ldh: L-lactate dehydrogenase; CBP: Consolidated bioprocessing; 5-FOA: 5-Fluoroorotic aicd; HPLC: High-performance liquid chromatography; NMR: Nuclear magnetic resonance; LOD: Low osmolarity defined growth medium; LOC: Low osmolarity complex growth medium; CDW: Cell dry weight; RI: Refractive index; GC: Gas chromatography.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MC and DC designed and carried out the genetic and growth experiments, analyzed results and participated in the writing of the manuscript. JGE and AMG designed and carried out the analysis of fermentation products and participated in writing the manuscript. JW participated in design of the study, coordination of the work, and writing of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Minseok Cha, Email: mcha@uga.edu.
Daehwan Chung, Email: chung301@gmail.com.
James G Elkins, Email: elkinsjg@ornl.gov.
Adam M Guss, Email: gussam@ornl.gov.
Janet Westpheling, Email: janwest@uga.edu.
Acknowledgements
We thank Jennifer Copeland for outstanding technical assistance, Bob Kelly and Sara Blumer-Schuette for providing the Caldicellulosiruptor species, Maria Pena and William York for NMR analysis, Li Tan and Debra Mohnen for assistance with the HPLC analysis and Jonathan Mielenz for providing the switchgrass used in this study. The BioEnergy Science Center is a U.S. Department of Energy Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science.
References
- Himmel ME, Ding SY, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- Wilson DB. Three microbial strategies for plant cell wall degradation. Ann N Y Acad Sci. 2008;1125:289–297. doi: 10.1196/annals.1419.026. [DOI] [PubMed] [Google Scholar]
- McCann MC, Carpita NC. Designing the deconstruction of plant cell walls. Curr Opin Plant Biol. 2008;11:314–320. doi: 10.1016/j.pbi.2008.04.001. [DOI] [PubMed] [Google Scholar]
- Zhang YH, Ding SY, Mielenz JR, Cui JB, Elander RT, Laser M, Himmel ME, McMillan JR, Lynd LR. Fractionating recalcitrant lignocellulose at modest reaction conditions. Biotechnol Bioeng. 2007;97:214–223. doi: 10.1002/bit.21386. [DOI] [PubMed] [Google Scholar]
- Negro MJ, Manzanares P, Ballesteros I, Oliva JM, Cabanas A, Ballesteros M. Hydrothermal pretreatment conditions to enhance ethanol production from poplar biomass. Appl Biochem Biotechnol. 2003;105–108:87–100. doi: 10.1385/abab:105:1-3:87. [DOI] [PubMed] [Google Scholar]
- Wyman CE. What is (and is not) vital to advancing cellulosic ethanol. Trends Biotechnol. 2007;25:153–157. doi: 10.1016/j.tibtech.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Klinke HB, Thomsen AB, Ahring BK. Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl Microbiol Biotechnol. 2004;66:10–26. doi: 10.1007/s00253-004-1642-2. [DOI] [PubMed] [Google Scholar]
- Barakat A, Monlau F, Steyer JP, Carrere H. Effect of lignin-derived and furan compounds found in lignocellulosic hydrolysates on biomethane production. Bioresour Technol. 2012;104:90–99. doi: 10.1016/j.biortech.2011.10.060. [DOI] [PubMed] [Google Scholar]
- Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton-Brehm SD, Mosher JJ, Vishnivetskaya T, Podar M, Carroll S, Allman S, Phelps TJ, Keller M, Elkins JG. Caldicellulosiruptor obsidiansis sp. nov., an anaerobic, extremely thermophilic, cellulolytic bacterium isolated from Obsidian Pool, Yellowstone National Park. Appl Environ Microbiol. 2010;76:1014–1020. doi: 10.1128/AEM.01903-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer-Schuette SE, Kataeva I, Westpheling J, Adams MW, Kelly RM. Extremely thermophilic microorganisms for biomass conversion: status and prospects. Curr Opin Biotechnol. 2008;19:210–217. doi: 10.1016/j.copbio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Yang SJ, Kataeva I, Hamilton-Brehm SD, Engle NL, Tschaplinski TJ, Doeppke C, Davis M, Westpheling J, Adams MW. Efficient degradation of lignocellulosic plant biomass, without pretreatment, by the thermophilic anaerobe “Anaerocellum thermophilum” DSM 6725. Appl Environ Microbiol. 2009;75:4762–4769. doi: 10.1128/AEM.00236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumer-Schuette SE, Giannone RJ, Zurawski JV, Ozdemir I, Ma Q, Yin Y, Xu Y, Kataeva I, Poole FL 2nd, Adams MW. Caldicellulosiruptor core and pangenomes reveal determinants for noncellulosomal thermophilic deconstruction of plant biomass. J Bacteriol. 2012;194:4015–4028. doi: 10.1128/JB.00266-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D, Farkas J, Huddleston JR, Olivar E, Westpheling J. Methylation by a unique alpha-class N4-cytosine methyltransferase is required for DNA transformation of caldicellulosiruptor bescii DSM6725. PLoS One. 2012;7:e43844. doi: 10.1371/journal.pone.0043844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanisho S, Kamiya N, Wakao N. Hydrogen evolution of Enterobacter aerogenes depending on culture pH: mechanism of hydrogen evolution from NADH by means of membrane-bound hydrogenase. Biochim Biophys Acta. 1989;973:1–6. doi: 10.1016/S0005-2728(89)80393-7. [DOI] [PubMed] [Google Scholar]
- Yoshida A, Nishimura T, Kawaguchi H, Inui M, Yukawa H. Enhanced hydrogen production from glucose using ldh- and frd-inactivated Escherichia coli strains. Appl Microbiol Biotechnol. 2006;73:67–72. doi: 10.1007/s00253-006-0456-9. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhu Y, Yang ST. Construction and characterization of ack deleted mutant of Clostridium tyrobutyricum for enhanced butyric acid and hydrogen production. Biotechnol Prog. 2006;22:1265–1275. doi: 10.1021/bp060082g. [DOI] [PubMed] [Google Scholar]
- Collet C, Girbal L, Peringer P, Schwitzguebel JP, Soucaille P. Metabolism of lactose by Clostridium thermolacticum growing in continuous culture. Arch Microbiol. 2006;185:331–339. doi: 10.1007/s00203-006-0098-4. [DOI] [PubMed] [Google Scholar]
- Chin HL, Chen ZS, Chou CP. Fedbatch operation using Clostridium acetobutylicum suspension culture as biocatalyst for enhancing hydrogen production. Biotechnol Prog. 2003;19:383–388. doi: 10.1021/bp0200604. [DOI] [PubMed] [Google Scholar]
- Kanai T, Imanaka H, Nakajima A, Uwamori K, Omori Y, Fukui T, Atomi H, Imanaka T. Continuous hydrogen production by the hyperthermophilic archaeon, Thermococcus kodakaraensis KOD1. J Biotechnol. 2005;116:271–282. doi: 10.1016/j.jbiotec.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Kadar Z, de Vrije T, van Noorden GE, Budde MA, Szengyel Z, Reczey K, Claassen PA. Yields from glucose, xylose, and paper sludge hydrolysate during hydrogen production by the extreme thermophile Caldicellulosiruptor saccharolyticus. Appl Biochem Biotechnol. 2004;114:497–508. doi: 10.1385/ABAB:114:1-3:497. [DOI] [PubMed] [Google Scholar]
- Kadar Z, de Vrije T, Mars AE, Budde MA, Lai MH, Dijkema C, de Waard P, Claassen PA. Glycolytic pathway and hydrogen yield studies of the extreme thermophile Caldicellulosiruptor saccharolyticus. Appl Microbiol Biotechnol. 2007;74:1358–1367. doi: 10.1007/s00253-006-0783-x. [DOI] [PubMed] [Google Scholar]
- Schroder C, Selig M, Schonheit P. Glucose fermentation to acetate, CO2 and H2 in the anaerobic hyperthermophilic eubacterium thermotoga-maritima - involvement of the embden-meyerhof pathway. Arch Microbiol. 1994;161:460–470. [Google Scholar]
- Willquist K, Zeidan AA, van Niel EW. Physiological characteristics of the extreme thermophile Caldicellulosiruptor saccharolyticus: an efficient hydrogen cell factory. Microb Cell Fact. 2010;9:89. doi: 10.1186/1475-2859-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schicho RN, Ma K, Adams MW, Kelly RM. Bioenergetics of sulfur reduction in the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1993;175:1823–1830. doi: 10.1128/jb.175.6.1823-1830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schut GJ, Adams MW. The iron-hydrogenase of Thermotoga maritima utilizes ferredoxin and NADH synergistically: a new perspective on anaerobic hydrogen production. J Bacteriol. 2009;191:4451–4457. doi: 10.1128/JB.01582-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D, Cha M, Farkas J, Westpheling J. Construction of a stable replicating shuttle vector for caldicellulosiruptor species: Use for extending genetic methodologies to other members of this genus. PLoS One. 2013;8:e62881. doi: 10.1371/journal.pone.0062881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung D, Farkas J, Westpheling J. Overcoming restriction as a barrier to DNA transformation in Caldicellulosiruptor species results in efficient marker replacement with non-replicating plasmid vectors. Biotech biofuels. 2013;6:82. doi: 10.1186/1754-6834-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas J, Chung D, Cha M, Copeland J, Grayeski P, Westpheling J. Improved growth media and culture techniques for genetic analysis and assessment of biomass utilization by Caldicellulosiruptor bescii. J Ind Microbiol Biotechnol. 2013;40:41–49. doi: 10.1007/s10295-012-1202-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AJ, Podkaminer KK, Desai SG, Bardsley JS, Rogers SR, Thorne PG, Hogsett DA, Lynd LR. Metabolic engineering of a thermophilic bacterium to produce ethanol at high yield. Proc Natl Acad Sci USA. 2008;105:13769–13774. doi: 10.1073/pnas.0801266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyros DA, Tripathi SA, Barrett TF, Rogers SR, Feinberg LF, Olson DG, Foden JM, Miller BB, Lynd LR, Hogsett DA, Caiazza NC. High ethanol titers from cellulose by using metabolically engineered thermophilic, anaerobic microbes. Appl Environ Microbiol. 2011;77:8288–8294. doi: 10.1128/AEM.00646-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi SA, Olson DG, Argyros DA, Miller BB, Barrett TF, Murphy DM, McCool JD, Warner AK, Rajgarhia VB, Lynd LR. Development of pyrF-based genetic system for targeted gene deletion in Clostridium thermocellum and creation of a pta mutant. Appl Environ Microbiol. 2010;76:6591–6599. doi: 10.1128/AEM.01484-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan M, Yilmaz EI, Lynd LR, Ozcengiz G. Cloning and expression of the Clostridium thermocellum L-lactate dehydrogenase gene in Escherichia coli and enzyme characterization. Can J Microbiol. 2004;50:845–851. doi: 10.1139/w04-071. [DOI] [PubMed] [Google Scholar]
- Thauer RK, Jungermann K, Decker K. Energy conversion in chemotrophic anaerobic bacteria. Bacteriol Rev. 1971;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetlichnyi VA, Svetlichnaya TP, Chernykh NA, Zavarzin GA. Anaerocellum-thermophilum Gen-Nov Sp-Nov - an extremely thermophilic cellulolytic eubacterium isolated from Hot-springs in the valley of geysers. Microbiol. 1990;59:598–604. [Google Scholar]