Abstract
The X-ray crystal structure, NMR binding studies, and enzyme activity of silver(I) metallated hen egg white lysozyme are presented. Primary bonding of silver is observed through His15 with secondary bonding interactions coming from nearby Arg14 and Asp87. A covalently bound nitrate completes a four coordinate binding pocket.
Although the therapeutic properties of silver(I) have been known for centuries its use in modern medicine remains largely limited to the topical treatment of burn wounds.1 Recently there has been a surge in research related to expanding the use of silver(I) beyond its typical mode of use. Such research includes the use of silver(I) N-heterocyclic carbene complexes as nebulized antibiotics for the treatment of cystic fibrosis lung infections2–5 and the systemic delivery of metallic silver nanoparticles and silver(I) complex loaded nanoparticles for the treatment of cancers.6–11 The application of silver based compounds for systemic diseases reveals a general lack of knowledge on how silver(I) interacts with biological systems. In order to understand the biochemical mechanism of silver it is imperative to identify the way that the metal interacts with biomolecules including what chemical motifs in proteins are able to bind silver(I), their specificity and affinity. As a first approach investigation of the biological interaction of silver(I) with proteins we report the metallation of hen egg white lysozyme (HEWL) with silver(I).
Lysozyme was selected as a model for probing the structural interactions of silver(I) ions with proteins due to its stability, relative propensity to crystallize under a range of conditions, and previous use in similar studies utilizing medicinal metal complexes.12–15 A prior study investigating the interaction of silver ions with chemically crosslinked lysozyme crystals has been reported, however no X-ray structure was deposited.16 Perhaps the greatest challenge in the crystallization of silver with HEWL, and proteins in general, is identifying conditions free of halogens and phosphates in order to avoid precipitation of the metal prior to its coordinating to the protein. Despite avoiding chloride containing salts as precipitants, initial protein crystallization screens with silver proved problematic due to intrinsically associated chloride present in commercially obtained HEWL (EMD OmniPur®). In order to address this issue, a bulk recrystallization of HEWL from a nitrate containing solution was performed prior to direct co-crystallization with silver.17 Crystals of silver(I) metallated HEWL (Fig. 1) suitable for X-ray diffraction studies (PDB 3RU5) were grown via sitting drop vapor diffusion by mixing a 10 μL solution of 5 mM nitrate recrystallized HEWL (unbuffered at pH between 4 and 5) with an equal volume of reservoir solution containing 5 mM silver(I) 3-hydroxybutyrate, 3% w/v NaNO3 and 25% v/v ethylene glycol. Colorless tetragonal crystals averaging 0.3 mm in their maximum dimension were obtained after three days growth in the absence of light. The position of the silver(I) ion was confirmed by comparison with a structure of metal free nitrate recrystallized HEWL.
Fig. 1.
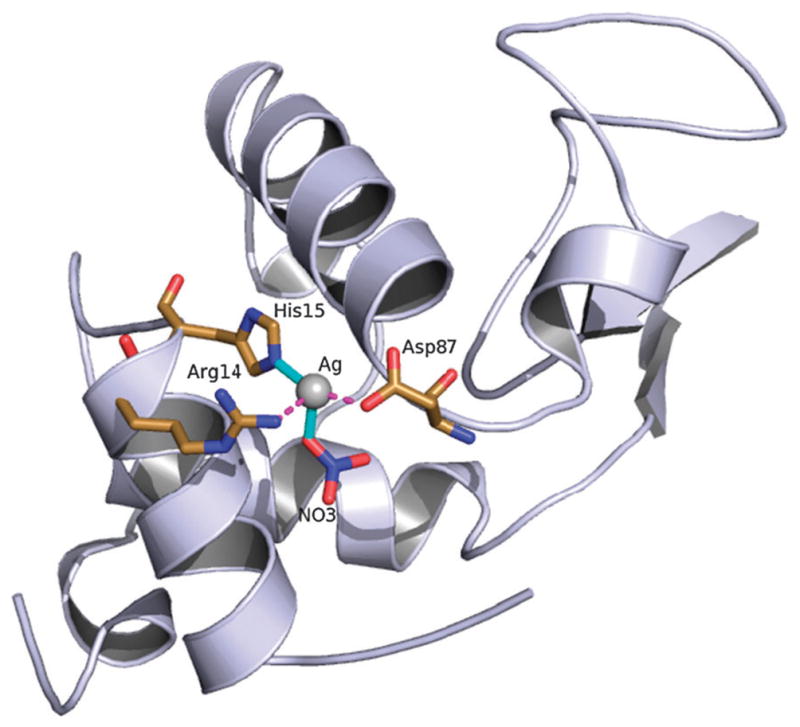
Full view of silver(I) metallated HEWL. Sodium atoms, nitrates, and ethylene glycols found from the difference map as well as solvent waters have been removed for clarity.
X-Ray data were collected at 1.54 Å using a Bruker Kappa APEX II Duo equipped with an ImuS micro-focus source with QUAZAR optics to a resolution of 1.35 Å. Data integration and scaling were performed using the APEX II software suite.18 An initial solution was obtained by molecular replacement using phaser and PDB 3KAM as a model.13 Additional model building and refinement were accomplished using Coot and Refmac5.19–23
Elucidation of the selected crystal shows that silver(I) binds with high occupancy to HEWL primarily through residue His15 with a Ag-Nε2 distance of ~2.1 Å. Of the 24 metal containing HEWL structures deposited on the PDB just over half include metals bound to lysozyme through His15. These 13 structures only represent six transition metals (Au(I), Mn(II), Pt(II), Re(I), Ru(II) and Rh(III)) the majority of which were achieved through metal soaks. From our studies we have shown that silver(I) will form a complex with HEWL via direct crystallization with the metal.11–14,24 The structure of Ag(I)-HEWL also contains secondary bonding interactions through residues Arg14 and Asp87 at ~2.5 and 2.3 Å respectively. The orientation of the side chains of both of these residues are significantly different than observed in the metal free nitrate recrystallized HEWL demonstrating the affinity of the protein for silver(I). However, given the low pH of the crystal drop (pH between 4 and 5) and a pKa value of ~12.5 for the arginine side chain it is likely that the proximity of Arg14 to the silver(I) ion is due more to an ionic interaction with the covalently bound nitrate (Ag–O distance of ~2.0Å) that completes the four coordinate binding site (Fig. 2).
Fig. 2.
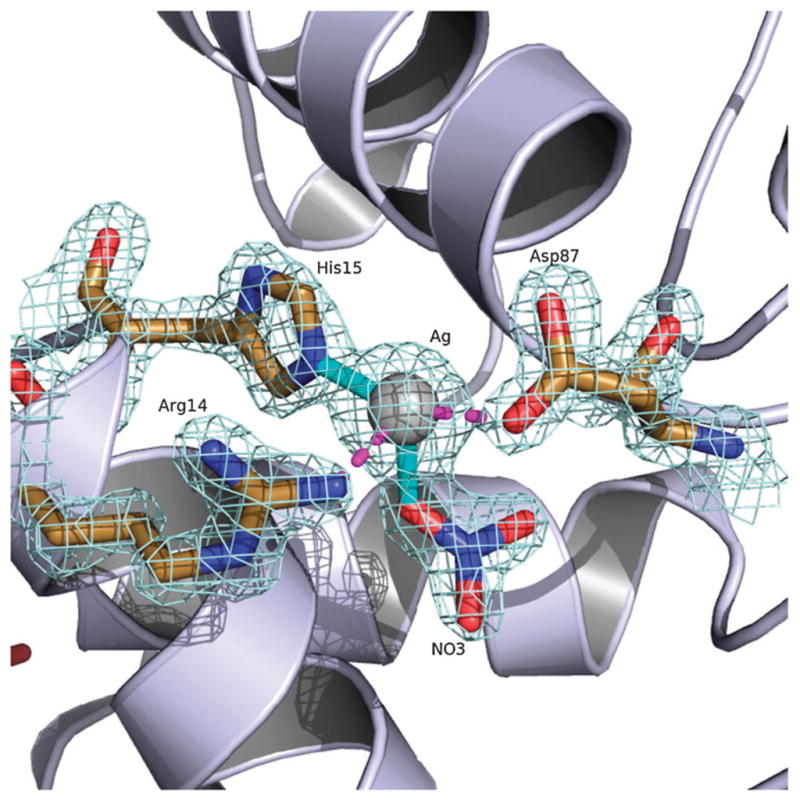
The silver(I) binding site of HEWL. Covalent bonds are shown in solid cyan and secondary interactions shown in dashed magenta. 2Fo–Fc electron density map is contoured at 1.5 σ.
Validation of the HEWL-silver ligation state in solution was performed by monitoring NMR chemical shift values of the protein upon metal titration. Natural abundance 13C and 15N HSQC25 spectra were collected on nitrate recrystallized HEWL at ~10 mM at 30 °C in 100% D2O. Under these conditions good quality 2D 13C and 15N spectra can be collected in 4 and 24 h respectively at 750 MHz with a triple resonance cold-probe (see supporting information). Resonances of the free protein were assigned based upon known chemical shifts for HEWL.26 Only His15 Cε1 and Cδ2 experience significant chemical shift changes in the aromatic 13C HSQC, consistent with the strong covalent bond to His15 Nε2 observed in the crystal structure.
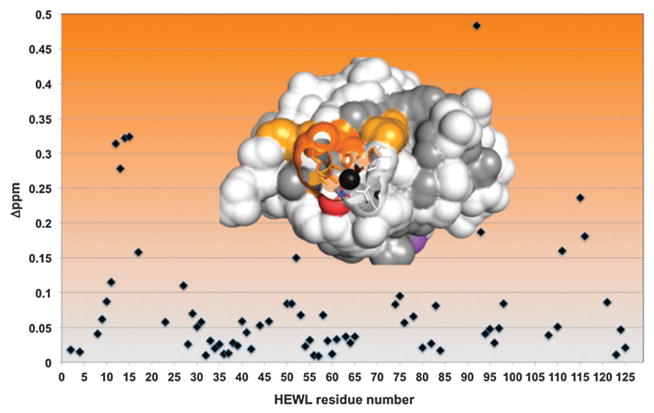
Good quality natural abundance 15N HSQC spectra could be collected on nitrate recrystallized HEWL, but only when the protein was dissolved in 100% D2O rather than water. This is presumably due to interference from the residual solvent associated baseline artifacts when the sample is in 10% D2O. However, due to the stable structure of the protein, many of the exchangeable resonances remain protonated when the protein is dissolved in D2O, even at 30 °C. As a result, good quality 1D and 15N HSQC spectra could be collected on this subset of resonances. Several chemical shift changes were observed and found to be on the border between intermediate and fast exchange on the chemical shift timescale. The 15N HSQC showed a fairly limited region of changes correlated to residues Arg14, His15 and V92 consistent with the crystal structure. No large changes were observed in regions outside of the binding site although about a third of the resonances were exchanged for deuteriums and thus unobservable. Asp87, at the end of helix 4, exchanges rapidly and was unfortunately unable to be confirmed as perturbed by NMR. Mapping these changes onto the surface of HEWL confirms that they cluster around the crystallographically observed metal binding site (Fig. 3). The strength of this association was also characterized by 1D NMR and found to give a Kd of ~40 μM. Thus, both in the solid state and in solution, HEWL binds silver via His15.
Fig. 3.
15N HSQC chemical shift changes in silver(I)-HEWL. Chemical shift changes ≥ 0.25 (dark orange) and ≥ 0.15 (yellow) are plotted on the inset structure. Unobservable amides are grey.
HEWL has been shown to stabilize silver and gold nanoparticles. 27,28 The behaviour of HEWL under these conditions has been probed by surface enhanced Raman spectroscopy (SERS). Such studies unambiguously demonstrate a strong signal characteristic of silver-nitrogen stretch upon HEWL deposition onto the nanoparticle surface.29 Some authors have gone on to propose that this is due to tryptophan indole nitrogen interaction with silver.30 We mildly dispute this claim because we see no evidence of silver-indole nitrogen bonding in our structure even when crystals are grown under significant excess of silver. However, it is also possible that the apparent discrepancy may result from the differences in chemical environment between solution conditions and the conditions at the silver nanoparticle surface. Additional experiments, such as solid state NMR, are required to resolve these possibilities.
Regarding the possible roles of this metal and other metallated HEWL structures, a number of studies have demonstrated that several differentmetals can bind to HEWL via His15 asmentioned earlier. However, a simple ClustalW alignment of available HEWL sequences reveals absolutely no significant conservation of this metal binding residue and most frequently hydrophobic residues tyrosine, phenylalanine, or leucine are present at this position. However, the nearby Arg14 and Asp87 are universally conserved and implicate their role for salt bridge formation. In an attempt to determine if the silver metallation of HEWL had an effect on the normal enzymatic function, an assay for activity was performed (see supporting information). There seemed to be no significant affect to HEWL activity when treated with various equivalents of silver while varying the concentration of the enzyme. At constant enzyme concentrations and varying silver concentrations there again appeared to be no significant effect from silver on HEWL activity. If chickens have evolved this site for metal binding, it has not been convergently evolved to any significant extent in other organisms and appears to have no role in enzymatic function. For this reason, we suggest that this binding site resulted from coincidence rather than by natural selection.
In conclusion, it was our original assumption that one or more binding sites could be significantly populated by silver(I) in HEWL because of the presence of multiple favourable binding residues within the protein structure. HEWL contains six tryptophan residues that are viable as binding sites based upon the aforementioned SERS, however with the exception of Trp62 the majority of these have little or no solvent accessibility. Additionally, the eight cysteines present are involved in disulfide bonds and not available for metal binding. There are also two methionines that could serve as preferred soft lone-pair ligands relative to harder nitrogen and oxygen containing residues. While we have observed weak density for silver near Met105 in other studies, such cases require a nearly tenfold excess silver hydroxybutyrate (40 mM). Also, there is no evidence that this site is bound in solution. Lastly there are two carboxylate containing residues in HEWL’s active site groove (Glu35 and Asp52) that are freely accessible to silver ligation and yet they are not metallated even at the highest concentrations studied. This was further confirmed by a lack of reduction in enzymatic activity upon titration with silver. Thus, despite the possibility of multiple binding residues present within HEWL we note that it is the surface accessibility of His15 and the proximity of neighbouring Arg14 and Asp87 that make this site preferentially populated by silver(I).
Supplementary Material
Acknowledgments
This work was supported by The University of Akron and The National Institute of General Medical Sciences (1 R01 GM86895-01). We wish to thank the the National Science Foundation for funds used to purchase the Bruker Apex II Duo CCD X-ray diffrac-tometer (CHE-0840446) used in this research. We thank the Kresge Foundation and donors to the Kresge Challenge Program at The University of Akron for funds used to purchase the NMR instrument used in this work. We also wish to thank the Center for Silver Therapeutics Research at The University of Akron.
Notes and references
- 1.Melaiye A, Youngs WJ. Expert Opin Ther Pat. 2005;15(2):125–130. [Google Scholar]
- 2.Kascatan-Nebioglu A, Melaiye A, Hindi K, Durmus S, Panzner MJ, Hogue LA, Mallett RJ, Hovis CE, Coughenour M, Crosby SD, Milsted A, Ely DL, Tessier CA, Cannon CL, Youngs WJ. J Med Chem. 2006;49(23):6811–6818. doi: 10.1021/jm060711t. [DOI] [PubMed] [Google Scholar]
- 3.Hindi KM, Ditto AJ, Panzner MJ, Medvetz DA, Han DS, Hovis CE, Hilliard JK, Taylor JB, Yun YH, Cannon CL, Youngs WJ. Biomaterials. 2009;30(22):3771–3779. doi: 10.1016/j.biomaterials.2009.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Panzner MJ, Hindi KM, Wright BD, Taylor JB, Han DS, Youngs WJ, Cannon CL. Dalton Trans. 2009:7308–7313. doi: 10.1039/b907726j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Hindi K, Watts KM, Taylor JB, Zhang K, Li Z, Hunstad DA, Cannon CL, Youngs WJ, Wooley KL. Chem Commun. 2010;46:121–123. doi: 10.1039/b916559b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medvetz DA, Hindi KM, Panzner MJ, Ditto AJ, Yun YH, Youngs WJ. Met-Based Drugs. 2008;2008:1–7. doi: 10.1155/2008/384010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Siciliano TJ, Deblock MC, Hindi KM, Durmus S, Panzner MJ, Tessier CA, Youngs WJ. J Organomet Chem. 2011;696(5):1066–1071. [Google Scholar]
- 8.Hindi KM, Panzner MJ, Tessier CA, Cannon CL, Youngs WJ. Chem Rev. 2009;109(8):3859–3884. doi: 10.1021/cr800500u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eby DM, Luckarift HR, Johnson GR. ACS Appl Mater Interfaces. 2009;1(7):1553–1560. doi: 10.1021/am9002155. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Y, Wang X. Nano Biomed Eng. 2010;2(4):208–213. [Google Scholar]
- 11.Patil S, Deally A, Gleeson B, Hackenberg F, Müller-Bunz H, Paradisi F, Tacke MZ. Z Anorg Allg Chem. 2011;637:386–396. [Google Scholar]
- 12.Razavet M, Artero V, Cavazza C, Oudart Y, Leburn C, Fontecilla-Camps JC, Fontecave M. Chem Commun. 2007:2805–2807. doi: 10.1039/b703887a. [DOI] [PubMed] [Google Scholar]
- 13.Binkley SL, Ziegler CJ, Herrick RS, Rowlett RS. Chem Commun. 2010;46:1203–1205. doi: 10.1039/b923688k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Santos-Silva T, Mukhopadkyay A, Seixas JD, Bernardes GJL, Romao CC, Romao MJ. J Am Chem Soc. 2011;133:1192–1195. doi: 10.1021/ja108820s. [DOI] [PubMed] [Google Scholar]
- 15.Casini A, Mastrobuoni G, Temperini C, Gabbiani C, Francese S, Moneti G, Supuran CT, Scozzafavaa A, Messori L. Chem Commun. 2007:156–158. doi: 10.1039/b611122j. [DOI] [PubMed] [Google Scholar]
- 16.Guli M, Lambert EM, Li M, Mann S. Angew Chem Int Ed. 2010;49:520–523. doi: 10.1002/anie.200905070. [DOI] [PubMed] [Google Scholar]
- 17.Alderton G, Fevold HL. J Biol Chem. 1946;164:1–5. [PubMed] [Google Scholar]
- 18.APEX2. Bruker2007. Bruker AXS Inc; Madison, Wiscon-sin, USA: [Google Scholar]
- 19.Emsley P, Lohkamp B, Scott W, Cowtan K. Acta Crystallogr, Sect D: Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lohkamp B, Emsley P, Cowtan K. CCP4 Newsletter. 2005;42 Contribution 7. [Google Scholar]
- 21.Collaborative Computational Project, Number 4. Acta Cryst. 1994;D50:757–759. [Google Scholar]
- 22.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr, Sect D: Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 23.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei H, Wang Z, Zhang J, House S, Gao YG, Yang L, Robinson H, Tan LH, Xing H, Hou C, Robertson IM, Zuo JM, Lu Y. Nat Nanotechnol. 2011;6:93–97. doi: 10.1038/nnano.2010.280. [DOI] [PubMed] [Google Scholar]
- 25.Bodenhausen G, Ruben DJ. Chem Phys Lett. 1980;69:185–189. [Google Scholar]
- 26.Schwalbe H, Grimshaw SB, Spencer A, Buck M, Boyd J, Dobson CM, Redfield C, Smith LJ. Protein Sci. 2001;10(4):677–688. doi: 10.1110/ps.43301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ebys M, Schaeblin N, Farrington K, Hussain S, Johnson G. ACS Nano. 2009;3(4):984–994. doi: 10.1021/nn900079e. [DOI] [PubMed] [Google Scholar]
- 28.Yang T, Li Z, Wang L, Guo C, Sun Y. Langmuir. 2007;23(21):10533–10538. doi: 10.1021/la701649z. [DOI] [PubMed] [Google Scholar]
- 29.Hu J, Sheng RS, Xu ZS, Zeng Y. Spectrochimica Acta. 1995;51A(6):1087–1096. [Google Scholar]
- 30.Chandra G, Ghosh KS, Dasgupta S, Roy A. Int J Biol Macromol. 2010;47:361–365. doi: 10.1016/j.ijbiomac.2010.05.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.