Abstract
Rhodopsin is the first G-protein-coupled receptor (GPCR) with its three-dimensional structure solved by X-ray crystallography. The crystal structure of rhodopsin has revealed the molecular mechanism of photoreception and signal transduction in the visual system. Although several other GPCR crystal structures have been reported over the past few years, the rhodopsin structure remains an important model for understanding the structural and functional characteristics of other GPCRs. This review summarizes the structural features, the photoactivation, and the G protein signal transduction of rhodopsin.
Keywords: G-protein-coupled receptor (GPCR), rhodopsin, crystal structure, photoactivation, transducin
Introduction
Rhodopsin, a visual pigment found in the rod photoreceptor cells of the retina, is responsible for converting photons into chemical signals that stimulate biological processes in the nervous systems of humans and other vertebrate animals, allowing them to sense light1. Rhodopsin is a member of class A of the GPCR superfamily2, which is a large group of cell surface signaling receptors that transduce extracellular signals into intracellular pathways through the activation of heterotrimeric G proteins. The human GPCR superfamily, comprised of about 900 members, is involved in many aspects of human physiology and diseases, and it represents the most important protein targets for pharmaceutical drug discovery2.
The crystal structure of ground-state bovine rhodopsin containing the reverse agonist 11-cis-retinal was the first high-resolution GPCR structure solved by X-ray crystallography3, 4, 5, 6. Recently, crystal structures have been published for both opsin and rhodopsin in active conformations, with or without the binding of a peptide derived from the C-terminal helix α5 of the α subunit of G protein transducin7, 8, 9, 10, 11. The crystal structures of rhodopsin and the results of related biochemical and biophysical studies12 have revealed the molecular mechanisms of photoactivation and visual signal transduction, leading to a significant progress in understanding the signaling pathways of the entire GPCR superfamily13.
As the first solved GPCR crystal structure, bovine rhodopsin has been used as a structural template in many efforts of molecular modeling and in designing therapeutic drugs for human diseases related to GPCR signaling pathways14. More recently, crystal structures have been published for β1-adrenergic receptor15, A2A adenosine receptor16, β2-adrenergic receptor17, 18, 19, 20, 21, and the complex of β2-adrenergic receptor with a trimeric G protein21. These results have further enriched our understanding of ligand-induced activation and downstream signal transduction by GPCRs.
In this review we summarize the structural features, the photoactivation, and the G protein signal transduction of rhodopsin.
Overall structure of bovine rhodopsin
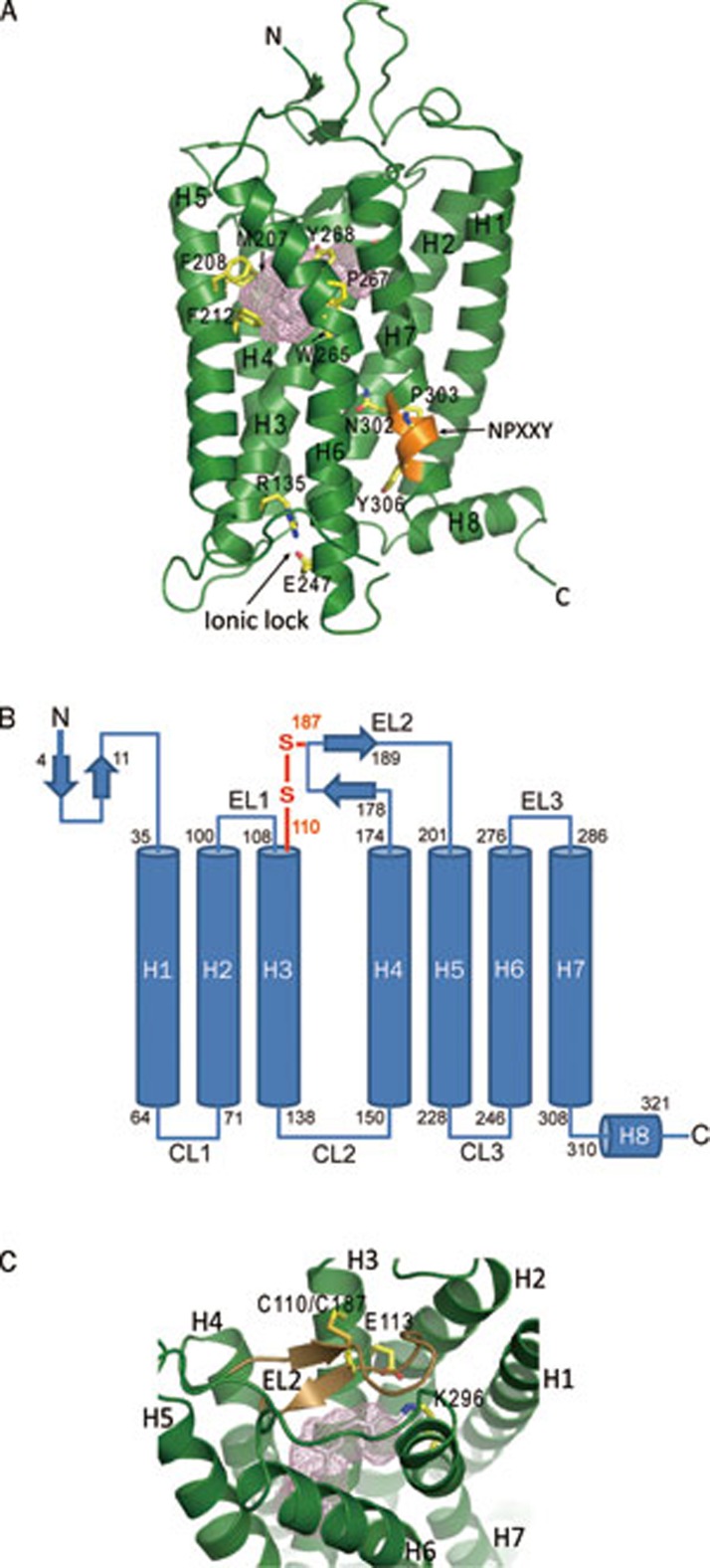
The crystal structure of inactive, 11-cis-retinal-bound bovine rhodopsin was first determined by Palczewski et al in 20003 and later by a few other groups4, 5, 6. The bovine rhodopsin structure features a seven-transmembrane (7TM) helix core architecture with three loop regions on both the extracellular and the cytoplasmic side of the membrane (Figure 1A and 1B). The N-terminus of rhodopsin, located on the extracellular side, consists of a two-stranded β sheet stretching from Gly4 to Pro11, followed by a loop region of about 24 amino acid residues (Figure 1A and 1B). The residues Asn2 and Asn15 are glycosylation sites for the receptor, and mutations that replace these residues with alanine lower the receptor's light-sensing activity22. The Thr4, Asn5, Thr17, Pro23, and Asn28 residues in this region have been proved to be crucial for the correct folding of the receptor, and mutations in these residues can lead to autosomal dominant retinitis pigmentosa (ADRP)23, 24.
Figure 1.
Overall structure of ground sate bovine rhodopsin and its key features (PDB: 1F88). (A) The seven-transmembrane helix domain with the retinal in gray stick and the ligand binding pocket shown as a pink mesh. Major ligand binding residues around the ligand binding pocket are shown as yellow sticks and are labeled. Other features include the ionic lock (yellow sticks) and the NPXXY motif (orange). (B) Two-dimensional sequence of bovine rhodopsin with the starting and ending residues of secondary structural elements indicated. The disulfide bond connecting EL2 to helix 3 is shown in orange. N, amino terminus; C, carboxyl terminus; EL, extracellular loop; CL, cytoplasmic loop. (C) The ligand binding pocket (pink mesh) of rhodopsin with EL2 (the lid of the pocket) shown in dark brown. The disulfide bond between C110 and C187 is labeled.
The second extracellular loop region (EL2), between helix 4 and helix 5 or from Gly175 to Asn200, is the longest among three extracellular loop regions, and it is essential for ligand binding of the receptor (Figure 1C). Biochemical data have shown that the interaction of EL2 with the helix bundle is important for the correct folding and biological function of the receptor25. The majority of this loop region, from Tyr178 to Ile189, forms a twisted two-stranded β sheet that is positioned at the opening of the ligand binding pocket, serving as a “lid” to block rapid exit of the ligand from the pocket (Figure 1C). The position of this β sheet is stabilized by hydrophobic interactions between residues Tyr178, Pro180, Met183 (Leu in humans), Cys185, and Cys187 of EL2, and surrounding residues of the helix bundle, especially those from helices 1, 2, 3, and 7. It is also stabilized by a disulfide bond between Cys187 of the β sheet and Cys110 of helix 3, and by a hydrogen bonding network among Glu181 and Tyr192 of EL2, Tyr268 of helix6 and a few water molecules. Between the two strands of this β sheet is a salt bridge formed by Asp190 and Arg177, which is a key interaction for maintaining the functional conformation of this lid over the retinal binding site (Figure 1C)26.
The architecture of the 7TM domain is a common feature across the GPCR superfamily with many conserved residues16, 17, 18, 19, 20. Among the seven helices, helix 6, from Lys246 to His276 in the ground-state model, bends about 36° at Pro267, with both ends facing away from the core of the 7TM domain (Figure 1A)3. Upon photoactivation, the cytoplasmic end of this helix shifts further away from the 7TM core, and this shift creates a crevice in the cytoplasmic side of the receptor for binding the α subunit of G protein transducin8, 11.
The ligand binding site of rhodopsin is a pocket on the extracellular side of the transmembrane bundle. It is surrounded by hydrophobic residues that stabilize the polyene backbone of the retinal, among which are Met207, Phe208, and Phe212 from helix 5, and Trp265 and Tyr268 from helix 6 (Figure 1A and 1C)3, 4, 6. The positions and the conformations of those hydrophobic residues undergo dynamic changes during the photoactivation of the retinal and the conformational changes in the receptor7, 8, 11.
Another important feature of this receptor is the “ionic lock”, a salt bridge between Arg135 of helix 3 and Glu247 of helix 6 (Figure 1A)3, 4, 6. This salt bridge blocks the G protein binding site of the receptor in its inactive conformation. Upon photoactivation, the transmembrane bundle undergoes a conformational change: the cytoplasmic side of helix 6 bends further away from the 7TM core and the ionic lock breaks, resulting in an opening on the cytoplasmic side of the receptor for G protein interaction7, 8, 11.
A conserved NPXXY motif on helix 7 is also common to all the GPCR family members based on sequence alignment and crystal structures3, 16, 17, 18. It has an important role in receptor activation: this motif shifts toward helix 6 and the key residue Tyr306 on this motif flips toward helix 6, helping breaking the ionic lock and pushing helix 6 away from the transmembrane bundle (Figure 1A).
The C-terminus of rhodopsin is on the cytoplasmic side of the membrane, extending from residue Met309 at the C-terminal end of helix 7 to the last residue of the receptor, and featuring a short amphipathic helix (helix 8) perpendicular to helix 7 (Figure 1A and 1B)3, 4, 6. It forms hydrophobic interactions by its residues Phe313, Cys316, and Met317 with residues Leu57, Val61 of helix 1 and His65 in the loop following helix 1, and is covalently anchored to the membrane by palmitoylation of residues Cys322 and Cys323 in the loop following helix 8. The C-terminal loop following residue Asn326 is disordered in all the crystal structures. In this region there are serine residues Ser334, Ser338, and Ser343, whose phosphorylation is important for arrestin binding to terminate the cycle of G protein activation27.
The loop region CL3 between helices 5 and 6 on the cytoplasmic side is flexible and largely disordered in ground-state rhodopsin crystal structures, but is structured in most activated rhodopsin models8, 9, 10, 11 (Figure 1A and 1B). Upon activation, the majority of this loop region becomes part of the elongated helix 5 that provides more interface for G protein interaction.
Ligand-induced conformational change and activation of rhodopsin
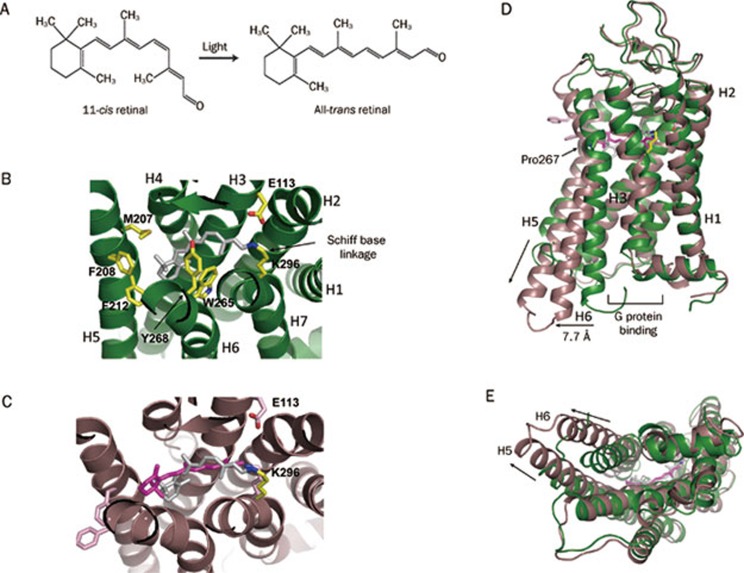
Retinal is one of the vitamin A compounds derived from carotenoids (Figure 2A). It is the photoactive moiety of rhodopsin that captures light and converts photons into chemical signals. Vision starts with the absorption of photons, and the photon-triggered isomerization of the retinal from the 11-cis to the all-trans state, followed by conformational changes in the 7TM domain of rhodopsin to accommodate the binding of G proteins, leading to the downstream signal transduction28.
Figure 2.
Ligand binding and conformational changes in rhodopsin. (A) Chemical structures of 11-cis- and all-trans-retinal. (B) 11-cis-retinal (in gray) in the ligand binding pocket is associated with the surrounding residues of the protein moiety (green, PDB: 1F88). (C) Conformational changes in retinal and the protein moiety of rhodopsin upon photoactivation. The photoactivated all-trans-retinal (PDB: 3PQR) is magenta and the ground-state 11-cis-retinal (PDB: 1F88, gray) is superposed on the activated all-trans-retinal for comparison. The protein moiety of activated rhodopsin (PDB: 3PQR) is dark brown. (D) The key conformational changes in rhodopsin upon photoactivation are the outward tilting of the cytoplasmic end of helix 6 (indicated by the horizontal arrow), creating a crevice for G-protein binding, and the elongation of the cytoplasmic end of helix 5 (indicated by the vertical arrow) that provides more interface for G-protein interaction. Green shows the ground-state conformation (PDB: 1F88), and brown shows the activated conformation (PDB: 3PQR). (E) Bottom view of panel D.
The ligand binding pocket of rhodopsin, with a volume of about 352 Å3, is located on the luminal side of the receptor (Figure 1A, 1C, and 2B). Prior to photoactivation, 11-cis-retinal in the ligand binding pocket is covalently bound to Lys296 of helix 7 through a Schiff base linkage. The Schiff base is protonated and the linkage is stabilized by charge delocalization over the π-electron system of the polyene backbone of the retinal chromophore, and by an electrostatic interaction with the carboxyl group of Glu113 of helix 3 (Figure 2B)3, 4, 6. The retinal in the ligand binding pocket is also stabilized by the surrounding hydrophobic residues including Met207, Phe208 and Phe212 from helix 5 and Trp265 and Tyr268 from helix 6 (Figure 2B). The Schiff base linkage and its electrostatic interaction are essential for maintaining the ground-state conformation of rhodopsin, and a mutation that alters either Lys296 or Glu113 results in constitutive activation of the receptor29.
Upon light absorption, the π-electron system of the retinal chromophore undergoes a π→π* transition that lowers the potential energy barrier for cis-trans isomerization. The isomerization changes the retinal from a bending cis configuration to a straight trans configuration. It flips the aldehyde group and the C20 methyl group about 180° around the C11=C12 double bond, and tilts the β-ionone group toward the space between helices 5 and 6 (Figure 2C). The photon-induced cis-trans transition in the retinal leads to a sterically strained excited-state rhodopsin that thermally decays through a series of intermediates, photorhodopsin, bathorhodopsin, lumirhodopsin, and metarhodopsin I, and forms the active-state metarhodopsin II7, 8, 11, 30, 31, 32. During the thermal relaxation, the protein moiety of rhodopsin undergoes a series of conformational changes that lead to the formation of a G protein binding site on the cytoplasmic side of the 7TM bundle7, 8, 9, 10, 11 (Figure 2D and 2E). The major conformational changes are the bending of helix 6 at Pro267, and the eight-residue elongation of the C-terminus of helix 5, the addition coming from the CL3 loop (Figure 2D and 2E). The bending of helix 6 results in the cytoplasmic end of this helix tilting away from the transmembrane core, and creates an opening with a diameter of about 14 Å8, 10, 11. The cytoplasmic end of helix 6 in all-trans-retinal bound rhodopsin is about 7.7 Å away from that of the cis-retinal bound conformation. The Pro267 residue of helix 6 is highly conserved among GPCR family members, and it is believed that it serves as a hinge for the bending because it has no hydrogen bond between its backbone nitrogen atom and the carbonyl group of the residue one helical turn upstream. The elongation of the cytoplasmic side of helix 5 expands the interface for G protein interaction, and thus contributes the binding affinity of rhodopsin for the G protein transducin (Figure 2D and 2E).
Rhodopsin-G protein interaction
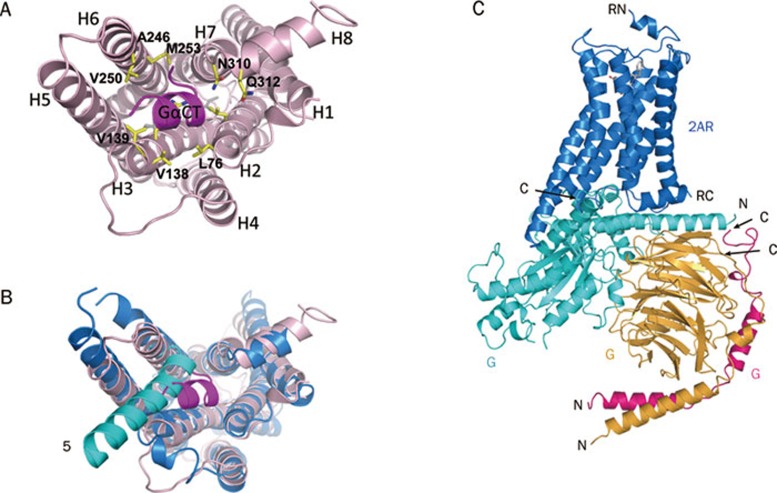
The binding of the C-terminus of the α subunit of transducin to rhodopsin was defined by the crystal structure of rhodopsin in complex with a synthetic peptide, GαCT (Gα C-terminus), derived from the C-terminus of helix α5 of the Gα subunit8, 10, 11 (Figure 3A). The outward movement of helix 6 of rhodopsin breaks the ionic lock between Arg135 on helix 3 and Glu247 on helix 6, thus creating an opening that accommodates the C-terminal helix of the α subunit of transducin. The binding of the Gα peptide on the cytoplasmic side of rhodopsin is largely facilitated by two sets of interactions: a) hydrophobic interactions between the peptide residues Leu341, Leu344, Val347, Leu349, and Phe350, and the receptor residues Ala246, Val250, and Met253 on helix 6, Val138 and Val139, and the backbone of Arg135 on helix 3, and Leu72 and Leu76 on helix 2, and 6) polar interactions between the carbonyl groups of residues Gly348 and Lys345 of the peptide, and the residues Asn310 and Q312 of helix 8 (Figure 3A). The binding of the α subunit of transducin to rhodopsin induces the exchange of GDP for GTP on the α subunit, and this exchange triggers the dissociation of the GTP-bound Gα subunit from the Gβγ dimer and the receptor. Both the dissociated GTP-bound α subunit and the βγ dimer of transducin then activate downstream signaling pathways33.
Figure 3.
Rhodopsin-G protein interface. (A) Rhodopsin in complex with a synthetic peptide derived from helix α5 of the Gα subunit of transducin (PDB: 3PQR). The rhodopsin residues interacting with the Gα peptide are labeled. (B) Comparison of the binding mode of rhodopsin with the synthetic peptide and that of the β2-adrenergic receptor with intact G protein (PDB: 3SN6). The β2-adrenergic receptor is blue and the Gα subunit is cyan. For clarity, only helix α5 of the Gα subunit is shown. (C) The whole-complex model of the β2-adrenergic receptor with intact G protein (PDB: 3SN6). The β2-adrenergic receptor is blue, the Gα subunit is cyan, the Gβ subunit is brown, and the Gγ subunit is pink. RN, N-terminus of the receptor; RC, C-terminus of the receptor; αN, N-terminus of the Gα subunit; αC, C-terminus of the Gα subunit; βN, N-terminus of the Gβ subunit; βC, C-terminus of the Gβ subunit; γN, N-terminus of the Gγ subunit; γC, C-terminus of the Gγ subunit.
Comparison of the rhodopsin-Gα peptide structure with the recently published crystal structure of the β2-adrenergic receptor in complex with its trimeric G protein21 revealed noteworthy differences. Helix α5 of the intact Gα subunit binds to the receptor in a position tilted about 38° toward helix 6 and 2 Å away from the bottom of the crevice relative to the position of the Gα peptide in the active rhodopsin-peptide complex (Figure 3B and 3C). The intact G protein α subunit does not fit deeply in the crevice of the receptor as the peptide does, probably because the whole Gα subunit is much larger, and therefore is not able to fit in the deep crevice in the transmembrane bundle of the receptor. Also in the β2-AR-G protein complex, helix 6 is bent outward about 26° more than that in the activated rhodopsin structure, and the opening of the crevice is larger than that of rhodopsin, to accommodate the intact C-terminal helix of the Gα subunit in the β2-AR-G protein complex (Figure 3B and 3C). Because the crystal structure of a rhodopsin-full length G protein complex is not available, we still do not have the whole picture of the interface of rhodopsin with the Gα subunit as a full length protein in the context of intact trimeric transducin.
Structural understanding of disease-related rhodopsin mutations
Rhodopsin is central to the process of vision. Mutations in rhodopsin are major causes of vision diseases or disorders. More than 120 point mutations have been identified in human gene of rhodopsin, many of which lead to vision diseases such as ADRP and congenital stationary night blindness (see http://www.retina-international.org/sci-news/rhomut.htm for most disease-linked mutations). Crystal structures have provided us molecular basis for understanding how mutations in rhodopsin influence the protein folding, stability and/or biological functions of the receptor.
Retinal is the photoactive chromophore in rhodopsin, and is the key to the entire light signal transduction. Crystal structures have revealed that 11-cis-retinal is covalently bound by Lys296 of helix 7 in ground state rhodopsin (Figure 2A and 2B). Mutations of this retinal binding residue, K296M and K296E, cause severe ADRP34. These mutants have been found to be folded properly, but constitutively activated35. Substitutions of other residues interacting with or at proximity of retinal can change retinal binding affinity of the receptor. It has been identified that mutations of those residues M44T36, G114D37, G114V38, L125R39, C167R40, Y178N41, Y178C42, E181K40, S186P40, S186W43, G188R44, G188E45, M207R46, M216R47, and M216K48 (Lue216 in bovine), cause different levels of ADRP disease. Mutations of G90D49 and A292E50, two other retinal binding residues, have been found in patients with congenital stationary night blindness.
The conserved residue Pro267 is at the kink in helix 6 and serves as a hinge for the bending of helix 6 to create the G protein binding site at cytoplasmic side of rhodopsin. Replacement of this residue with other non-proline residues affects the conformation of the G protein binding site of rhodopsin and the kinetics of transducin activation51, 52. Mutations of P267L and P267R have been reported to cause ADRP41, 52, 53.
Based on the crystal structure of rhodopsin in complex with peptide GαCT8, 10, 11, the binding of the α subunit of transducin to rhodopsin is facilitated by hydrophobic residues at the interface of the two proteins. Cys140 in CL2 is an important residue at rhodopsin side for G protein binding54. Mutation C140S that lowers the hydrophobicity of the residue has been found associated with ADRP45.
Mutations of residue Cys110 to Arg, Phe, or Tyr37, 55, or Cys187 to Tyr56, have been identified in ADRP patients. The conserved residues Cys110 and Cys187 forms a disulfide bond that constrains EL2, the lid of the ligand binding pocket, and is required for the biological function of the receptor (Figure 1B)57. Mutations of those residues break the disulfide bond, thus affect the folding and biological function of the receptor.
GPCRs are membrane proteins and their interactions with the phospholipid bilayer membrane have great influence on their physiological functions58. Mutations at the membrane interaction interface of rhodopsin can change the affinity of rhodopsin to the phospholipid bilayers and influence the functions of the receptor. Lue46 is a hydrophobic residue at the membrane interaction interface of rhodopsin, and mutation of L46R largely affects the membrane-receptor interaction and causes severe ADRP59. Other disease-linked mutations at the receptor-membrane interface are F45L60, P53R61, and V209M45.
Some disease-causing mutations in rhodopsin occur at posttranslational modification residues including glycosylation or phosphorylation sites. Glycosylation is important for correct folding and stability of most eukaryotic proteins. N-terminal glycosylation sites of rhodopsin are located at Asn2, Thr4, Asn15 and Thr17, and mutations of those residues, T4K62, N15S63, and T17M60, are responsible for ADRP. Rhodopsin phosphorylation is a key step of the regulatory mechanism of light signal transduction. Activated rhodopsin is phosphorylated by rhodopsin kinase at multiple sites in its C-terminal region, and the phosphorylation facilitates the association of arrestin that completely terminates light signal transduction. Mutations of a rhodopsin kinase binding residue Arg135 to Gly45, Trp60, Pro64, or Leu60, and the mutation of a phosphorylation site Thr342 to Met, which affect phosphorylation and rhodopsin-arrestin interaction, lead to ADRP65.
Comparison of the activation of GPCRs and nuclear hormone receptors
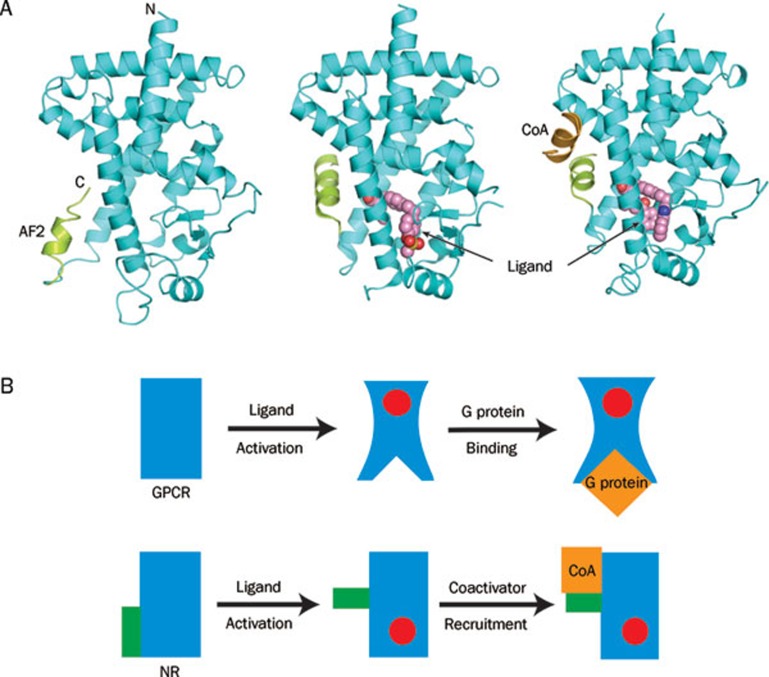
Nuclear hormone receptors (NRs) and GPCRs are two different groups of protein receptors that sense extracellular signals and transduce those signals to the inside of cells. GPCRs transmit signals by coupling ligand-receptor interactions to G protein activation. Activated G proteins then trigger the downstream signaling cascades in the cytoplasm to regulate various biological events in the cells. In contrast, ligand binding converts NRs into transcriptional activators that directly bind target genes and stimulate their expression. A typical nuclear receptor ligand binding domain forms a conserved helical sandwich fold that harbors a ligand binding pocket adjacent to its C-terminal activation function helix AF2. Ligand binding does not change the helical sandwich scaffold but induces the rearrangement of the receptor's AF2 helix and the formation of a coactivator binding cleft for the recruitment of transcriptional coactivators, which further facilitate the formation of transcription complexes and subsequent gene expression66 (Figure 4A). In the cascade of NR activation, the conformation of the receptor core structure remains unchanged. That is a major difference from the activation of a GPCR, in which the core 7TM domain is rearranged upon ligand binding and receptor activation and the core domain rearrangement creates a crevice at the cytoplasmic side of the receptor for G protein interaction and signal transduction (Figure 4B)10, 11, 21.
Figure 4.
A comparison of the ligand-induced activation modes of GPCRs and nuclear receptors. (A) Ligand-induced rearrangement of the C-terminal AF2 helix and coactivator recruitment of a peroxisome proliferator-activated receptor ligand binding domain (PPAR LBD). At left is an apo PPAR LBD (PDB: 1PRG)69; middle, the LBD upon the ligand binding-induced conformational change in the AF2 helix and the formation of the coactivator binding site (PDB: 1I7G)70; at right, the LBD upon the subsequent coactivator recruitment (PDB: 1K7L)71. The LBD core structure is cyan; the AF2 helix, green; and the coactivator motif, brown. (B) Cartoon presentation showing that ligand activation induces conformational changes in the core domain of GPCRs but not in that of NRs. Blue are the core domains of both receptors; Red are the ligands; Orange are the G protein binding to the GPCR and the coactivator binding to the NR.
While GPCRs and NRs are functionally comparable, the GPCR group has many more members than the NR family, and senses a wide variety of environmental signals. It is interesting that the GPCR group members, in spite of their highly diverse ligands, share a highly conserved 7TM core architecture for ligand binding and G protein interaction, whereas the NRs share a conserved three-layered helix bundle for ligand binding and coactivator recruitment. In summary, ligand binding and activation in GPCRs involve a much more extensive rearrangement of the 7TM helical core than the helical sandwich of NRs.
Rhodopsin as a molecular model for GPCR studies
During the long period before the second GPCR crystal structure was published in 200717, 18, rhodopsin was the only GPCR crystal structure available, and it has been used extensively as a model for understanding the structural and functional characteristics of other GPCRs67, 68. Whereas more GPCR crystal structures have been published and the importance of the rhodopsin structure as a molecular template for modeling other GPCRs has been correspondingly diminished, rhodopsin still remains a prototype of the GPCR superfamily and a model system for all 7TM domain proteins.
All GPCR structures have a conserved transmembrane core domain followed by helix 8 on the cytoplasmic side. The root mean square deviation (RMSD) of the residues of the 7TM core structures between rhodopsin and other GPCRs whose structures have been solved are among 1.2 to 1.4 Å, indicating a close similarity and high level of conservation. The most significant structural differences between rhodopsin and other GPCRs are in the ligand binding pocket and the lid covering the pocket, the EL2 loop region. The EL2 of rhodopsin adopts a β-sheet fold, which tightly plugs into the entrance of the pocket, while the EL2 regions of other solved GPCR structures are loosely positioned above the ligand binding pocket and can be more easily opened for ligands to move in or out. This is consistent with the observation that rhodopsin is activated by the photon-triggered isomerization of retinal in the ligand binding pocket, which requires the ligand not only being bound but also being tightly hold in the pocket, while most other GPCRs are activated by simply binding to the ligands. Although the mechanism of activation differs, rhodopsin and other solved GPCR structures share conserved core residues that define the conformation of the seven transmembrane domain and the molecular basis of the conformational change upon ligand activation, G protein interaction, and downstream signal transduction.
Acknowledgments
We thank D NADZIEJKA for editing the manuscript. This work was supported in part by the Jay and Betty Van Andel Foundation, and National Institute of Health grant GM087413 (H Eric XU).
References
- Hubbard R, Kropf A. The action of light on rhodopsin. Proc Natl Acad Sci U S A. 1958;44:130–9. doi: 10.1073/pnas.44.2.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson R, Lagerström MC, Lundin LG, Schiöth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:17. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–45. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–38. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- Standfuss J, Xie G, Edwards PC, Burghammer M, Oprian DD, Schertler GF. Crystal structure of a thermally stable rhodopsin mutant. J Mol Biol. 2007;372:1179–88. doi: 10.1016/j.jmb.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J Mol Biol. 2004;342:571–83. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- Salom D, Lodowski DT, Stenkamp RE, Le Trong I, Golczak M, Jastrzebska B, et al. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc Natl Acad Sci U S A. 2006;103:16123–8. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Standfuss J, Edwards PC, D'Antona A, Fransen M, Xie G, Oprian DD, et al. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–60. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–7. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- Choe HW, Kim YJ, Park JH, Morizumi T, Pai EF, Krauss N, et al. Crystal structure of metarhodopsin II. Nature. 2011;471:651–5. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- Brown MF, Martinez-Mayorga K, Nakanishi K, Salgado GF, Struts AV. Retinal conformation and dynamics in activation of rhodopsin illuminated by solid-state H NMR spectroscopy. Photochem Photobiol. 2009;85:442–53. doi: 10.1111/j.1751-1097.2008.00510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jastrzebska B, Tsybovsky Y, Palczewski K. Complexes between photoactivated rhodopsin and transducin: progress and questions. Biochem J. 2010;428:1–10. doi: 10.1042/BJ20100270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch L, Iarriccio L, Garriga P. New prospects for drug discovery from structural studies of rhodopsin. Curr Pharm Des. 2005;11:2243–56. doi: 10.2174/1381612054367436. [DOI] [PubMed] [Google Scholar]
- Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, et al. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–91. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, Lane JR, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–7. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–7. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–65. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–80. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, et al. Structure and function of an irreversible agonist: beta(2) adrenoceptor complex. Nature. 2011;469:236–40. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, Devree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, et al. Crystal structure of the beta(2) adrenergic receptor–Gs protein complex. Nature. 2011;477:549–55. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal S, Ridge KD, Khorana HG. Structure and function in rhodopsin: the role of asparagine-linked glycosylation. Proc Natl Acad Sci U S A. 1994;91:4024–8. doi: 10.1073/pnas.91.9.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung CH, Davenport CM, Hennessey JC, Maumenee IH, Jacobson SG, Heckenlively JR, et al. Rhodopsin mutations in autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci U S A. 1991;88:6481–5. doi: 10.1073/pnas.88.15.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Maghtheh M, Gregory C, Inglehearn C, Hardcastle A, Bhattacharya S. Rhodopsin mutations in autosomal dominant retinitis pigmentosa. Hum Mutat. 1993;2:249–55. doi: 10.1002/humu.1380020403. [DOI] [PubMed] [Google Scholar]
- Karnik SS, Khorana HG. Assembly of functional rhodopsin requires a disulfide bond between cysteine residues 110 and 187. J Biol Chem. 1990;265:17520–4. [PubMed] [Google Scholar]
- Janz JM, Fay JF, Farrens DL. Stability of dark state rhodopsin is mediated by a conserved ion pair in intradiscal loop E-2. J Biol Chem. 2003;278:16982–91. doi: 10.1074/jbc.M210567200. [DOI] [PubMed] [Google Scholar]
- Ohguro H, Rudnicka-Nawrot M, Buczylko J, Zhao X, Taylor JA, Walsh KA, et al. Structural and enzymatic aspects of rhodopsin phosphorylation. J Biol Chem. 1996;271:5215–24. doi: 10.1074/jbc.271.9.5215. [DOI] [PubMed] [Google Scholar]
- Chabre M. Trigger and amplification mechanisms in visual phototransduction. Annu Rev Biophys Biophys Chem. 1985;14:331–60. doi: 10.1146/annurev.bb.14.060185.001555. [DOI] [PubMed] [Google Scholar]
- Rao VR, Oprian DD. Activating mutations of rhodopsin and other G protein-coupled receptors. Annu Rev Biophys Biomol Struct. 1996;25:287–314. doi: 10.1146/annurev.bb.25.060196.001443. [DOI] [PubMed] [Google Scholar]
- Nakamichi H, Okada T. Crystallographic analysis of primary visual photochemistry. Angew Chem Int Ed Engl. 2006;45:4270–3. doi: 10.1002/anie.200600595. [DOI] [PubMed] [Google Scholar]
- Nakamichi H, Okada T. Local peptide movement in the photoreaction intermediate of rhodopsin. Proc Natl Acad Sci U S A. 2006;103:12729–34. doi: 10.1073/pnas.0601765103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruprecht JJ, Mielke T, Vogel R, Villa C, Schertler GF. Electron crystallography reveals the structure of metarhodopsin I. EMBO J. 2004;23:3609–20. doi: 10.1038/sj.emboj.7600374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge KD, Palczewski K. Visual rhodopsin sees the light: structure and mechanism of G protein signaling. J Biol Chem. 2007;282:9297–301. doi: 10.1074/jbc.R600032200. [DOI] [PubMed] [Google Scholar]
- Yang T, Snider BB, Oprian DD. Synthesis and characterization of a novel retinylamine analog inhibitor of constitutively active rhodopsin mutants found in patients with autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci U S A. 1997;94:13559–64. doi: 10.1073/pnas.94.25.13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen TJ, Inglehearn CF, Lester DH, Bashir R, Jay M, Bird AC, et al. Autosomal dominant retinitis pigmentosa: four new mutations in rhodopsin, one of them in the retinal attachment site. Genomics. 1991;11:199–205. doi: 10.1016/0888-7543(91)90119-y. [DOI] [PubMed] [Google Scholar]
- Reig C, Antich J, Gean E, Garcia-Sandoval B, Ramos C, Ayuso C, et al. Identification of a novel rhodopsin mutation (Met-44-Thr) in a simplex case of retinitis pigmentosa. Hum Genet. 1994;94:283–6. doi: 10.1007/BF00208284. [DOI] [PubMed] [Google Scholar]
- Vaithinathan R, Berson EL, Dryja TP. Further screening of the rhodopsin gene in patients with autosomal dominant retinitis pigmentosa. Genomics. 1994;21:461–3. doi: 10.1006/geno.1994.1301. [DOI] [PubMed] [Google Scholar]
- Dryja TP, McEvoy JA, McGee TL, Berson EL. Novel rhodopsin mutations Gly114Val and Gln184Pro in dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2000;41:3124–7. [PubMed] [Google Scholar]
- Dryja TP. Doyne Lecture. Rhodopsin and autosomal dominant retinitis pigmentosa. Eye (Lond) 1992;6:1–10. doi: 10.1038/eye.1992.2. [DOI] [PubMed] [Google Scholar]
- Dryja TP, Hahn LB, Cowley GS, McGee TL, Berson EL. Mutation spectrum of the rhodopsin gene among patients with autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci U S A. 1991;88:9370–4. doi: 10.1073/pnas.88.20.9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souied E, Gerber S, Rozet JM, Bonneau D, Dufier JL, Ghazi I, et al. Five novel missense mutations of the rhodopsin gene in autosomal dominant retinitis pigmentosa. Hum Mol Genet. 1994;3:1433–4. doi: 10.1093/hmg/3.8.1433. [DOI] [PubMed] [Google Scholar]
- Farrar GJ, Kenna P, Redmond R, Shiels D, McWilliam P, Humphries MM, et al. Autosomal dominant retinitis pigmentosa: a mutation in codon 178 of the rhodopsin gene in two families of Celtic origin. Genomics. 1991;11:1170–1. [PubMed] [Google Scholar]
- Matias-Florentino M, Ayala-Ramirez R, Graue-Wiechers F, Zenteno JC. Molecular screening of rhodopsin and peripherin/RDS genes in Mexican families with autosomal dominant retinitis pigmentosa. Curr Eye Res. 2009;34:1050–6. doi: 10.3109/02713680903283169. [DOI] [PubMed] [Google Scholar]
- Liu X, Garriga P, Khorana HG. Structure and function in rhodopsin: correct folding and misfolding in two point mutants in the intradiscal domain of rhodopsin identified in retinitis pigmentosa. Proc Natl Acad Sci U S A. 1996;93:4554–9. doi: 10.1073/pnas.93.10.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macke JP, Davenport CM, Jacobson SG, Hennessey JC, Gonzalez-Fernandez F, Conway BP, et al. Identification of novel rhodopsin mutations responsible for retinitis pigmentosa: implications for the structure and function of rhodopsin. Am J Hum Genet. 1993;53:80–9. [PMC free article] [PubMed] [Google Scholar]
- Farrar GJ, Findlay JB, Kumar–Singh R, Kenna P, Humphries MM, Sharpe E, et al. Autosomal dominant retinitis pigmentosa: a novel mutation in the rhodopsin gene in the original 3q linked family. Hum Mol Genet. 1992;1:769–71. doi: 10.1093/hmg/1.9.769. [DOI] [PubMed] [Google Scholar]
- Haim M, Grundmann K, Gal A, Rosenberg T. Novel rhodopsin mutation (M216R) in a Danish family with autosomal dominant retinitis pigmentosa. Ophthalmic Genet. 1996;17:193–7. doi: 10.3109/13816819609057893. [DOI] [PubMed] [Google Scholar]
- Al-Maghtheh M, Inglehearn C, Lunt P, Jay M, Bird A, Bhattacharya S. Two new rhodopsin transversion mutations (L40R; M216K) in families with autosomal dominant retinitis pigmentosa. Hum Mutat. 1994;3:409–10. doi: 10.1002/humu.1380030417. [DOI] [PubMed] [Google Scholar]
- Sieving PA, Richards JE, Naarendorp F, Bingham EL, Scott K, Alpern M. Dark-light: model for nightblindness from the human rhodopsin Gly-90 → Asp mutation. Proc Natl Acad Sci U S A. 1995;92:880–4. doi: 10.1073/pnas.92.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryja TP, Berson EL, Rao VR, Oprian DD. Heterozygous missense mutation in the rhodopsin gene as a cause of congenital stationary night blindness. Nat Genet. 1993;4:280–3. doi: 10.1038/ng0793-280. [DOI] [PubMed] [Google Scholar]
- Nakayama TA, Khorana HG. Mapping of the amino acids in membrane-embedded helices that interact with the retinal chromophore in bovine rhodopsin. J Biol Chem. 1991;266:4269–75. [PubMed] [Google Scholar]
- Hwa J, Garriga P, Liu X, Khorana HG. Structure and function in rhodopsin: packing of the helices in the transmembrane domain and folding to a tertiary structure in the intradiscal domain are coupled. Proc Natl Acad Sci U S A. 1997;94:10571–6. doi: 10.1073/pnas.94.20.10571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield VC, Fishman GA, Beck JS, Kimura AE, Stone EM. Identification of novel rhodopsin mutations associated with retinitis pigmentosa by GC-clamped denaturing gradient gel electrophoresis. Am J Hum Genet. 1991;49:699–706. [PMC free article] [PubMed] [Google Scholar]
- Natochin M, Gasimov KG, Moussaif M, Artemyev NO. Rhodopsin determinants for transducin activation: a gain-of-function approach. J Biol Chem. 2003;278:37574–81. doi: 10.1074/jbc.M305136200. [DOI] [PubMed] [Google Scholar]
- Fuchs S, Kranich H, Denton MJ, Zrenner E, Bhattacharya SS, Humphries P, et al. Three novel rhodopsin mutations (C110F, L131P, A164V) in patients with autosomal dominant retinitis pigmentosa. Hum Mol Genet. 1994;3:1203. doi: 10.1093/hmg/3.7.1203. [DOI] [PubMed] [Google Scholar]
- Richards JE, Scott KM, Sieving PA. Disruption of conserved rhodopsin disulfide bond by Cys187Tyr mutation causes early and severe autosomal dominant retinitis pigmentosa. Ophthalmology. 1995;102:669–77. doi: 10.1016/s0161-6420(95)30972-4. [DOI] [PubMed] [Google Scholar]
- Hwa J, Klein-Seetharaman J, Khorana HG. Structure and function in rhodopsin: Mass spectrometric identification of the abnormal intradiscal disulfide bond in misfolded retinitis pigmentosa mutants. Proc Natl Acad Sci U S A. 2001;98:4872–6. doi: 10.1073/pnas.061632798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber T, Botelho AV, Beyer K, Brown MF. Membrane model for the G-protein-coupled receptor rhodopsin: hydrophobic interface and dynamical structure. Biophys J. 2004;86:2078–100. doi: 10.1016/S0006-3495(04)74268-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez JA, Herrera CA, Birch DG, Daiger SP. A leucine to arginine amino acid substitution at codon 46 of rhodopsin is responsible for a severe form of autosomal dominant retinitis pigmentosa. Hum Mutat. 1993;2:205–13. doi: 10.1002/humu.1380020309. [DOI] [PubMed] [Google Scholar]
- Sung CH, Schneider BG, Agarwal N, Papermaster DS, Nathans J. Functional heterogeneity of mutant rhodopsins responsible for autosomal dominant retinitis pigmentosa. Proc Natl Acad Sci U S A. 1991;88:8840–4. doi: 10.1073/pnas.88.19.8840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglehearn CF, Keen TJ, Bashir R, Jay M, Fitzke F, Bird AC, et al. A completed screen for mutations of the rhodopsin gene in a panel of patients with autosomal dominant retinitis pigmentosa. Hum Mol Genet. 1992;1:41–5. doi: 10.1093/hmg/1.1.41. [DOI] [PubMed] [Google Scholar]
- Bunge S, Wedemann H, David D, Terwilliger DJ, van den Born LI, Aulehla-Scholz C, et al. Molecular analysis and genetic mapping of the rhodopsin gene in families with autosomal dominant retinitis pigmentosa. Genomics. 1993;17:230–3. doi: 10.1006/geno.1993.1309. [DOI] [PubMed] [Google Scholar]
- Kranich H, Bartkowski S, Denton MJ, Krey S, Dickinson P, Duvigneau C, et al. Autosomal dominant 'sector' retinitis pigmentosa due to a point mutation predicting an Asn-15-Ser substitution of rhodopsin. Hum Mol Genet. 1993;2:813–4. doi: 10.1093/hmg/2.6.813. [DOI] [PubMed] [Google Scholar]
- Chuang JZ, Vega C, Jun W, Sung CH. Structural and functional impairment of endocytic pathways by retinitis pigmentosa mutant rhodopsin-arrestin complexes. J Clin Invest. 2004;114:131–40. doi: 10.1172/JCI21136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KM, Sieving PA, Bingham E, Bhagat VJ, Sullivan J, Alpern M, et al. Rhodopsin mutations associated with autosomal dominant retinitis pigmentosa. Am J Hum Genet. 1993;53:147. [Google Scholar]
- Li Y, Lambert MH, Xu HE. Activation of nuclear receptors: a perspective from structural genomics. Structure. 2003;11:741–6. doi: 10.1016/s0969-2126(03)00133-3. [DOI] [PubMed] [Google Scholar]
- Jacobson KA, Gao ZG, Liang BT. Neoceptors: reengineering GPCRs to recognize tailored ligands. Trends Pharmacol Sci. 2007;28:111–6. doi: 10.1016/j.tips.2007.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi S. On the applicability of GPCR homology models to computer-aided drug discovery: a comparison between in silico and crystal structures of the beta2-adrenergic receptor. J Med Chem. 2008;51:2907–14. doi: 10.1021/jm800044k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, et al. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–43. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- Cronet P, Petersen JF, Folmer R, Blomberg N, Sjoblom K, Karlsson U, et al. Structure of the PPARalpha and -gamma ligand binding domain in complex with AZ 242; ligand selectivity and agonist activation in the PPAR family. Structure. 2001;9:699–706. doi: 10.1016/s0969-2126(01)00634-7. [DOI] [PubMed] [Google Scholar]
- Xu HE, Lambert MH, Montana VG, Plunket KD, Moore LB, Collins JL, et al. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proc Natl Acad Sci U S A. 2001;98:13919–24. doi: 10.1073/pnas.241410198. [DOI] [PMC free article] [PubMed] [Google Scholar]