Abstract
Rationale and Objectives
Landmark point-pairs provide a strategy to assess deformable image registration (DIR) accuracy in terms of the spatial registration of the underlying anatomy depicted in medical images. In this study, we propose to augment a publicly available database (www.dir-lab.com) of medical images with large sets of manually identified anatomic feature pairs between breath-hold computed tomography (BH-CT) images for DIR spatial accuracy evaluation.
Materials and Methods
10 BH-CT image pairs were randomly selected from the COPDgene study cases. Each patient had received CT imaging of the entire thorax in the supine position at 1/4th dose normal expiration and maximum effort full dose inspiration. Using dedicated in-house software, an imaging expert manually identified large sets of anatomic feature pairs between images. Estimates of inter- and intra-observer spatial variation in feature localization were determined by repeat measurements of multiple observers over subsets of randomly selected features.
Results
7298 anatomic landmark features were manually paired between the 10 sets of images. Quantity of feature pairs per case ranged from 447 to 1172. Average 3D Euclidean landmark displacements varied substantially among cases, ranging from 12.29 (SD: 6.39) to 30.90 (SD: 14.05) mm. Repeat registration of uniformly sampled subsets of 150 landmarks for each case yielded estimates of observer localization error, which ranged in average from 0.58 (SD: 0.87) to 1.06 (SD: 2.38) mm for each case.
Conclusions
The additions to the online web database (www.dir-lab.com) described in this work will broaden the applicability of the reference data, providing a freely available common dataset for targeted critical evaluation of DIR spatial accuracy performance in multiple clinical settings. Estimates of observer variance in feature localization suggest consistent spatial accuracy for all observers across both 4D CT and COPDgene patient cohorts.
Keywords: deformable image registration, clinical validation, COPDgene, landmark feature pairs
INTRODUCTION
Deformable image registration (DIR) is an emerging technology with diagnostic and therapeutic medical applications. DIR algorithms were first developed in computer vision research to estimate motion by warping a source image onto a target, producing an estimated image that visually appeared similar to the target image [1]. Though their spatial accuracy was undetermined, DIR algorithms previously have been applied to extract physiologic information such as cardiac wall motion [2] and ventilation [3, 4] from medical images. For medical applications, the goal in applying DIR is to obtain accurate spatial registration of the underlying anatomy, and not simply quantitative image similarity. The fundamental difficulty associated with objective evaluation of DIR for use in the clinical setting is due to the lack of a known reference solution against which calculations objectively can be compared. Prior to clinical implementation of applications contingent upon accurate spatial registration between images, thorough performance evaluation should be conducted utilizing image data that will be encountered in practice. The best standard, therefore, is one derived from actual patient images, for which absolute “ground truth” is not known.
Expert identified anatomic landmark feature-pairs offer a direct measurement of the overall DIR spatial accuracy by sampling voxel registration errors throughout the target image. These measurements are limited to discrete voxel positions for which an estimate of the true correspondence is available. Anatomic feature-pairs from patient images have been used directly for DIR algorithm evaluation and for validation of their applied use (e.g., [5-9]). Others have reported publicly available reference datasets and services to provide evaluation of submitted DIR outputs. In 2007, Sarrut et al. reported on the public availability of a reference dataset for DIR spatial accuracy evaluation, which consists of >500 manually identified reference positions distributed over four component 4D CT phases and three patients [10-12]. Referred to as the POPI-model (a point-validated pixel-based breathing thorax model), the dataset has since grown to include three additional 4D CT images for which 100 feature-pairs are identified on the maximum inhalation and exhalation component phases. The “submission” paradigm has been employed by others, such as reported in a 2011 study describing the public EMPIRE10 platform [13]. In that work, small (≤100) sets of feature correspondences are identified on 30 lung CT images, comprised of the extreme phases from 4D CT, BH-CT, and synthetically deformed images.
Previously we described a practical framework and operative procedure for rapidly identifying and managing large numbers of anatomic feature pairs for objective evaluation of DIR spatial accuracy using clinically acquired radiotherapy treatment planning four-dimensional computed tomography (4D CT) images [14]. As part of that work, we constructed and made publicly available (www.dir-lab.com) a reference image library consisting of 5 4D CT image sets with >1100 landmark feature points identified across maximum inhalation and exhalation component phase images obtained during resting tidal breathing. In subsequent work, we made additions to the image library to include a total of 10 4D CT image sets, with feature points identified across the 6 expiratory phase images from maximum tidal inhalation to maximum tidal exhalation [15]. Over 200 academic, commercial, and government research groups from around the world have acquired our reference library data for their research. The reference library (Castillo et al. [14]) represents the first of its kind, with >300 anatomic feature pairs identified for each image set, each identified with repeated localization by multiple observers to estimate inter- and intra-observer variance. The practical utility of the reference library is demonstrated by the quantity of subscribers, and their citations in the peer-reviewed literature (e.g., from 2012: [16-29]). In this work, we propose to augment our existing database to include reference images representing independent and substantially different imaging protocols. These additions will greatly enhance applicability of the reference data, providing a publicly available common dataset for targeted critical evaluation of DIR spatial accuracy performance in multiple clinical settings. The availability of a common dataset for algorithm performance assessment that is broadly applicable will facilitate streamlined comparative evaluation and meta-analysis of the scientific literature, and further provide a foundation upon which to develop a rigorous and standardized evaluation methodology that is presently lacking.
Chronic obstructive pulmonary disease (COPD) is the fourth leading cause of death in the United States and is projected to be the third leading cause of death worldwide by the year 2020 [30]. The COPDgene study (www.copdgene.org) is a National Heart Lung Blood Institute (NHLBI) funded cross-sectional study, with a recruitment goal of over 10,000 patients, designed to discover what genetic factors contribute to the development of COPD. Disease severity is documented using pulmonary function testing and inspiratory & expiratory breath-hold CT (i & eBH-CT) imaging. The iBH-CT is acquired at maximum effort and the eBH-CT at end normal expiration, or functional residual capacity (FRC). The eBH-CT images are acquired at one-fourth the CT tube current as the iBH-CT in order to reduce the radiation dose to the study subjects. As a consequence, the eBH-CT image noise is doubled. Reports on diagnostic evaluation of the BH-CT image pairs for COPD primarily have been limited to evaluation of each of the eBH- & iBH-CT images separately (e.g., [31-34]). This is most likely due to the fact that there is not currently a validated algorithm that has been shown to accurately register these image pairs for their joint analysis. Despite this, recent reports have demonstrated the potential utility of quantitative methods to facilitate regional characterization of COPD based on deformable registration of the BH-CT image pairs. Murphy et al. [35] described a methodological framework to incorporate quantitative ventilation images derived from deformable registration to supplement traditional global spirometry measures of pulmonary function. 216 subjects obtained from the COPDgene study archive were used to demonstrate their approach. Evaluation of DIR was performed based on subjective visual scoring by multiple observers, with 4 performance metrics ranging from “Excellent” to “Poor”. More recently, Galban et al. [36] demonstrated proof-of-concept for a novel imaging biomarker derived from the spatially registered BH-CT images, however, there is no description of DIR algorithm performance testing specific to the COPDgene dataset. Availability of a DIR reference dataset for targeted evaluation specific to the COPD setting will facilitate further investigation and validation of advanced image analysis techniques for novel application in diagnosis and phenotyping in COPD.
Deformable registration of the COPDgene BH-CT image pairs is subject to numerous additional confounding factors that significantly increase the complexity of the appropriate model formulation relative to traditionally successful methods for spatial registration of treatment planning 4D CT. Those challenges are due to the large displacements, change in density and CT value, the difference in image noise, the highly non-uniform mechanical properties of lung tissue with COPD, and the extreme changes in anatomic shape of the pulmonary vasculature due to the large volume change. These pronounced differences are illustrated in Figures 1 and 2. There are currently no validated DIR tools available to provide a link between the COPDgene BH-CT image pairs. As such, new algorithms are required to achieve high spatial accuracy.
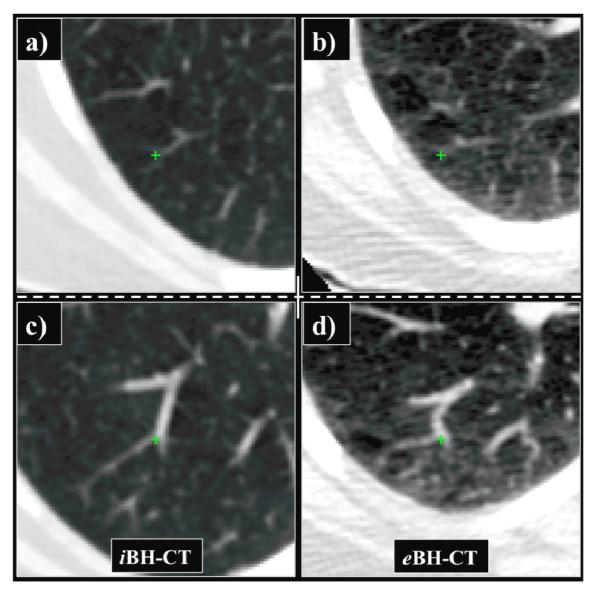
Figure 1. Breath-hold CT image pair from the COPDgene study.
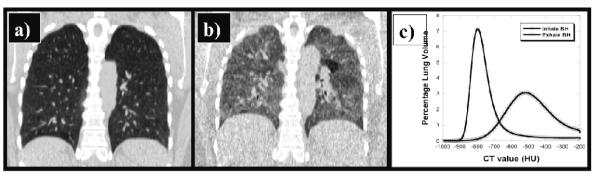
a) The inhale BH-CT shown in coronal section was acquired at high tube current (200 mA) resulting in low image noise. b) The corresponding exhale BH-CT coronal section was acquired at low tube current (50 mA) resulting in a two-fold increase in the relative image noise. Note the mottled appearance in the soft tissue regions compared to the iBH-CT. c) Plot of the histogram distribution of lung CT values in Hounsfield Units (HU) from the BH-CT image pair shown in (a) and (b).
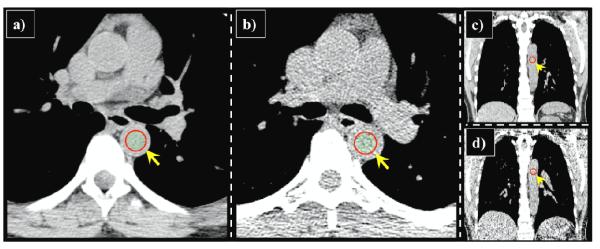
Figure 2. Anatomic deformation.
Two example vessel bifurcations, which were selected as reference landmark features, are shown in transverse section with green crosshair indicating corresponding voxel positions in (a, c) iBH- and (b, d) eBH-CT. There is substantial change in the shape of the structures that make-up the bifurcations. On the iBH-CT the blood vessels are straight and the lung tissue is dark. On the eBH-CT the blood vessels are bent, due to the substantially reduced lung volume, and the surrounding lung tissue is lighter. The bifurcations shown are located much closer to the lung border on the eBH-CT. These changes make automated estimation of these point-pairs difficult using cross-correlation.
MATERIALS AND METHODS
COPDgene CT Images
The COPDgene study protocol indicates the acquisition of dual inspiratory and expiratory chest CT scans for all study subjects to identify and quantify airway abnormalities, emphysema, and expiratory air trapping [37]. The inspiratory BH-CT is to provide thorough assessment of small airway wall thickness and emphysema, whereas the expiratory BH-CT is performed to assess for air trapping. To facilitate image-based measurements to assess small airway wall thickness and abnormalities, images are reconstructed using a high-resolution recon kernel. The algorithm used is vendor-specific (i.e., GE: BONE, SIEMENS: B46f, PHILIPS: Detail (D)), and represents a balance between enhancement of image resolution via higher spatial frequency cutoff, and increase in image noise. Since each algorithm previously has been selected as meeting the COPDgene study criteria, it is not expected that there will be CT vendor specific issues with the quantitative image analysis.
In this work, ten BH-CT image pairs were randomly selected from the COPDgene study cases. Each patient had received CT imaging of the entire thorax in the supine position at normal expiration and maximum effort full inspiration. The CT imaging was performed with a GE VCT 64-slice scanner (GE Healthcare Technologies, Waukesha, WI) with a pitch of 1.375 mm, speed of 13.75 mm per rotation, 120 kVp, 0.5 sec per rotation, 400 mA per rotation for inhale BH, and 100 mA per rotation for exhale BH. The images used in this study were reconstructed with high-resolution (BONE) recon kernel, with lung diameter setting the field of view, and with contiguous 2.5 mm slice spacing. Each image set was reconstructed to image dimensions 512 × 512 × N (N∈[102 135]), with in-plane voxel dimensions ranging from (0.590 × 0.590) – (0.652 × 0.652) mm2. The image characteristics of the 10 COPD cases utilized in this study are given in Table 1.
TABLE 1. CT Imaging & Landmark Summary Characteristics.
The image and voxel dimensions are shown for all cases included in this study. Also shown is the number of reference feature pairs for each case, along with the corresponding average (and standard deviation) magnitude displacements. Estimates of observe variance in target feature localization were obtained by repeat registration as described, and are also shown as mean (and standard deviation), combined for the set of multiple observers. All measurements of distance are reported in units of millimeters.
| Case Number |
Image Dimensions |
Voxel Dimensions (mm) |
# Landmarks |
Avg. Displacement (mm) |
Observer Error (mm) |
|---|---|---|---|---|---|
| 1 | 512×512×121 | 0.625×0.625×2.5 | 773 | 25.90 (11.57) | 0.65 (0.73) |
| 2 | 512×512×102 | 0.645×0.645×2.5 | 612 | 21.77 (6.46) | 1.06 (1.51) |
| 3 | 512×512×126 | 0.652×0.652×2.5 | 1172 | 12.29 (6.39) | 0.58 (0.87) |
| 4 | 512×512×126 | 0.590×0.590×2.5 | 786 | 30.90 (13.49) | 0.71 (0.96) |
| 5 | 512×512×131 | 0.647×0.647×2.5 | 1029 | 30.90 (14.05) | 0.65 (0.87) |
| 6 | 512×512×119 | 0.633×0.633×2.5 | 633 | 28.32 (9.20) | 1.06 (2.38) |
| 7 | 512×512×112 | 0.625×0.625×2.5 | 575 | 21.66 (7.66) | 0.65 (0.78) |
| 8 | 512×512×115 | 0.586×0.586×2.5 | 791 | 25.57 (13.61) | 0.96 (3.07) |
| 9 | 512×512×116 | 0.664×0.664×2.5 | 447 | 14.84 (10.01) | 1.01 (2.54) |
| 10 | 512×512×135 | 0.742×0.742×2.5 | 480 | 22.48 (10.64) | 0.87 (1.65) |
CT Image Analysis
Additional quantitative analyses were performed characterizing lung volume change and relative image noise between BH-CT pairs. To approximate lung volume from each BH-CT image, the set of voxels with CT number in the range [−1000 −200] was initially selected. Two-dimensional region growing was applied slice-by-slice in the transvers plain to remove background objects defined by connectivity to the image border [14]. A voxel erosion and subsequent growing technique was applied to remove extraneous voxels. The final segmented regions were visually assessed and manually modified as necessary. The volume of each segmented lung region was calculated and the difference between corresponding iBH- & eBH-CT determined for each case. Volume changes were determined as absolute magnitude change (in ml) and percent change relative to the segmented eBH-CT lung volume.
Estimates of relative noise between BH-CT pairs were determined by characterization of image intensity distributions within manually placed regions of interest (ROIs). For each case, an expert in thoracic imaging placed a fixed 1.5 cm diameter sphere on corresponding iBH- & eBH-CT within the aorta, approximately mid-position relative to the superior-inferior lung volume extent (Figure 3). The choice of aorta for ROI placement was based on the availability of centrally located, uniform measurements within the greater lung volume of interest, and the expected constant blood image intensity between inhale and exhale breathing states. Care was taken to avoid regions with prominent shadowing effects by calcifications in the immediate vicinity of the major vessel. For each ROI, the coefficient of variation (COV) was determined as standard deviation divided by mean CT number within the sphere. The ratio of eBH- to iBH- COVs was determined for each case as a surrogate measure of the relative noise content within corresponding image pairs.
Figure 3. CT noise estimation.
Estimates of the relative noise ratio between (a, c) iBH- and (b, d) eBH-CT were determined by quantitative assessment of the CT numbers within manually placed 1.5 cm diameter spherical ROIs (indicated by yellow arrow). Visually, the eBH-CT clearly demonstrates increased mottled appearance within soft-tissue structures compared to corresponding iBH-CT. The COV ratio indicates average relative noise increase of 2.2× for the set of 10 cases included in this study. The increase is due to the (4:1) acquisition tube current ratio, and the use of edge-enhancing BONE reconstruction kernel.
Landmark Selection
A MATLAB-based software interface named APRIL (Assisted Point Registration of Internal Landmarks), which has been previously described [14], was utilized to facilitate manual selection of landmark feature pairs between the volumetric eBH- & iBH-CT images. The software incorporates basic image processing and visualization tools that are common to most commercial treatment planning systems, including independent window and level settings for all displays, digital magnification and pan, visualization of equivalent voxel positions in the principal orthogonal plains, transparent volume rendering, and interactive tools for segmentation of lung voxels from the image data. To determine corresponding features, the user initially must manually delineate a candidate voxel in the designated “source” image via mouse click over the desired position. The target image voxel corresponding to the same underlying anatomy is then similarly selected to complete the feature pairing. The process is repeated until the desired sample size and uniformity of distribution have been achieved. No implanted fiducials or added contrast agents were used to aid in the selection of landmark features, which typically included vessel and bronchial bifurcations.
To assist in localization of the anatomic features within the target image space, the software provides an optional feature localization tool based on normalized cross-correlation of the local feature neighborhood depicted in the source image with a larger, user-adjustable target window in the eBH-CT [14, 38, 39]. The image content within the feature neighborhoods is converted to a binary mask according to a user-defined intensity threshold, such that the cross correlation is computed only over the local structural content. The intensity threshold controls the level of structural detail included in the feature masks. The target voxel representing the maximum of the three-dimensional correlation function is highlighted with a crosshair within the target display, and represents an estimate of the feature correspondence. Previously we found that localization estimates based on the cross-correlation coefficient were suitable for expediting the manual feature pair registration process using treatment planning 4D CT. However, preliminary investigation as part of this work demonstrated little to no utility for use with the COPDgene imaging data. Most notably, this is due to the drastic deformation of prominent vasculature structures commonly identified as anatomic landmark features. In addition, the relatively large displacements, which require excessively large search windows for both images, further increase the probability that the correlation function maximum does not correspond to accurate approximation of the underlying physical displacement.
As an alternative, to facilitate maximally efficient workflow, we have upgraded the APRIL software to include an additional feature localization capability, which incorporates the current set of registered feature pairs into moving least squares (MLS) interpolation applied to the current source feature position to determine the corresponding target estimate. MLS is a highly generic and versatile tool for approximating an unknown function by fitting polynomials to function samples given at uniform or non-uniform locations [40, 41]. Though most commonly employed to reconstruct surfaces from noisy, unstructured point cloud data, MLS has recently found utility in DIR [15, 25, 42]. For our purposes, ther MLS function operates on the current source voxel position , for which we would like to obtain an estimate of the corresponding target position. An affine function is determined by minimizing the expression:
| (1.1) |
where and are the ith source and target feature pair positions, respectively. The scalar weights ωi are of the form:
| (1.2) |
In this way, each newly selected feature-pair provides additional contribution to estimates of subsequent feature locations. While an initial set of registered feature pairs is required before the MLS estimation procedure provides non-trivial support, in practice we have found that approximately 4 input pairs are necessary before the estimates contribute to improved target feature localization. This significantly reduces the time spent navigating the target volume space, and is independent of the magnitude displacements or image content.
In this work, both cross-correlation and MLS-based estimation procedures were available to all users. Cross correlation and MLS-based estimates of target features are presented as a crosshair in the target image. In practice, multiple cross correlations varying both the feature neighborhood dimension and/or intensity threshold may be performed for a given feature. The corresponding target estimate will vary according to the particular search criteria. Similarly, the MLS-based estimates will vary according to the current set of previously matched feature pairs. As more features are matched, the estimates become more robust. However, for both estimation procedures, the user ultimately must designate the feature correspondence via mouse click within the target image. The reader visually determines every point; there is not a mechanism to automatically accept a feature position based on either automated assistance operation.
An expert in thoracic imaging, beginning at the apex of the lung, systematically selected the source feature points on the 10 image pairs. The manual registration process consists of initial feature localization via mouse click within the source image. The feature position is highlighted with a crosshair, and the user is prompted for confirmation. Following confirmation, the optional estimation tools become enabled to provide computer assistance for target feature localization based on either cross correlation or MLS operations. The reader is free to navigate the target volume along each of the three principal orthogonal axes, with the option to display the source feature coordinate via crosshair within the target image space for reference. The matched target feature position is identified via mouse click and indicated with a crosshair. Optionally, the user may overlay the discretized image voxel grid for assistance in precise localization of the intended anatomic feature. Following confirmation, the coordinate position is stored. At any time, the reader is free to enable a review mode to scroll through the current set of matched feature pairs. The reader may specify a feature pair index, or sequentially review the current set via scroll bar. Previously matched features are presented in both source and target displays, with crosshair centered according to the current magnification setting in each window. Previously accepted source and target positions can be modified as necessary.
In this study, the iBH-CT was designated as the “source” image, which served as the primary dataset within which anatomic features were initially selected. This was due to the better image quality (Table 3) and the higher contrast of vessels relative to the inflated lung background. Points were selected with an initial goal of >5 for each lung per axial image slice. This approach ensured the collection of ≥447 validation point pairs for each case distributed throughout the lungs. Following feature selection for a given case, all landmark pairs were visually reviewed by the primary reader a second time and the locations adjusted on either image as necessary. The verification step was a required part of the initial registration process performed by the primary reader.
TABLE 3. Relative Noise Estimation.
Estimates of relative noise between corresponding iBH- and eBH-CT were determined by regional assessment of the distribution of image intensities within manually placed ROIs. The ratio of COVs was calculated from these measurements as a surrogate for the relative noise between image pairs. On average, the estimated eBH-CT noise is 2.2× that of the corresponding iBH-CT.
| Case Number |
Inhale COV | Exhale COV | Ratio (Exhale / Inhale) |
|---|---|---|---|
| 1 | 0.011 | 0.018 | 1.64 |
| 2 | 0.028 | 0.042 | 1.50 |
| 3 | 0.015 | 0.031 | 2.07 |
| 4 | 0.012 | 0.024 | 2.00 |
| 5 | 0.016 | 0.031 | 1.94 |
| 6 | 0.008 | 0.019 | 2.37 |
| 7 | 0.014 | 0.037 | 2.64 |
| 8 | 0.012 | 0.026 | 2.17 |
| 9 | 0.014 | 0.039 | 2.78 |
| 10 | 0.013 | 0.037 | 2.85 |
|
|
|||
| Average Ratio | 2.20 | ||
A subset of landmark points was re-registered by the primary reader, to estimate intra-observer variance in target localization, and by secondary readers, to estimate the inter-observer variance. For each case, the APRIL software was used to extract a random sample of 150 feature pairs from the corresponding complete set generated by the primary reader. For each of the 150 sampled pairs, the reader was presented with the primary iBH-CT feature position via crosshair within the source image space. The re-registration process consists of locating the corresponding point within the eBH-CT. All feature estimation and visualization tools were available in both primary and secondary registration processes. For all secondary readings, observers were blinded to the primary target selections; all secondary readings were performed independently and without prior knowledge of the primary point matches. Observer variation in target point selections for a given image pair were estimated based on the three-dimensional Euclidean distance in units of millimeters between the primary and secondary target positions. The mean distance, with standard deviation (SD), between corresponding targets was determined over the range of subsampled data. Inter- and intra-observer variance in feature localization was determined on a case-by-case basis.
Software Familiarity and Training
Proper and efficient utilization of the assisted landmark registration software is a learning process that requires a certain degree of practice and experience to realize. In general, care should be taken to ensure that variations in target point selection both within and between users are true reflections of variability in individual judgment and selection criteria, rather than reflections of variation in time-specific aptitude or experience with the registration software. To address this concern, all observers were required to complete a minimum training set of 1000 independent feature-pair registrations, using image pairs not included in the study, prior to repeat measurements over the reference data. Additionally, potential degradation in CT image quality due to noise or image acquisition/reconstruction artifacts is likely to have some impact on the quantity of prominent features an observer is able to match between images. Rather than filter the image data, we will accept all clinically acquired cases, such that the reference library will accurately reflect those images that are likely to be encountered in routine clinical application.
RESULTS
Quantitative Image Analysis
Estimated lung volumes, with corresponding magnitude and relative changes are shown in Table 2 for the set of BH-CT pairs included in this study. Magnitude volume change between image pairs varied considerably among cases, with range from 974 – 2819 ml. To provide context for these measurements, we previously reported estimates for resting tidal volumes derived from 7 treatment planning 4D CTs using an analogous image segmentation-based approach, which yielded measured volume changes in the range 462 – 926 ml [43]. The extreme discrepancy is expected, and reflective of the markedly different clinical scenarios investigated in each study (i.e., tidal breathing versus maximum effort breath-hold). Relative changes between BH-CT pairs ranged from 25% - 106%, indicating in some instances greater than two-fold increase in lung volume between corresponding inhale and exhale breath-holds. Figure 4 provides visual context for these measurements, illustrating an example image pair (case #5) for which the relative change was >100%. Maximum distance between the inferior contoured lung boundaries is on the order of 40 image voxels.
TABLE 2. Measured Volume Change.
Volume change measurements were estimated from semi-automated lung voxel segmentation, and are shown expressed as absolute difference (ml) and percent difference relative to the eBH-CT lung segmentation. Note that cases #4 and #6 more than doubled in relative volume between exhale and inhale breathing states.
| Case Number |
Inhale Lung Volume (ml) |
Exhale Lung Volume (ml) |
Absolute Difference (ml) |
Relative Difference (% Exhale) |
|---|---|---|---|---|
| 1 | 6852 | 4268 | 2584 | 61% |
| 2 | 4230 | 3051 | 1179 | 39% |
| 3 | 5332 | 4260 | 1072 | 25% |
| 4 | 4332 | 2105 | 2227 | 106% |
| 5 | 5203 | 2521 | 2682 | 106% |
| 6 | 4550 | 2784 | 1766 | 63% |
| 7 | 3879 | 2905 | 974 | 34% |
| 8 | 4798 | 2759 | 2039 | 74% |
| 9 | 4042 | 2954 | 1088 | 37% |
| 10 | 7181 | 4362 | 2819 | 65% |
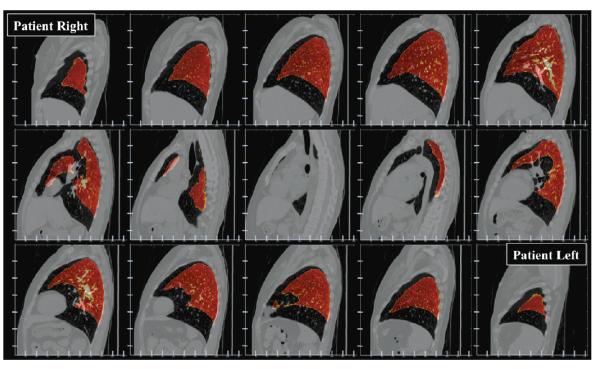
Figure 4. Extreme volume chang.
Fusion image is shown depicting the iBH-CT in sagittal section, with overlay of the contoured eBH-CT lung volume (orange color-scale) in 25-slice increments. Vertical-axis tick marks are spaced in 20-voxel (50 mm) increments from the top of the axis. Horizontal-axis tick marks are spaced in 50-voxel (32.35 mm) increments from the left. The figure illustrates considerable magnitude lung motion in AP and SI component directions. Maximum distance between the inferior contoured lung boundaries is on the order of 40 image voxels.
Estimates of the relative noise between corresponding iBH- & eBH-CT image pairs are summarized in Table 3, in terms of the COV ratio within manually placed spherical ROIs (Figure 3). The measurements indicate relative noise greater than the theoretical (2:1) given by consideration of the relative acquisition tube currents alone. However, the high-resolution BONE reconstruction filter, which will increase the noise in greater relative proportion in the exhale image, also contributed to the increase in the overall comparison ratio [44]. Figure 3 shows representative transverse and coronal views through an example iBH- & eBH-CT image pair (case #7), with cross-section of the spherical ROI also indicated. The example views illustrate the obvious increase in mottled appearance of soft-tissue structures depicted in the eBH-CT.
Feature Pair Datasets
The reference landmark characteristics for all cases included in this study are summarized in Table 1. A total of 7298 anatomic landmark features were manually paired between the 10 sets of images, while the registered feature pairs per case ranged from 447 to 1172. Approximately 10-12 hours were required to complete the primary feature pair dataset for each case. For secondary readers, approximately 6-8 hours were required to re-register the 150 sampled source features onto the eBH-CT. Average three-dimensional Euclidean landmark displacements varied substantially among cases, ranging from 12.29 (SD: 6.39) to 30.90 (SD: 14.05) mm. Average magnitude displacements in component right-left (RL), anterior-posterior (AP), and superior-inferior (SI) directions ranged from 2.11 (SD: 1.67) to 6.06 (SD: 3.97), 6.11 (SD: 3.50) to 24.57 (SD: 6.25), and 6.20 (SD: 5.45) to 21.20 (SD: 14.75) mm, respectively. The component displacement magnitudes clearly indicate that the largest displacements occur along the AP axis, which is unlike the previously reported 4D CT reference images, for which the most significant tidal motion occurred along the SI axis [14]. Repeat registration of uniformly sampled subsets of 150 landmarks for each case yielded estimates of observer localization error, which ranged in average from 0.58 (SD: 0.87) to 1.06 (SD: 2.38) mm for each case. Figure 5 graphically summarizes each individual observer’s localization errors per case. Figure 6 shows the set of reference displacement vector field projections for each case. Transparent isosurface renderings of the maximum effort iBH-CT lung fields are shown overlain in oblique projection. The color scale shown at right is fixed for each projection, illustrating the substantial variability in motion field characteristics among the 10 cases, which is due in part to the variable extent of their COPD disease.
Figure 5. Observer target localization variability.
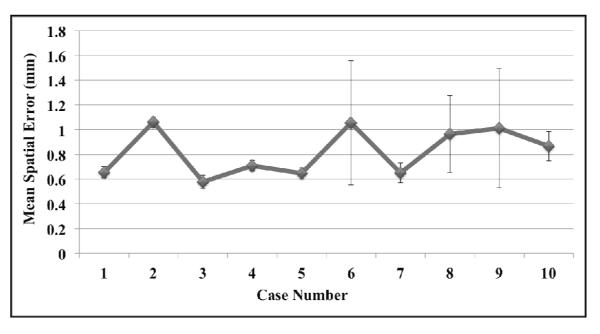
The average (± standard deviation) of the 3 mean observer localization errors is shown for each case. For cases 6, 8, and 9 the highest observer average was approximately twice that of the lower observer, resulting in the relatively large standard deviations. Combining the set of observer measurements for each case yields average repeat registration errors ≤1.06 mm over the set of 10 cases utilized in this study.
Figure 6. COPDgene BH-CT reference data.
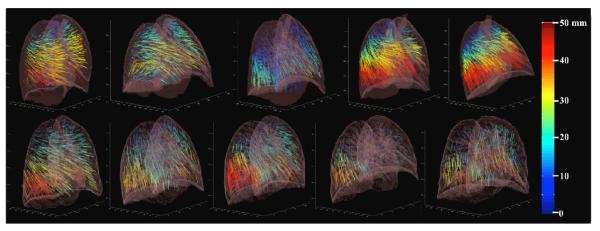
Landmark point pairs were selected on exhale and inhale BH-CT images from 10 COPDgene cases. The number of pairs ranged from 447-1172 for each case. Vector field projections are shown using the color map for displacement length (0 to 50 mm) shown at right. Those vectors with >50 mm displacement are mapped to the same red-colored maximum. The range of displacements varies for each case, reflecting the extent of their COPD disease.
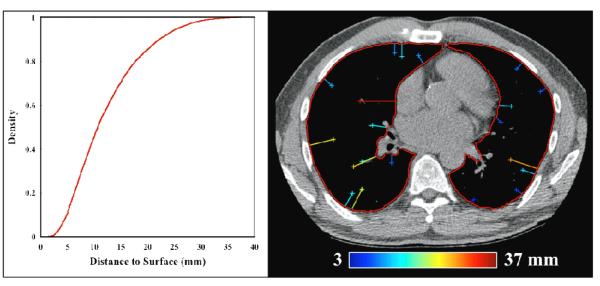
Of notable concern for feature-based DIR evaluation is the density of anatomical landmarks with respect to the organ surface. Analyses based on samples that are biased towards high contrast, centrally located structures are likely to misrepresent algorithm performance at the periphery where the landmark density is low. To characterize the overall feature pair distribution with respect to the lung surface, the minimum distance to the segmented lung contour was determined for each landmark in the set of eBH-CTs. The cumulative distribution of distance measurements was compiled for all cases and shown in Figure 7. For the dataset generated in this study, approximately 11% of the reference features are within 5 mm of the contoured exhale lung surface, while approximately 46% are within 1 cm. The measurement process is illustrated in Figure 7 for an example transverse section in which 20 features are identified ranging in distance to surface from 3 – 37 mm.
Figure 7. Distribution of landmarks.
For each anatomic feature identified in this work, the minimum distance to the respective contoured eBH-CT lung surface was determined. The set of distance measurements was compiled to generate the overall cumulative distribution shown at left. An illustration of the measurement process is depicted at right for an example transverse section. Reference features are identified by crosshair, with adjoining line segment indicating the minimum distance to the contoured surface (overlain in red). For this example slice, 20 features (12 right lung; 8 left lung) were identified and are shown color-coded according to magnitude distance to surface, ranging from 3 – 37 mm.
DISCUSSION
Landmark point-pairs provide a strategy to assess the accuracy of DIR spatial registration of the underlying anatomy depicted in medical images. Others have demonstrated the utility of implanted fiducial markers for precise real-time motion management in the clinical radiotherapy setting [45]. However, application to DIR evaluation is generally confounded by limited sample size, and perturbation of the natural anatomical state imposed by the fiducial markers. Although expert-determined feature-pair correspondences have become a widely adopted reference for evaluating DIR spatial accuracy, there is still great variability in their use. The ongoing DIR-Lab web project (www.dir-lab.com) was initiated in 2009 as a means to facilitate: 1) algorithm development and performance characterization, 2) comparative evaluation between multiple algorithms or implementations, as well as 3) the development and standardization of procedures for objective evaluation of DIR spatial accuracy performance that are applicable to acceptance testing and quality assurance protocols for use in the clinical setting. Prior to clinical deployment, DIR-based applications should be optimized and rigorously tested using patient images that are specific to each application. The basic utility of any available reference library is therefore largely contingent upon the range of clinical scenarios that are adequately represented by the patient images comprising it.
The focus of the present study was to expand the utility of our current reference database by incorporating an additional and independent imaging cohort. Three major multi-site NHLBI-sponsored studies (Lung Tissue Research Consortium (LTRC), COPDgene, and Sub-populations and Intermediate Outcome Measures in COPD Study (SPIROMICS)) collect and archive inhale and exhale BH-CT image pairs from thousands of research study subjects along with clinical, tissue, and genomic data [37, 46-48]. These studies were each designed with an imaging archival core to make the study CT images available to other investigators who may develop novel image derived biomarkers to characterize COPD disease or its consequences. Presently, reports on diagnostic evaluation of the BH-CT image pairs for COPD are limited to the evaluation of each of the eBH- & iBH-CT images separately [31-34], as there is no validated algorithm that has been shown to accurately register these image pairs for their joint analysis. The dataset generated in this study is directly applicable to facilitate development in this area.
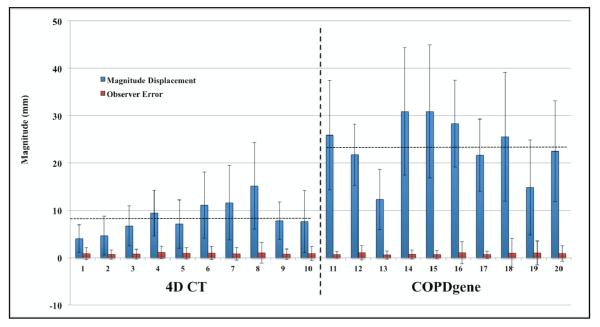
Figure 8 provides a graphical summary of the reference library data that is currently available online. In the figure, cases 1-10 represent clinically acquired treatment planning 4D CT, while cases 11-20 represent those obtained from the COPDgene archive and described in this work. For each case, mean (± standard deviation) magnitude landmark displacement is shown adjacent to the corresponding mean (± standard deviation) observer localization error obtained from all repeat measures over the subsampled set of features. For the 4D CT cases, magnitude displacements are given between the maximum inhalation and exhalation component phase images, although feature-pairs are available over the set of exhalation phase images (i.e., on each of T00, T10, T20, T30, T40, T50). By visual inspection alone, it is clear the independent datasets demonstrate markedly different magnitude motion characteristics; the data ranges from 4.01 (2.91) – 15.16 (9.11) mm for 4D CT, and 12.29 (6.39) – 30.90 (14.05) mm for COPDgene. These large differences are expected, and reflective of the variable acquisition protocol and clinical indication for the imaging studies represented by the two independent cohorts. In contrast, the mean observer localization errors are relatively consistent across datasets, in the range: 0.70 (0.99) – 1.13 (1.27) mm for 4D CT, and 0.58 (0.87) – 1.06 (2.38) mm for COPDgene. Based on these data, it is reasonable to surmise that the increased image noise [in the eBH-CT] relative to 4D CT did not impart a significant deleterious effect on the manual feature registration process. Previous work investigated the spatial accuracy of dense DIR displacement fields determined via MLS interpolation of input point pairs [49]. In that study, between 256-368 visually verified feature pairs comprised the input interpolation data. Spatial registration error was determined using a subset of the DIR-lab reference images, with average (standard deviation) three-dimensional error in the range 0.94 (1.16) – 2.55 (1.92) mm. Comparison versus the average observer localization errors determined in this study suggests that the manual selections were reflections of individual perception and search criteria, which provided better feature registration accuracy than the MLS algorithm alone.
Figure 8. DIR-Lab reference library data.
Mean (± standard deviation) magnitude landmark displacement (blue) is shown adjacent to the corresponding mean (± standard deviation) observer localization error (red). For the 4D CT cases, magnitude displacements are given between the maximum inhalation and exhalation component phase images. Horizontal lines indicate the overall average landmark displacement magnitude for the two imaging cohorts (4D CT: 8.52 mm, COPDgene: 23.46 mm).
On average, time required to complete feature registrations per case, including final verification once all point pairs had been initially registered, for both primary and secondary readers was approximately the same as we previously found for treatment planning 4D CT datasets [14]. This is further reflection of the independent and distinctive characteristics of the two study cohorts. Although in this study relatively fewer points were selected overall, the landmark features were more challenging to match between images. This was the result of a combination of effects, including the inconsistent image noise and the considerable variation in anatomic shape of the pulmonary vasculature due to the large volume change (Figures 1-4). For each source feature, the MLS estimation procedure provided rapid and effective guidance towards the corresponding feature neighborhood within the target image space. However, readers were instructed to spend as much time as deemed necessary to manually delineate and verify precise feature localization at the voxel level.
Alternative approaches for computer assisted feature identification schemes in the DIR evaluation setting have been described. Murphy et al. [50] reported a novel software application for semi-automated construction of image feature pairs, which was intended to minimize excessive consumption of time and personnel resources required to complete the task. This was achieved first by implementing an automated feature identification process based on magnitude image gradients, and secondly by allowing automated feature registration across images via thin-plate splines following manual matching of an initial feature subset. Although the authors report considerable reduction in time requirements (~30 min to completion per scan pair), the improvement is afforded by reliance on automated procedures to identify and subsequently define the reference standard feature correspondences. In the present study, all feature matching was manually performed to prevent the use of any singular DIR model to introduce potential bias into the reference data.
The pioneering work by Sarrut et al. [12] has since motivated further investigation into the feasibility, practical utility, and statistical necessities surrounding the use of large samples of anatomic feature-pairs for critical and objective evaluation of DIR [14]. Previously we reported our findings in this area, which subsequently led to the development and public deployment of the DIR-Lab reference dataset. By making the reference data freely and publicly available, the present framework allows for in-house algorithm development and optimization, without the need for submission of data to a larger overseeing body for performance evaluation [13]. Additionally, the available data mitigates the potential logistical concerns associated with generation of the feature-pair datasets, which may be impractical without sufficient resources [50]. To our knowledge, no datasets exist with comparable wealth of spatial accuracy sampling, inclusion of images derived from the COPDgene study archive, with explicit quantification of relative noise content and magnitude volume change between BH-CTs. Additionally, the reference dataset is further supplemented with additional measurements of feature trajectories provided for the set of expiration phase images on 4D CT, and explicit quantification for estimates of observer variance in feature localization.
CONCLUSIONS
The availability of a common dataset for algorithm performance assessment that is broadly applicable will facilitate streamlined comparative evaluation and meta-analysis of the scientific literature, and further provide a foundation upon which to develop a rigorous and standardized evaluation methodology that is presently lacking. The additions to the DIR-lab web database described in this work will greatly enhance applicability of the reference data, providing a freely available common dataset for targeted and state of the art critical evaluation of DIR spatial accuracy performance in multiple clinical settings.
ACKNOWLEDGEMENTS
We wish to thank the COPDgene Ancillary Studies Committee and Executive Committee who approved this study. This work was partially funded by the National Institutes of Health (NIH) through a National Cancer Institute Grant R21CA141833 and through an NIH Director’s New Innovator Award DP2OD007044.
LITERATURE REFERENCES
- 1.Horn BKP, Schunck BG. Determining optical flow. Artificial Intelligence. 1981;17:185–203. [Google Scholar]
- 2.Song SM, Leahy RM. Computation of 3-D velocity fields from 3-D cine CT images of a human heart. IEEE Trans Med Imaging. 1991;10(1):295–306. doi: 10.1109/42.97579. [DOI] [PubMed] [Google Scholar]
- 3.Guerrero TM, Sanders K, Noyola-Martinez J, Castillo E, Zhang Y, Tapia R, Guerra R, Borghero Y, Komaki R. Quantification of regional ventilation from treatment planning CT. Int J Radiat Oncol Biol Phys. 2005;62(3):630–634. doi: 10.1016/j.ijrobp.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 4.Guerrero T, Sanders K, Castillo E, Zhang Y, Bidaut L, Pan T, Komaki R. Dynamic ventilation imaging from four-dimensional computed tomography. Phys Med Biol. 2006;51(4):777–91. doi: 10.1088/0031-9155/51/4/002. [DOI] [PubMed] [Google Scholar]
- 5.Kaus MR, Brock KK, Pekar V, Dawson LA, Nichol AM, Jaffray DA. Assessment of a Model-Based Deformable Image Registration Approach for Radiation Therapy Planning. Int J Radiat Oncol Biol Phys. 2007;68(2):572–580. doi: 10.1016/j.ijrobp.2007.01.056. [DOI] [PubMed] [Google Scholar]
- 6.Boldea V, Sharp GC, Jiang SB, Sarrut D. 4D-CT lung motion estimation with deformable registration: Quantification of motion nonlinearity and hysteresis. Med Phys. 2008;35(3):1008–1018. doi: 10.1118/1.2839103. [DOI] [PubMed] [Google Scholar]
- 7.Al-Mayah A, Moseley J, Brock KK. Contact surface and material nonlinearity modeling of human lungs. Phys Med Biol. 2008;53(1):305–317. doi: 10.1088/0031-9155/53/1/022. [DOI] [PubMed] [Google Scholar]
- 8.Wolthaus JWH, Sonke JJ, van Herk M, Damen MF. Reconstruction of a time-averaged midposition CT scan for radiotherapy planning of lung cancer patients using deformable registration. Med Phys. 2008;35(9):3998–4011. doi: 10.1118/1.2966347. [DOI] [PubMed] [Google Scholar]
- 9.Li P, Malsch U, Bendl R. Combination of intensity-based image registration with 3D simulation in radiation therapy. Phys Med Biol. 2008;53:4621–4637. doi: 10.1088/0031-9155/53/17/011. [DOI] [PubMed] [Google Scholar]
- 10.Vandemeulebroucke J, Sarrut D, Clarysse P. The POPI-Model, a point validated pixel-based breathing thorax model. International Conference on the Use of Computers in Radiation Therapy (ICCR); Toronto, Canada. 2007. [Google Scholar]
- 11.Vandemeulebroucke J, et al. Spatiotemporal motion estimation for respiratory-correlated imaging of the lungs. Medical Physics. 2011;38(1):166–178. doi: 10.1118/1.3523619. [DOI] [PubMed] [Google Scholar]
- 12.Sarrut D, Delhay S, Villard PF, Boldea VA, Beuve MA, Clarysse PA. A Comparison Framework for Breathing Motion Estimation Methods From 4-D Imaging. IEEE Trans Med Imaging. 2007;26(12):1636–1648. doi: 10.1109/tmi.2007.901006. [DOI] [PubMed] [Google Scholar]
- 13.Murphy K, et al. Evaluation of registration methods on thoracic CT: The EMPIRE10 challenge. IEEE Transactions on Medical Imaging. 2011;30(11):1901–1920. doi: 10.1109/TMI.2011.2158349. [DOI] [PubMed] [Google Scholar]
- 14.Castillo R, Castillo E, Guerra R, Johnson VE, McPhail T, Garg AK, Guerrero T. A framework for evaluation of deformable image registration spatial accuracy using large landmark point sets. Phys Med Biol. 2009;54:1849–1870. doi: 10.1088/0031-9155/54/7/001. [DOI] [PubMed] [Google Scholar]
- 15.Castillo E, Castillo R, Martinez J, Shenoy M, Guerrero T. Four-dimensional deformable image registration using trajectory modeling. Phys Med Biol. 2010;55:305–327. doi: 10.1088/0031-9155/55/1/018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muenzing SEA, et al. Supervised quality assessment of medical image registration: Application to intra-patient CT lung registration. Medical Image Analysis. 2012;16(8):1521–1531. doi: 10.1016/j.media.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 17.Gaidhane VH, Hote YV, Singh V. Nonrigid image registration using efficient similarity measure and Levenberg-Marquardt optimization. Biomedical Engineering Letters. 2012;2(2):118–123. [Google Scholar]
- 18.Heinrich MP, et al. MIND: Modality independent neighborhood descriptor for multi-modal deformable registration. Medical Image Analysis. 2012;16:1423–1435. doi: 10.1016/j.media.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Yan C, et al. A method to evaluate dose errors introduced by dose mapping processes for mass conserving deformations. Medical Physics. 2012;39:2119–2028. doi: 10.1118/1.3684951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vandemeulebroucke J, et al. Automated segmentation of a motion mask to preserve sliding motion in deformable registration of thoracic CT. Medical Physics. 2012;39:1006–1015. doi: 10.1118/1.3679009. [DOI] [PubMed] [Google Scholar]
- 21.Gorbunova V, et al. Mass preserving image registration for lung CT. Medical Image Analysis. 2012;16(4):786–795. doi: 10.1016/j.media.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Bender ET, Hardcastle N, Tome WA. On the dosimetric effect and reduction of inverse consistency and transitivity errors in deformable image registration for dose accumulation. Medical Physics. 2012;39:272–280. doi: 10.1118/1.3666948. [DOI] [PubMed] [Google Scholar]
- 23.Castillo R, et al. Spatial correspondence of 4D CT ventilation and SPECT pulmonary perfusion defects in patients with malignant airway stenosis. Physics in Medicine and Biology. 2012;57(7):1855–1871. doi: 10.1088/0031-9155/57/7/1855. [DOI] [PubMed] [Google Scholar]
- 24.Shusharina N, Sharp G. Analytic regularization for landmark-based image registration. Physics in Medicine and Biology. 2012;57(6):1477–1498. doi: 10.1088/0031-9155/57/6/1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castillo E, et al. Least median of squares filtering of locally optimal point matches for compressible flow image registration. Physics in Medicine and Biology. 2012;57(15):4827–4833. doi: 10.1088/0031-9155/57/15/4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shusharina N, Sharp G. Image registration using radial basis functions with adaptive radius. Medical Physics. 2012;39:6542–6549. doi: 10.1118/1.4756932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mencarelli A, et al. Validation of deformable registration in head and neck cancer using analysis of variance. Medical Physics. 2012;39:6879–6884. doi: 10.1118/1.4760990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu G, Lian J, Shen D. Improving image-guided radiation therapy of lung cancer by reconstructing 4DCT from a single free-breathing 3DCT on the treatment day. Medical Physics. 2012;39(12):7694–7709. doi: 10.1118/1.4768226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sommer S, Nielsen M, Pennec X. Sparse multi-scale diffeomorphic registration: the kernel bundle framework Journal of Mathematical Imaging and Vision. ARTICLE IN PRESS; 2012. [Google Scholar]
- 30.Lopez A.D.a.M.C.C.J.L. The global burden of disease, 1990-2020. Nature Medicine. 1998;4(11):1241–1243. doi: 10.1038/3218. [DOI] [PubMed] [Google Scholar]
- 31.Akira M, et al. Quantitative CT in chronic obstructive pulmonary disease: Inspiratory and expiratory assessment. Am J Roentgenol. 2009;192:267–272. doi: 10.2214/AJR.07.3953. [DOI] [PubMed] [Google Scholar]
- 32.Kubo K, et al. Expiratory and inspiratory chest computed tomography and pulmonary function tests in cigarette smokers. Eur Respir J. 1999;13:252–256. doi: 10.1034/j.1399-3003.1999.13b06.x. [DOI] [PubMed] [Google Scholar]
- 33.Yamashiro T, et al. Collapsibility of lung volume by paired inspiratory and expiratory CT scans: Correlations with lung function and mean lung density. Academic Radiology. 2010;17:489–495. doi: 10.1016/j.acra.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaporozhan J, Ley S, Eberhardt R, Weinheimer O, Iliyushenko S, Herth F, Kauczor HU. Paired inspiratory/expiratory volumetric thin-slice CT scan for emphysema analysis: Comparison of different quantitative evaluations and pulonary function test. Chest. 2005;128:3212–3220. doi: 10.1378/chest.128.5.3212. [DOI] [PubMed] [Google Scholar]
- 35.Murphy K, et al. Toward automatic regional analysis of pulmonary function using inspiration and expiration thoracic CT. Medical Physics. 2012;39(3):1650–1662. doi: 10.1118/1.3687891. [DOI] [PubMed] [Google Scholar]
- 36.Galban CJ, et al. Computed tomography-based biomarker provides unique signature for diagnosis of COPD phenotypes and disease progression. Nature Medicine. 2012;18:1711–1715. doi: 10.1038/nm.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Regan EA, et al. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis JP. Fast Template Matching. Vision Interface. 1995:120–123. [Google Scholar]
- 39.Gonzalez RC, Woods RE. Digital Image Processing. 3 ed Prentice-Hall, Inc.; Upper Saddle River, NJ: 2008. p. 954. [Google Scholar]
- 40.Bos LP, Salkauskas K. Moving least-squares are Backus-Gilbert optimal. Journal of Approximation Theory. 1989;59:267–275. [Google Scholar]
- 41.Lancaster P, Salkauskas K. Surfaces generated by moving least squares methods. Math Comp. 1981;37:141–158. [Google Scholar]
- 42.Schaefer S, McPhail T, Warren J. ACM SIGGRAPH 2006 Papers. ACM Press; Boston, MA: 2006. Image deformation using moving least squares. [Google Scholar]
- 43.Castillo R, Castillo E, Martinez J, Guerrero T. Ventilation from four-dimensional computed tomography: density versus Jacobian methods. Phys Med Biol. 2010;55(16):4661–4685. doi: 10.1088/0031-9155/55/16/004. [DOI] [PubMed] [Google Scholar]
- 44.Goldman LW. Principles of CT: Radiation dose and image quality. J Nucl Med Technol. 2007;35:213–225. doi: 10.2967/jnmt.106.037846. [DOI] [PubMed] [Google Scholar]
- 45.Seppenwoolde Y, et al. Precise and real-time measurement of 3D tumor motion in lung due to breathing and heartbeat, measured during radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2002;53(4):822–34. doi: 10.1016/s0360-3016(02)02803-1. [DOI] [PubMed] [Google Scholar]
- 46.Beek EJR, Hoffman EA. Imaging in COPD. Imaging Decisions MRI. 2009;13:11–17. [Google Scholar]
- 47.Holmes DR, III, et al. The Lung Tissue Research Consortium: An extensive open database containing histological, clinical, and radiological data to study chronic lung disease. The Insight Journal, 2006 MICCAI Open Science Workshop. 2006:1–5. [Google Scholar]
- 48.Punturieri A, et al. Chronic obstructive pulmonary disease: A view from the NHLBI. Am J Respir Crit Case Med. 2008;178:441–443. doi: 10.1164/rccm.200807-1128OE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castillo R, et al. Interior landmarks improve deformable image registration spatial accuracy in the lung. Int. J. Radiation Oncology Biol. Phys. 2008;72(1S):S452. [Google Scholar]
- 50.Murphy K, et al. Semi-automatic construction of reference standards for evaluation of image registration. Medical Image Analysis. 2011;15:71–84. doi: 10.1016/j.media.2010.07.005. [DOI] [PubMed] [Google Scholar]