Abstract
The brain tumor glioblastoma multiforme (GBM) is among the most lethal forms of human cancer. Here, we report that a small subset of GBMs (3.1%; 3 of 97 tumors examined) harbors oncogenic chromosomal translocations that fuse in-frame the tyrosine kinase coding domains of fibroblast growth factor receptor (FGFR) genes (FGFR1 or FGFR3) to the transforming acidic coiled-coil (TACC) coding domains of TACC1 or TACC3, respectively. The FGFR-TACC fusion protein displays oncogenic activity when introduced into astrocytes or stereotactically transduced in the mouse brain. The fusion protein, which localizes to mitotic spindle poles, has constitutive kinase activity and induces mitotic and chromosomal segregation defects and triggers aneuploidy. Inhibition of FGFR kinase corrects the aneuploidy, and oral administration of an FGFR inhibitor prolongs survival of mice harboring intracranial FGFR3-TACC3–initiated glioma. FGFR-TACC fusions could potentially identify a subset of GBM patients who would benefit from targeted FGFR kinase inhibition.
Chromosomal translocations leading to production of oncogenic fusion proteins are critical events in the pathogenesis of human cancer (1–3). To examine whether such alterations are present in the tumor glioblastoma multiforme (GBM), we used massively parallel, paired-end sequencing of expressed transcripts (RNA-seq) to detect gene fusions in short-term cultures of glioma stemlike cells (GSCs) freshly isolated from nine patients with primary GBMs. Using TX-Fuse, a methodology that detects split reads and split inserts (see supplementary materials and methods section and fig. S1A), we discovered six rearrangements (all of which were intrachromosomal) that gave rise to in-frame fusion transcripts (table S1). We validated five in-frame fusion predictions by direct sequencing of polymerase chain reaction (PCR) products spanning the fusion breakpoint (Fig. 1 and fig. S1, B to E).
Fig. 1.
FGFR3-TACC3 gene fusion identified by whole-transcriptome sequencing of GSCs. (A) Here, 76 split-reads are shown aligning on the breakpoint. The predicted reading frame at the breakpoint is shown at the top with FGFR3 sequences in red and TACC3 in blue. (B) (Left) FGFR3-TACC3–specific PCR from cDNA derived from GSCs and GBMs. M, 1-kb DNA ladder. (Right) Sanger sequencing chromatogram showing the reading frame at the breakpoint and putative translation of the fusion protein in the positive samples. T, threonine; S, serine; D, aspartic acid; F, phenylalanine; E, glutamic acid. (C) Schematics of the FGFR3-TACC3 protein. Regions corresponding to FGFR3 or TACC3 are shown in red or blue, respectively. The fusion protein joins the tyrosine kinase domain of FGFR3 to the TACC domain of TACC3. (D) Genomic fusion of FGFR3 exon 17 with intron 7 of TACC3. In the fused mRNA, exon 16 of FGFR3 is spliced 5′ to exon 8 of TACC3. Solid black arrows indicate the position of the fusion-genome primers, which generate fusion-specific PCR products in GSC-1123 and GBM-1123.
In Fig. 1, A and B, we show the prediction and cDNA sequence validation, respectively, for the fusion with the highest read support involving fibroblast growth factor receptor 3 (FGFR3) fused in-frame with transforming acidic coiled-coil 3 (TACC3) in GSC-1123 and GBM-1123 primary tumor. The cDNA contained an open reading frame coding for a protein of 1048 amino acids resulting from the in-frame fusion of the FGFR3 N terminus (residues 1 to 758) with the TACC3 C terminus (residues 549 to 838) (Fig. 1C and fig. S2A). FGFR3 is a member of the FGFR receptor tyrosine kinase (TK) family (4), whereas TACC3 belongs to the evolutionarily conserved TACC gene family, which also includes TACC1 and TACC2. The distinctive feature of TACC proteins is a coiled-coil domain at the C terminus, known as the TACC domain, which mediates localization to the mitotic spindle (5, 6). TACC proteins are hypothesized to be oncogenic in several human tumors, including GBMs (7, 8).
In the predicted fusion protein, the intracellular TK domain of FGFR3 is fused upstream of the TACC domain of TACC3 (Fig. 1C). Exon-specific gene expression analysis from the RNA-seq coverage in GSC-1123 and quantitative reverse transcription PCR showed that the expression of the fused FGFR3-TACC3 exons is higher in GSC-1123 than in other GSCs or the normal brain (80- to 130-fold) (fig. S2, B and C). The FGFR3-TACC3 fusion protein was abundantly expressed in GSC-1123 and GBM-1123, and immunoprecipitation followed by mass spectrometry revealed the presence of FGFR3 and TACC3 peptides, consistent with the cDNA translation prediction (fig. S2, D to F). We used PCR to map the genomic breakpoint coordinates to chromosome 4 (1,808,966 for FGFR3 and 1,737,080 for TACC3, genome build GRCh37/hg19), falling within FGFR3 exon 17 and TACC3 intron 7, which gives rise to a transcript in which the 5′ FGFR3 exon 16 is spliced to the 3′ TACC3 exon 8 (Fig. 1D). The DNA junctions of FGFR3 and TACC3 show microhomology within a 10-base region, an observation consistent with results previously reported for other chromosomal rearrangements in human cancer (Fig. 1D) (9, 10).
We next investigated whether FGFR3-TACC3 fusions are recurrent in GBMs. The analysis of the GBM data set from the Cancer Genome Atlas (TCGA) (11) revealed that four tumors display marked co-outlier expression of FGFR3 and TACC3 (fig. S3A). The same four tumors harbor microamplification events of the FGFR3 and TACC3 genes (fig. S3B). We modified the gene-fusion discovery method as a package called EXome-Fuse to detect split reads and split inserts from exome DNA sequences of the available GBM cases and matched constitutional DNA in TCGA (fig. S3C). From this analysis, FGFR3-TACC3 emerged as the sole recurrent somatic genomic rearrangement among the six fusions detected in GSCs. We found split reads and split inserts joining FGFR3 and TACC3 exons in the four GBM tumor DNAs carrying co-outlier expression and microamplification of the FGFR3 and TACC3 genes (fig. S4A and tables S2 and S3). Among the four positive TCGA GBM specimens, two were available for molecular analysis, and we confirmed the predicted fusion transcripts by Sanger sequencing (fig. S4, B and C).
The FGFR3 and TACC3 genes are located 48 kb apart on human chromosome 4p16. The other members of the FGFR and TACC families retain the close physical association, with FGFR1 and TACC1 paired on chromosome 8p11 and FGFR2 and TACC2 paired on chromosome 10q26 (12). To determine whether other intrachromosomal FGFR-TACC fusion combinations exist in human GBM, we screened cDNA from an independent panel of 88 primary GBMs and discovered two additional cases (one harboring FGFR1-TACC1 and one FGFR3-TACC3), corresponding to 3 of 97 total GBMs (3.1%), including the GBM-1123 case (fig. S4, D and E). None of the tumors harboring FGFR-TACC fusions had mutations in IDH1 or IDH2 genes (table S4). In all seven GBMs harboring FGFR-TACC rearrangements (four from TCGA and three from our tumor collection), the FGFR-TK domain is fused upstream of the TACC domain.
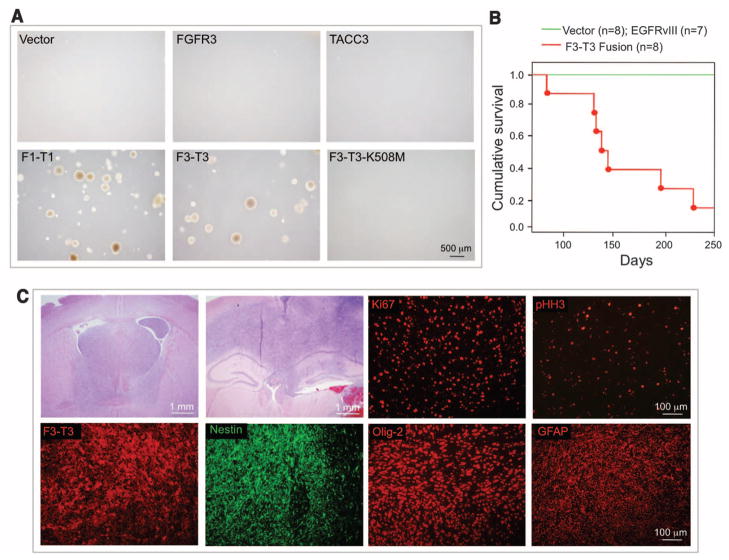
To explore whether FGFR-TACC fusions are oncogenic, we transduced Rat1A fibroblasts and Ink4A;Arf−/− astrocytes with a lentivirus expressing FGFR3-TACC3, which resulted in the expression of the fusion protein at levels comparable to those present in GSC-1123 (fig. S5, A to D). Rat1A cells expressing FGFR3-TACC3 (or FGFR1-TACC1) but not those expressing a kinase-dead FGFR3-TACC3 protein (FGFR3-TACC3-K508M), FGFR3, TACC3, or the empty lentivirus acquired the ability to grow in anchorage-independent conditions in soft agar (Fig. 2A and table S5). Transduction of the same lentiviruses in primary Ink4A;Arf−/− astrocytes followed by subcutaneous injection into immunodeficient mice revealed that only astrocytes expressing FGFR3-TACC3 or FGFR1-TACC1 formed tumors (table S5). The tumors were glioma-like lesions with strong positivity for Ki67, phosphohistone H3, nestin, glial fibrillary acidic protein (GFAP), and Olig2 (fig. S5E). To target a small number of cells with the fusion protein into the brain of immunocompetent animals, we stereotactically transduced the adult mouse hippocampus with purified lentivirus expressing FGFR3-TACC3 and short hairpin RNA (shRNA) against p53 (pTomo-FGFR3-TACC3-shp53) (13). Seven of eight mice (87.5%) transduced with FGFR3-TACC3 succumbed from malignant brain tumors within 240 days (Fig. 2B). None of the mice transduced with a lentivirus expressing epidermal growth factor receptor version III (EGFRvIII)/shp53 or the shp53 control lentivirus developed clinical signs of brain tumors or died. The FGFR3-TACC3 tumors were invasive, rapidly growing high-grade gliomas that stained positive for the glioma stem cell markers nestin and Olig2, the glial marker GFAP, and Ki67 and phosphohistone H3 (Fig. 2C). The FGFR3-TACC3 fusion protein was expressed in the xenograft and intracranial tumor models at levels comparable to those seen in human GSCs and GBMs (fig. S5, C, F, and G).
Fig. 2.
Transforming activity of FGFR-TACC fusion proteins. (A) FGFR1-TACC1 and FGFR3-TACC3 induce anchorage-independent growth in Rat1A fibroblasts. F1-T1, FGFR1-TACC1; F3-T3, FGFR3-TACC3. (B) Kaplan-Meier survival curves of mice injected intracranially with pTomo-shp53 (n = 8 animals) or pTomo-EGFRvIII-shp53 (n = 7) (green line) and pTomo-FGFR3-TACC3-shp53 (n = 8) (red line). Points on the curves indicate deaths (log-rank test, P = 0.00001, pTomo-shp53 versus pTomo-FGFR3-TACC3-shp53). (C) Representative microphotographs of hematoxylin and eosin staining of advanced FGFR3-TACC3-shp53–generated tumors showing histological features of high-grade glioma. Note the high degree of infiltration of the normal brain by the tumor cells. Immunofluorescence staining shows that glioma and stem cell markers (Nestin, Olig2, and GFAP), proliferation markers (Ki67 and pHH3), and the FGFR3-TACC3 protein are widely expressed in the FGFR3-TACC3-shp53 brain tumors.
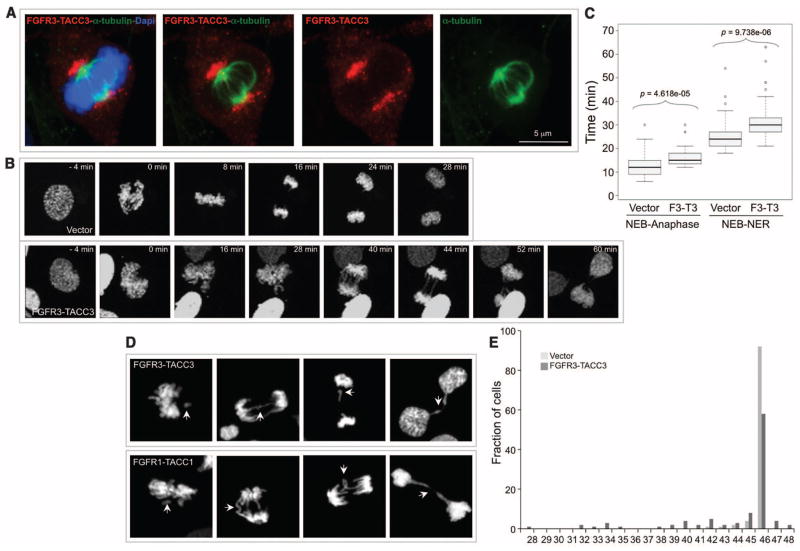
To investigate the mechanism by which the FGFR-TACC fusion drives oncogenesis, we explored whether it activates downstream FGFR signaling. FGFR3-TACC3 failed to hyperactivate the canonical signaling events downstream of FGFR (pERK and pAKT) in the presence or absence of the ligands FGF-1, FGF-2, or FGF-8 (fig. S6, A to C) (14). However, FGFR3-TACC3 displayed constitutive phosphorylation of its TK domain and the adaptor protein FRS2, both of which were abolished by PD173074, a compound that is a specific inhibitor of FGFR-associated TK activity (15), or by the Lys508→Met508 (K508M) mutation (fig. S6, D and E). Thus, FGFR3-TACC3 gains constitutive kinase activity that is essential for oncogenic transformation, but the downstream signaling of this aberrant activity is distinct from the canonical signaling events downstream to FGFR. We hypothesized that by driving the localization of the fusion protein, the TACC domain might create TK-dependent functions. Confocal imaging showed that FGFR3-TACC3 painted an arc-shaped structure, bending over and encasing the metaphase spindle poles, frequently displaying asymmetry toward one of the two poles and relocating to the midbody as cells progressed into the late stages of mitosis (telophase and cytokinesis) (Fig. 3A and figs. S7 and S8, A and B). Conversely, the localization of TACC3 was restricted to spindle microtubules; TACC3 did not relocalize to the mid-body (fig. S8C). Wild-type FGFR3 lacked discrete localization patterns in mitosis (fig. S8D).
Fig. 3.
FGFR3-TACC3 localizes to spindle poles, delays mitotic progression, and induces chromosome segregation defects and aneuploidy. (A) Confocal microscopy analysis of FGFR3-TACC3 (red) covering the spindle poles of a representative mitotic cell. α-tubulin, green; DNA [stained with 4′,6-diamidino-2-phenylindole (DAPI)], blue. (B) Representative fluorescence video microscopy for cells transduced with vector or FGFR3-TACC3. (C) Box-and-whisker plot representing the analysis of the time from nuclear envelope breakdown (NEB) to anaphase onset and from NEB to nuclear envelope reconstitution (NER). The duration of mitosis was measured by following 50 mitoses for each condition by time-lapse microscopy. (D) Representative images of cells with chromosome missegregation. Arrows point to chromosome misalignments, lagging chromosomes, and chromosome bridges. (E) Distribution of chromosome counts of human astrocytes transduced with control or FGFR3-TACC3–expressing lentivirus. Chromosomes were counted in 100 metaphase cells for each condition to determine the ploidy and diversity of chromosome counts within the cell population.
The mitotic localization of FGFR3-TACC3 suggests that the fusion protein might affect the fidelity of mitosis and generate aneuploidy. Time-lapse microscopy revealed that the average time from nuclear envelope breakdown to anaphase onset was increased in cells expressing FGFR3-TACC3 in comparison with control cells. The mitotic delay was exacerbated by delays in the completion of cytokinesis (Fig. 3, B and C). Quantitative analyses of mitoses revealed that cells expressing FGFR3-TACC3 or FGFR1-TACC1 exhibit three to five times more errors in chromosomal segregation compared with control cells. The most frequent mitotic aberrations triggered by the fusion proteins were misaligned chromosomes during metaphase, lagging chromosomes at anaphase, and chromosome bridges that impaired cytokinesis and generated micronuclei in the daughter cells (Fig. 3D, fig. S9A, and table S6). After treatment with the spindle poison colcemid, more than 18% of cells expressing FGFR3-TACC3 displayed prematurely separated sister chromatids, in contrast with less than 3% of control, FGFR3-, or TACC3-expressing cells (figs. S9, B and C). Accordingly, the fusion protein induced resistance to metaphase arrest after nocodazole treatment (fig. S9D). These findings suggest that the FGFR3-TACC3 fusion protein may induce aneuploidy. Karyotype analysis revealed that FGFR3-TACC3 increased the percentage of aneuploidy by more than 2.5-fold and led to the accumulation of cells with broad distribution of chromosome counts when compared with cells transduced with empty vector, FGFR3, or TACC3 (fig. S9E and table S7). Hence, GSC-1123 contained an aneuploid modal number of chromosomes (49) with broad distribution of chromosome counts (table S8).
To explore whether aneuploidy is a direct consequence of FGFR3-TACC3 expression and is induced in diploid neural cells, we analyzed primary human astrocytes 6 days after transduction with the FGFR3-TACC3 lentivirus. The transduced cells exhibited a fivefold increase of the rate of aneuploidy and a wider distribution of chromosome counts than controls (Fig. 3E, fig. S9F, and table S7). Consistent with the notion that aneuploidy is detrimental to cellular fitness, acute expression of FGFR3-TACC3 inhibited the proliferation of human astrocytes. However, continuous culture of FGFR3-TACC3–expressing astrocytes led to progressive gain of proliferative capacity that overrode that of control cells (fig. S10, A and B). Thus, acute expression of FGFR3-TACC3 in normal cells from the central nervous system causes chromosomal instability (CIN) and aneuploidy with an acute fitness cost manifested by slower proliferation.
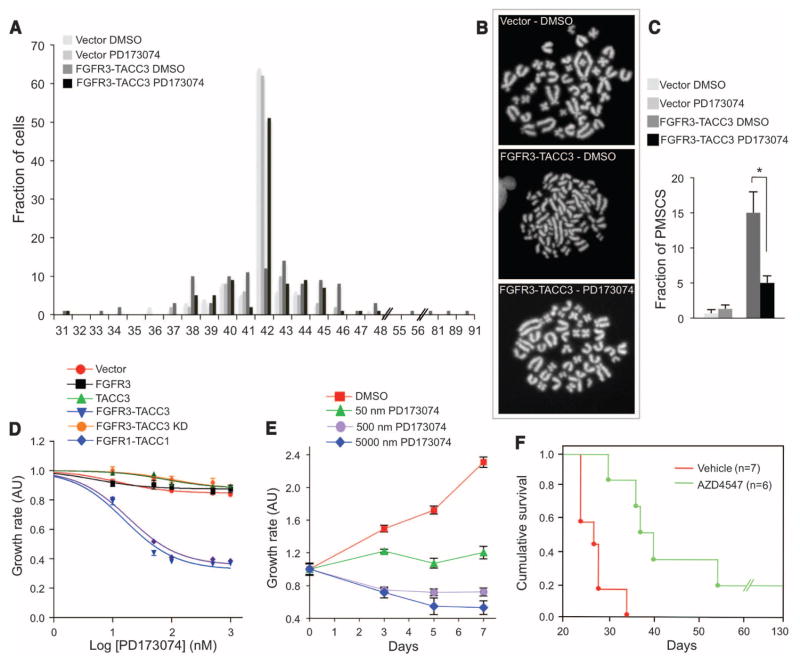
Next, we determined whether the CIN and aneuploidy caused by FGFR3-TACC3 requires TK activity and can be corrected. Treatment of Rat1A cells with PD173074 corrected FGFR3-TACC3–induced aneuploidy by more than 80%, restored the narrow distribution of chromosome counts typical of control cells, and largely corrected the cohesion defect (Fig. 4, A to C, and table S9). To determine whether FGFR-TACC–expressing Rat1A and GSC-1123 cells are dependent on FGFR-TK activity for their growth, we studied the effect of PD173074, AZD4547, or BGJ398. The latter two compounds are highly specific inhibitors of FGFR-TK under clinical investigation (16, 17). Each of the three drugs inhibited the growth of cells expressing FGFR3-TACC3 and FGFR1-TACC1 at concentrations <10 nM, whereas they were ineffective at concentrations as high as 1 μM in cells transduced with vector, FGFR3, TACC3, and the FGFR3-TACC3-K508M mutant (Fig. 4D and fig. S10, C and D). The growth of GSC-1123 was also abolished by nanomolar concentrations of these FGFR-TK inhibitors (Fig. 4E). Targeting of the fusion gene by FGFR3 shRNA inhibited the growth of cells ectopically expressing FGFR3-TACC3 and GSC-1123 in proportion to the silencing efficiency of FGFR3-TACC3 (fig. S10, E and F).
Fig. 4.
Inhibition of FGFR-TK activity corrects the aneuploidy and suppresses tumor growth initiated by FGFR3-TACC3. (A) Karyotype analysis of Rat1A cells transduced with control or FGFR3-TACC3 lentivirus and treated with vehicle [dimethyl sulfoxide (DMSO)] or PD173470 (100 nM) for 5 days. (B) Correction of premature sister chromatid separation (PMSCS) by PD173470 in cells expressing FGFR3-TACC3. Panels show representative metaphase spreads. (C) Quantitative analysis of metaphases with loss of sister chromatid cohesion (FGFR3-TACC3 treated with DMSO versus FGFR3-TACC3 treated with PD173470). P = 0.001; error bars indicate SD. (D and E) Growth-inhibition assays of Rat1A cells transduced with the indicated lentivirus (D) and GSC-1123 (E) treated with PD173470 at the indicated concentrations. Cells were treated for 3 days (D) or for the indicated time (E). Cell viability was determined by the MTT assay. Error bars show means ± SE (n = 4 culture wells). AU, arbitrary units. (F) Survival of glioma-bearing mice was tracked after intracranial implantation of Ink4A;Arf−/− astrocytes transduced with FGFR3-TACC3. After tumor engraftment, mice were treated with vehicle or AZD4547 (50 mg/kg) for 20 days (vehicle, n = 7 animals; AZD4547, n = 6; P = 0.001).
Finally, we studied mice bearing glioma xenografts of FGFR3-TACC3–transformed astrocytes and investigated whether PD173074 affected tumor growth. Twelve days after injection of tumor cells, mice were treated with PD173074 or vehicle (lactate buffer). Only the group treated with PD173074 showed inhibition of tumor growth (fig. S10G). We also tested a more clinically relevant FGFR inhibitor, AZD4547, in mice bearing intracranial luciferase-expressing glioma xenografts. Oral administration of AZD4547 prolonged survival of the mice by 28 days compared with mice treated with the vehicle control (Fig. 4F).
In summary, our functional characterization of the FGFR-TACC fusion genes found in a small subset of GBM patients indicates that the constitutively active FGFR-TK and the TACC domain are both essential for oncogenesis. It has long been thought that mutation of the genes that control chromosome segregation during mitosis may explain the high rate of CIN and aneuploidy, which is typical of most solid tumors, including GBMs (18). A few examples of mutational inactivation of candidate genes have been reported in human cancer (19, 20). However, gain-of-function mutations causally implicated in the control of mitotic fidelity have not been described. The absence of dominant mutations of CIN genes in human cancer clashes with the classic observation from cell-fusion experiments that the underlying mechanisms that cause CIN behave as dominant traits, indicating that the CIN phenotype results from gain-of-function events rather than gene inactivation (21, 22). The FGFR-TACC gene fusion is a mechanism for the initiation of CIN and provides a potential clue to the nature of dominant mutations responsible for aneuploidy in human cancer.
In itself, induction of aneuploidy is detrimental to cellular fitness (23). Oncogenic transformation requires cooperation between aneuploidy and genetic lesions that confer growth advantage and protect cells against the detrimental effects of aneuploidy (24–26). The tumor-initiating activity of the FGFR3-TACC3 fusion protein suggests that it has growth-promoting signaling functions that complement the loss of mitotic fidelity and aneuploidy to induce full-blown tumorigenesis (23).
There are now several well-known examples in which kinase inhibitors have been developed into effective therapies for patients whose tumors carry functional gene fusions that deregulate kinase activity (27, 28). The antitumor effects in mouse models and the correction of aneuploidy precipitated by FGFR-TK inhibition of glioma cells driven by FGFR-TACC fusions provide a strong rationale for clinical investigation of FGFR inhibitors in GBM patients whose tumors exhibit FGFR-TACC rearrangements.
Supplementary Material
Acknowledgments
This work was supported by National Cancer Institute grants R01CA101644 and R01CA131126 (A.L.), R01CA085628 and R01CA127643 (A.I.), U54 CA121852-05 (R.R.), National Library of Medicine grant 1R01LM010140-01 (R.R.), National Institute of Neurological Disorders and Stroke grant R01NS061776 (A.I.), a grant from Partnership for Cure (R.R., 7-78947), and a grant from The Chemotherapy Foundation (A.I.). P.Z. and F.N. are supported by fellowships from the Italian Ministry of Welfare/Provincia di Benevento. G.F. was supported by grants from the Associazione Italiana per la Ricerca sul Cancro and from the Italian Ministry of Health. Three of the authors (A.I., A.L., and R.R.) and Columbia University Medical Center have filed a patent application related to the diagnostic and therapeutic use of FGFR-TACC gene fusions. Next-generation RNA-sequencing data have been deposited into the Database of Genotypes and Phenotypes (accession no. phs000505.v1.p1).
Footnotes
References and Notes
- 1.Ablain J, Nasr R, Bazarbachi A, de Thé H. Cancer Discov. 2011;1:117. doi: 10.1158/2159-8290.CD-11-0087. [DOI] [PubMed] [Google Scholar]
- 2.Charest A, et al. Genes Chromosomes Cancer. 2003;37:58. doi: 10.1002/gcc.10207. [DOI] [PubMed] [Google Scholar]
- 3.Mitelman F, Johansson B, Mertens F. Nat Rev Cancer. 2007;7:233. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 4.Turner N, Grose R. Nat Rev Cancer. 2010;10:116. doi: 10.1038/nrc2780. [DOI] [PubMed] [Google Scholar]
- 5.Hood FE, Royle SJ. Bio Architecture. 2011;1:105. doi: 10.4161/bioa.1.3.16518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peset I, Vernos I. Trends Cell Biol. 2008;18:379. doi: 10.1016/j.tcb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Duncan CG, et al. Oncotarget. 2010;1:265. doi: 10.18632/oncotarget.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao R, et al. Oncogene. 2012;31:135. doi: 10.1038/onc.2011.235. [DOI] [PubMed] [Google Scholar]
- 9.Bass AJ, et al. Nat Genet. 2011;43:964. doi: 10.1038/ng.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephens PJ, et al. Nature. 2009;462:1005. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Cancer Genome Atlas Research Network. Nature. 2008;455:1061. [Google Scholar]
- 12.Still IH, Vince P, Cowell JK. Genomics. 1999;58:165. doi: 10.1006/geno.1999.5829. [DOI] [PubMed] [Google Scholar]
- 13.Marumoto T, et al. Nat Med. 2009;15:110. doi: 10.1038/nm.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wesche J, Haglund K, Haugsten EM. Biochem J. 2011;437:199. doi: 10.1042/BJ20101603. [DOI] [PubMed] [Google Scholar]
- 15.Mohammadi M, et al. EMBO J. 1998;17:5896. doi: 10.1093/emboj/17.20.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gavine PR, et al. Cancer Res. 2012;72:2045. doi: 10.1158/0008-5472.CAN-11-3034. [DOI] [PubMed] [Google Scholar]
- 17.Guagnano V, et al. J Med Chem. 2011;54:7066. doi: 10.1021/jm2006222. [DOI] [PubMed] [Google Scholar]
- 18.Gordon DJ, Resio B, Pellman D. Nat Rev Genet. 2012;13:189. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 19.Solomon DA, et al. Science. 2011;333:1039. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson SL, Bakhoum SF, Compton DA. Curr Biol. 2010;20:R285. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lengauer C, Kinzler KW, Vogelstein B. Nature. 1997;386:623. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 22.Lengauer C, Kinzler KW, Vogelstein B. Nature. 1998;396:643. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 23.Sheltzer JM, Amon A. Trends Genet. 2011;27:446. doi: 10.1016/j.tig.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coschi CH, Dick FA. Cell Mol Life Sci. 2012;69:2009. doi: 10.1007/s00018-011-0910-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holland AJ, Cleveland DW. Nat Rev Mol Cell Biol. 2009;10:478. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weaver BA, Cleveland DW. J Cell Biol. 2009;185:935. doi: 10.1083/jcb.200905098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Druker BJ. Nat Med. 2009;15:1149. doi: 10.1038/nm1009-1149. [DOI] [PubMed] [Google Scholar]
- 28.Gerber DE, Minna JD. Cancer Cell. 2010;18:548. doi: 10.1016/j.ccr.2010.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.