Cytoplasmic poly(A)-binding protein (PABP) C1 recruits different interacting partners to regulate mRNA fate. The majority of PABP-interacting proteins contain a PAM2 motif to mediate their interactions with PABPC1. However, little is known about the regulation of these interactions or the corresponding functional consequences. Through in silico analysis, the authors found that PAM2 motifs are generally embedded within an extended intrinsic disorder region (IDR) and are located next to cluster(s) of potential serine (Ser) or threonine (Thr) phosphorylation sites within the IDR. They hypothesized that phosphorylation at these Ser/Thr sites regulates the interactions between PAM2-containing proteins and PABPC1. In the present study, they have tested this hypothesis using complementary approaches to increase or decrease phosphorylation. The results provide a new framework for understanding the roles of intrinsically disordered proteins in the dynamic and signal-dependent control of cytoplasmic mRNA functions.
Keywords: PABP, PAM2 motif, intrinsically disordered region, GW182, miRNA function, translation, mRNA turnover, protein phosphorylation, deadenylation
Abstract
Cytoplasmic poly(A)-binding protein (PABP) C1 recruits different interacting partners to regulate mRNA fate. The majority of PABP-interacting proteins contain a PAM2 motif to mediate their interactions with PABPC1. However, little is known about the regulation of these interactions or the corresponding functional consequences. Through in silico analysis, we found that PAM2 motifs are generally embedded within an extended intrinsic disorder region (IDR) and are located next to cluster(s) of potential serine (Ser) or threonine (Thr) phosphorylation sites within the IDR. We hypothesized that phosphorylation at these Ser/Thr sites regulates the interactions between PAM2-containing proteins and PABPC1. In the present study, we have tested this hypothesis using complementary approaches to increase or decrease phosphorylation. The results indicate that changing the extent of phosphorylation of three PAM2-containing proteins (Tob2, Pan3, and Tnrc6c) alters their ability to interact with PABPC1. Results from experiments using phospho-blocking or phosphomimetic mutants in PAM2-containing proteins further support our hypothesis. Moreover, the phosphomimetic mutations appreciably affected the functions of these proteins in mRNA turnover and gene silencing. Taken together, these results provide a new framework for understanding the roles of intrinsically disordered proteins in the dynamic and signal-dependent control of cytoplasmic mRNA functions.
INTRODUCTION
Mammalian cytoplasmic poly(A)-binding protein (PABP) C1 is a highly conserved and abundant RNA-binding protein that binds to the 3′ poly(A) tails of mRNAs and has several roles in controlling the cytoplasmic fate of mRNA (Mangus et al. 2003; Kuhn and Wahle 2004). In the cytoplasm, PABPC1 proteins in complex with mRNA 3′ poly(A) tails facilitate formation of the “closed-loop” structure of the mRNA-ribonucleoprotein particle (mRNP) (Jacobson 1996; Wells et al. 1998). This closed-loop mRNP structure helps enhance translation initiation and termination, promote recycling of ribosomes, and influence the stability of the mRNA (Jacobson 1996; Sachs 2000; Wilusz et al. 2001; Coller and Parker 2004; Sonenberg and Hinnebusch 2009; Brook and Gray 2012). In these cytoplasmic processes, PABPC1 serves as a binding scaffold for other proteins involved in translation regulation and mRNA turnover. The recent discovery that interactions of GW182/Tnrc6 proteins with PABPC1 are critical for miRNA-mediated gene silencing (for review, see, e.g., Tritschler et al. 2010) further highlights the importance of studying the actions of interactions between PABPC1 and its partners for understanding the post-transcriptional control of gene expression.
Among mammalian PABPC1-interacting proteins, at least 16 contain the motif of ∼12 amino acids termed the PABP-interacting Motif 2 (PAM2) (Albrecht and Lengauer 2004; Tritschler et al. 2010). Structural data show that the PAM2 motif binds to the highly conserved MLLE domain in the C-terminal region of PABPC1 (Kozlov et al. 2004; Jinek et al. 2010; Kozlov and Gehring 2010). Among the PAM2-containing proteins are several well-studied translation and mRNA decay factors, including PAIP1 (Roy et al. 2002), PAIP2 (Khaleghpour et al. 2001), Tnrc6 (Eystathioy et al. 2003), translation termination factor 3 (eRF3) (Hoshino et al. 1999), PABP-dependent poly(A) nuclease subunit 3 (Pan3) (Uchida et al. 2004), and transducer of erbB-2 proteins (Tob) (Ikematsu et al. 1999). The number of PAM2-containing proteins and their varied roles in mRNA metabolism suggest several important questions. For example, how does the cell regulate interactions between PABPC1 and its binding partners to respond to different biological processes? What molecular mechanism controls association and dissociation between PABPC1 and individual binding partners?
In view of the observation that PAM2 motifs appear to occur outside globular protein domains (Albrecht and Lengauer 2004), we hypothesized that PAM2 motifs primarily reside in intrinsically disordered or unstructured regions (IDRs). IDRs can retain significant regions of disorder under physiological conditions, even when positioned near one or more structured domains (Radivojac et al. 2007; Brown et al. 2011; Uversky 2011). Interestingly, a recent study of mRNA interactomes found that proteins associated with mRNAs are highly enriched in IDRs compared to the human proteome overall (Castello et al. 2012). The dynamic nature of IDRs combines structural flexibility with a high functional density and multiple interaction interfaces (Dunker et al. 2002; Tompa and Csermely 2004; Dyson and Wright 2005). The major functions currently ascribed to disordered regions are protein–protein binding, protein–DNA/RNA binding, substrate–ligand binding, and phosphorylation targets (Dunker et al. 2002; Tompa and Csermely 2004; Dyson and Wright 2005).
Reversible protein phosphorylation provides a key regulatory mechanism for a plethora of signal transduction processes in eukaryotic cells (for review, see, e.g., Hunter 2007; Temporini et al. 2008; Gnad et al. 2011). Most phosphorylation occurs at serine or threonine residues (Sugiyama et al. 2008; Reiland et al. 2009). One characteristic feature of disordered segments is an enrichment in polar, uncharged amino acids such as serine and threonine (Iakoucheva et al. 2004). It has also been noted that the amino acid composition, sequence complexity, hydrophobicity, and charge of regions adjacent to phosphorylation sites resemble those of IDRs (Johnson and Lewis 2001; Iakoucheva et al. 2004). Based on these observations, IDRs are predicted to be “hotspots” for protein phosphorylation.
We have conducted a bioinformatics analysis of thirteen human PAM2-containing proteins and found that, with the exception of eRF3b, the PAM2 motif is embedded in an extended IDR and is close to a cluster or clusters of potential Ser/Thr phosphorylation sites. We hypothesized that reversible phosphorylation of these Ser/Thr clusters is required for modulating interactions between the PAM2-containing protein and PABPC1, thereby controlling mRNA fate. To test this hypothesis, in this study we used complementary approaches to change the phosphorylation states of three representative PAM2-containing proteins and determine the corresponding functional consequences. The experimental results not only support our hypothesis but also provide a new framework for designing future studies to investigate how the many distinct mRNA functions involving PABPC1 and its PAM2-containing interacting partners may be regulated and coordinated in eukaryotic cells.
RESULTS
PAM2 motifs are generally embedded within intrinsically disordered regions
As PAM2 motifs in general occur in protein regions that seem not to assume any known structure (Albrecht and Lengauer 2004), we used two online tools, “PONDR” and “Globplot,” to identify candidate IDRs within 13 human MLLE domain-interacting, or PAM2-containing, proteins (Supplemental Table S1). PONDR analysis predicts regions that are not rigid (including random coils, partially unstructured regions, and molten globules) based on local amino acid composition, flexibility, and hydropathy (Romero et al. 2004; Xue et al. 2010). The Globplot predicts regions with a high propensity for globularity on the Russell/Linding scale (propensities for secondary structures and random coils) (Linding et al. 2003). Analyses done by PONDR predict that all PAM2-containing proteins examined have an extensive IDR, with eRF3b predicted to be ∼25% IDR and the others predicted to be >32% IDR (Supplemental Table S1). Moreover, the PAM2 motifs in these proteins reside within the predicted IDRs (see representative examples schematically shown in Fig. 1).
FIGURE 1.
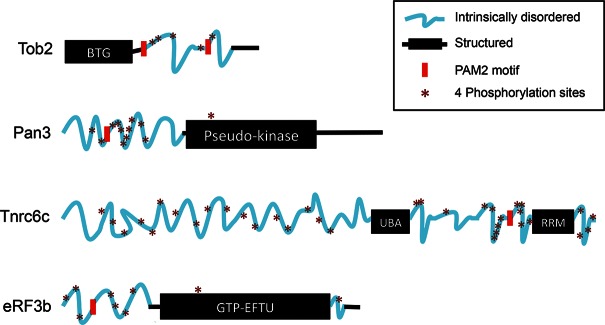
Diagram of four representative PAM2-containing PABPC1-interacting proteins showing PAM2 motifs (red blocks), IDRs (blue wavy lines), clusters of potential phosphorylation sites (asterisks; each asterisk denotes four Ser/Thr sites within a span of 10–20 aa), and structured regions (black rectangles and lines). Human Tob2 (Q14106), Pan3 (Q58A45), TNRC6c (Q9HCJ0), and eRF3b (Q8IYD1) amino acid sequences were analyzed by DISPHOS (DISorder-enhanced PHOSphorylation predictor; http://www.ist.temple.edu/DISPHOS).
Four of the PAM2-containing proteins analyzed by PONDR (Tob2, Pan3, eRF3b, and Tnrc6c, a paralog of Tnrc6 or GW182) are relatively well-characterized, with known cellular functions. We thus focused on these four proteins to perform additional analysis using Globplot. The results of this analysis (Supplemental Fig. S1A) corroborated the PONDR results (Supplemental Fig. S1B) and also highlight regions of low complexity (i.e., sequences with overrepresentation of a few particular residues), a feature of many proteins containing IDRs (Linding et al. 2003). The predicted disordered regions with embedded PAM2 motifs and structured regions of the four proteins are schematized in Figure 1.
PAM2 motifs in IDRs are generally located next to a cluster of phosphorylation sites
We used several web-based protein phosphorylation algorithms to predict the sites and extent of potential phosphorylation in the IDRs of the PAM2-containing proteins listed in Supplemental Table S1. The tools we used included DISPHOS (DISorder-enhanced PHOSphorylation predictor, http://www.ist.temple.edu/DISPHOS), Scansite (locates motifs associated with protein kinase phosphorylation; http://scansite.mit.edu/motifscan_seq.phtml), PhosphoNET (a repository of known and predicted information on human phosphorylation sites and their evolutionary conservation; http://www.phosphonet.ca), and NetPhos 2.0 Server (http://www.cbs.dtu.dk/services/NetPhos). These algorithms predicted that the IDRs in the PAM2-containing proteins are also the segments that contain clusters of potential phosphorylation sites. It is striking that, in all cases except eRF3b, there is at least one cluster of phosphorylation sites very close to PAM2 motifs (see representative examples schematically shown in Fig. 1). These bioinformatics results suggest that PAM2-containing proteins have the potential to be hyperphosphorylated by kinases at clusters within their IDRs. Based on this, we suspected that the interaction of a PAM2 motif with the MLLE domain of PABPC1 could be modulated via variable phosphorylation at these serine/threonine clusters. This regulatory scheme could form the basis by which cells differentially and selectively control the interaction of PABPC1 with distinct PAM2-containing partners during mRNA turnover, translation, and gene silencing.
Phosphorylation of Tob2 and its ability to interact with PABPC1
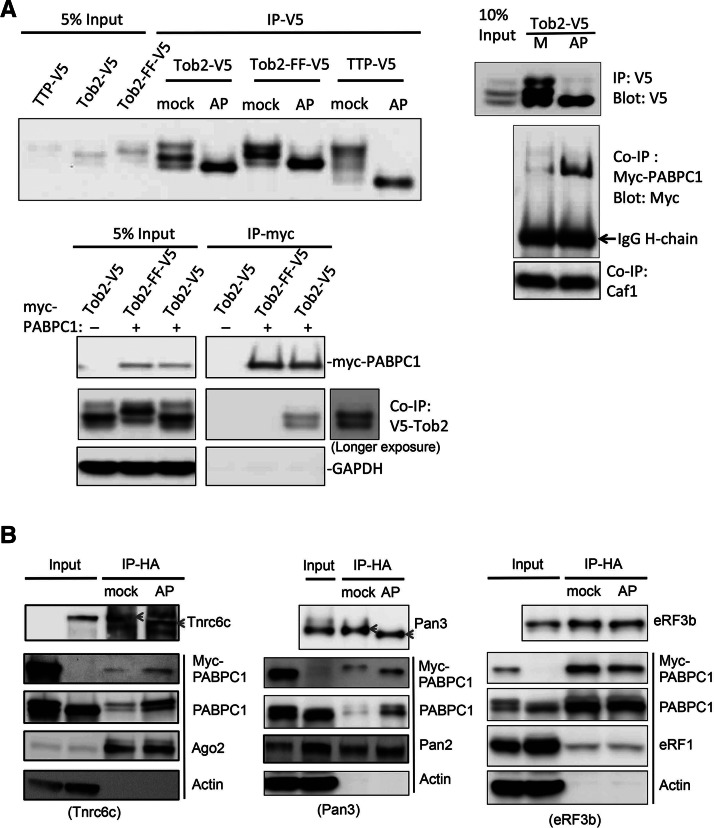
To test whether the interaction of a PAM2 motif with the MLLE domain of PABPC1 is modulated by the degree of phosphorylation, we first determined whether the extent of Tob2 phosphorylation affects its association with PABPC1. Tob2 is a known phosphoprotein that can simultaneously interact with Caf1 deadenylase via its structured N-terminal BTG domain and with PABPC1 via the two PAM2 motifs in the C-terminal IDRs (Okochi et al. 2005; Ezzeddine et al. 2007; Mauxion et al. 2008; Miyasaka et al. 2008). Point mutations that change the highly conserved phenylalanines to alanines in each of the two PAM2 motifs (Tob2-FF mutant) (Ezzeddine et al. 2007) abolish Tob2’s ability to interact with PABPC1. Ectopically expressed Tob2, either wild-type (WT) or FF mutant, displays three distinct bands by Western blot analysis, and in vitro alkaline phosphatase (AP) pretreatment collapsed the three bands into one major band (Fig. 2A, upper left), indicating that Tob2 proteins can be phosphorylated to different extents. As a control, we showed that AP pretreatment of TTP, an ARE-binding protein known to be highly phosphorylated (Tiedje et al. 2010; Sanduja et al. 2011), effectively removed its phosphates. It is worth noting that the relative mobilities and intensities of the three bands for Tob2-FF differ from those for WT Tob2, suggesting a link between its phosphorylation state and its ability to interact with PABPC1.
FIGURE 2.
Effects of alkaline phosphatase treatment on phosphorylation of four PAM-2 containing proteins and their interactions with PABPC1. (A) Upper left: Alkaline phosphatase (AP) or control (mock) treatment of immunoprecipitated Tob2-V5, Tob2-FF-V5, and TTP-V5 leads to a collapse of multiple higher bands into one band in SDS-PAGE. TTP, a highly phosphorylated protein, was used as a positive control for phosphatase treatment. Upper right: Western blot analysis showing that phosphatase-treated Tob2-V5 pulls down much more myc-PABPC1 than does mock-treated Tob2-V5, whereas both pull down an equal amount of Caf1. Lower left: Western blot analysis showing that myc-PABPC1 selectively pulled down the bottom two bands, corresponding to less-phosphorylated forms of Tob2-V5 and did not pull down Tob2-FF-V5 or GAPDH. (B) Effects of alkaline phosphatase treatment on phosphorylation of Tnrc6c (left), Pan3 (middle), or eRF3 (right) and their interactions with PABPC1 or other partners. Actin served as a negative control. Gray arrowheads denote migration change of HA-Tnrc6c or Pan3 bands following AP treatment. Cell extracts for IP experiments were prepared from NIH3T3 cells that were transiently transfected with the plasmids encoding the proteins indicated.
We then performed coimmunoprecipitation (co-IP) experiments to see whether phosphate removal from Tob2 affects its interaction with PABPC1. To rule out the possibility that coprecipitation of the proteins is due to their association with RNA, the co-IP experiments were performed in the presence of RNase A. Ectopically expressed Tob2-V5 was pulled down and divided into two equal parts, one with AP treatment and one with mock treatment. Both samples were then incubated with lysate from myc-PABPC1-expressing cells. The results showed that phosphatase-treated Tob2 pulled down more myc-tagged or endogenous PABPC1 than mock-treated Tob2 that remains phosphorylated (Fig. 2A, upper right; Supplemental Fig. S2). In contrast, there was no difference in the amount of Caf1 pull-down via the structured N-terminal domain of Tob2 (Fig. 2A, upper right). Together, these observations suggest that the extent of phosphorylation of Tob2 is inversely proportional to the strength of its interaction with PABPC1. To corroborate this result, we conducted a complementary co-IP experiment in which cells were cotransfected with myc-PABPC1 and Tob2-V5. The result (Fig. 2A, lower left) showed that myc-PABPC1 selectively pulls down the two less phosphorylated bands of WT Tob2. The control experiment showed that myc-PABPC1 did not pull down the Tob2-FF mutant, which lacks the ability to interact with PABPC1 (Ezzeddine et al. 2007). Collectively, we conclude that, while the two PAM2 motifs of Tob2 are essential for eliciting its interaction with PABPC1, the extent of Tob2 phosphorylation modulates the strength of this interaction.
Removal of phosphates from Pan3 and Tnrc6c enhances their interaction with PABPC1
We next tested whether removal of phosphates from three other PAM2-containing proteins (Pan3, Tnrc6c, and eRF3b) also enhances their interaction with PABPC1. Among the three proteins, Tnrc6c is predicted to have the highest proportion of IDR and potential phosphorylation sites, and eRF3b is predicted to have the least (Fig. 1). The results of the co-IP experiments showed that phosphatase-treated Tnrc6c pulled down appreciably more PABPC1 than the mock-treated one (Fig. 2B, left). As expected, phosphatase treatment resulted in a clear increase of HA-Tnrc6c migration on SDS-PAGE, indicative of removal of an appreciable amount of phosphates from this protein. In contrast, there was no obvious difference in the amount of coimmunoprecipitated Ago2 following phosphatase treatment.
Similarly, phosphatase treatment increased the migration of HA-Pan3 and increased the amount of coimmunoprecipitated PABPC1 (Fig. 2B, middle), whereas there was only a marginal increase in the amount of Pan2 deadenylase pull-down after phosphatase treatment. In contrast to Tnrc6c and Pan3, HA-eRF3b did not show any apparent change in its SDS-PAGE migration after phosphatase treatment (Fig. 2B, right), consistent with the prediction that this protein has fewer phosphorylation sites in its IDR compared to other PAM2-containing proteins (Fig. 1). Moreover, phosphatase treatment did not change the amount of PABPC1 or eRF1 pulled down by HA-eRF3b. It is worth noting that eRF3b coimmunoprecipitates much more PABPC1 than Tob2, Pan3, or Tnrc6c does (Fig. 2), suggesting that eRF3b has stronger intrinsic PABPC1-binding than the other three proteins. Together, these results support the notion that the extent of phosphorylation of PAM2-containing proteins modulates the strength of their interactions with PABPC1.
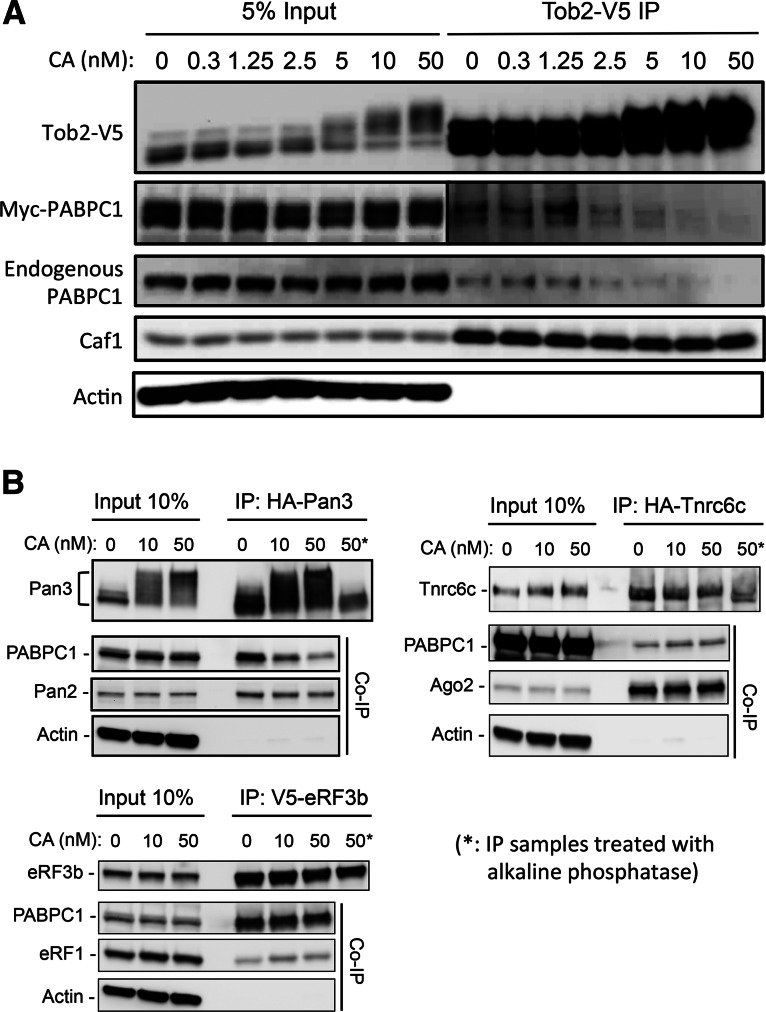
FIGURE 3.
Effects of calyculin A treatment on phosphorylation of PAM2-containing proteins and their interactions with PABPC1. (A) Western blot analysis showing that Tob2-V5 undergoes dosage-dependent increases in phosphorylation after calyculin A (CA) treatment. Myc-PABPC1, endogenous PABPC1, Caf1, and actin did not show discernable changes in migration. Immunoprecipitated Tob2-V5 from CA-treated cell lysates pulled down less myc-PABPC1 and endogenous PABPC1 as the CA concentration increased, whereas no such change was observed for co-IP of Caf1. (B) Co-IP and Western blotting experiments showing the effects of CA treatment of Pan3 (upper left), Tnrc6c (upper right), and eRF3 (lower left) on their interactions with endogenous PABPC1 and other partners. Asterisks denote IP samples that were further treated with alkaline phosphatase and served as unphosphorylated controls. Note that eRF3 did not show any discernable change in its migration. Cell extracts for IP experiments were prepared from NIH3T3 cells that were transiently transfected with the plasmids encoding the proteins indicated.
Effects of increased phosphorylation of Tob2 and Pan3 on their interactions with PABPC1
We then used a complementary approach to test whether increasing phosphorylation of the PAM2-containing proteins affects their interaction with PABPC1. We first treated cells transfected with Tob2 with increasing amounts of calyculin A, a potent inhibitor of the Ser/Thr phosphatase activities (Suganuma et al. 1990) of two major mammalian phosphatases, PP1 and PP2A (Brautigan 2012). The calyculin A treatment resulted in significantly increased phosphorylation of Tob2. This effect appears to be specific for Tob2 as it was not detected with PABPC1, Caf1, or actin (Fig. 3A). The results of co-IP experiments using these calyculin A-treated lysates showed that, when the phosphorylation of Tob2 increased, the amount of PABPC1 (both endogenous and myc-tagged) pulled down by Tob2 decreased (Fig. 3A). In contrast, calyculin A treatment had no effect on the ability of Tob2 to pull down Caf1. Together, these results show that increasing phosphorylation of Tob2 compromised its interaction with PABPC1.
We then tested the effect of calyculin A treatment on Pan3, Tnrc6, and eRF3b. The results showed that calyculin A treatment increased the phosphorylation of Pan3 and also diminished the amount of PABPC1 pulled down by Pan3 (Fig. 3B, upper left). On the other hand, Tnrc6 exhibited only a modest change in SDS-PAGE mobility after calyculin A treatment (Fig. 3B, upper right), suggesting that little increase in phosphorylation occurred. As expected, eRF3b showed no migration change (Fig. 3B, lower left), indicating that calyculin A did not elicit an increase in phosphorylation of eRF3b. The results of co-IP/Western experiments showed that the amounts of PABPC1 pulled down by Tnrc6c and eRF3b were not affected by calyculin A treatment (Fig. 3B, upper right and lower left). Thus, the extent of change in phosphorylation of PAM2-containing proteins correlated with the degree of change in their interaction with PABPC1. Calyculin A did not change the electrophoretic migration of PABPC1 (Fig. 3) or other interacting partners of Pan3, Tnrc6, and eRF3 (Pan2, Ago2, and eRF1, respectively) (Fig. 3B), confirming that phosphorylation of these proteins was unaltered by calyculin A. The control experiments showed that calyculin A treatment did not change the amount of Pan2, Ago2, or eRF1 pulled down by Pan3, Tnrc6c, or eRF3b, respectively (Fig. 3B). Thus, results from the two complementary approaches of decreasing and increasing phosphorylation (Figs. 2, 3) support our hypothesis that phosphorylation at the Ser/Thr clusters in their IDRs can modulate the interaction of PAM2-containing proteins with PABPC1.
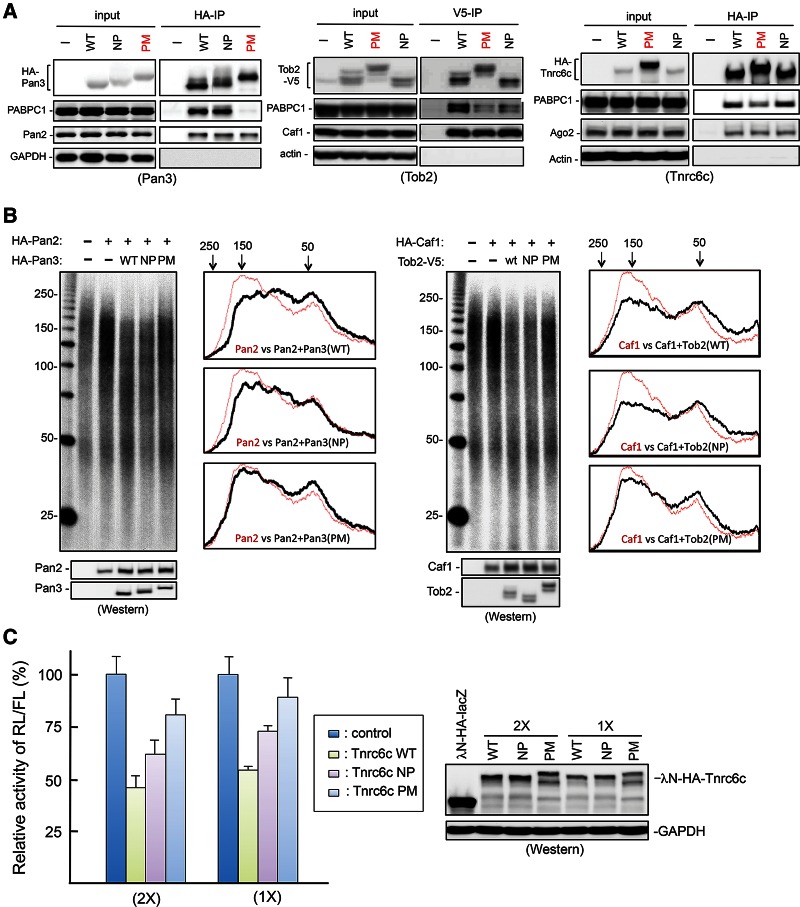
Phosphomimetic mutations in the IDRs of Tob2, Pan3, and Tnrc6c diminish their interactions with PABPC1
To further test our hypothesis, we created phosphomimetic mutants of Tob2, Pan3, and Tnrc6c. We targeted the Ser/Thr residues within IDRs that are predicted to be phosphorylation sites and are near the PAM2 motif in each of the three proteins (Fig. 1; also see Supplemental Fig. S3). The Ser/Thr residues were changed either to alanines to create nonphosphorylatable (NP) mutants or to Asp/Glu residues to create phosphomimetic (PM) mutants. Western blot analysis showed a discernable difference in the apparent molecular weights between the respective NP and PM mutants, as expected with different levels of phosphorylation (Fig. 4A). The results of co-IP experiments (Fig. 4A) showed that, in all three proteins, the PM mutants pulled down much less PABPC1 than did the corresponding WT proteins, indicating a diminished interaction of PM mutants with PABPC1. On the other hand, the amounts of PABPC1 pulled down by the NP mutants of Pan3 and Tnrc6c were similar to those pulled down by the corresponding WT proteins (Fig. 4A, left and right). The Tob2 NP mutant pulled down less PABPC1 than did WT Tob2 (Fig. 4A, middle). However, in all cases the NP mutant pulled down more PABPC1 than did the corresponding PM mutant (Fig. 4A). Moreover, neither PM nor NP mutations affected the amount of Caf1 pulled down by Tob2, Pan2 pulled down by Pan3, or Ago2 pulled down by Tnrc6c (Fig. 4A). Collectively, these results further support the notion that phosphorylation in the IDRs near PAM2 motifs specifically compromises the interaction of PAM2-containing proteins with PABPC1.
FIGURE 4.
(A) Co-IP and Western blotting experiments showing effects of nonphosphorylatable (NP) and phosphomimetic (PM) mutations of Pan3 (left), Tob2 (middle), and Tnrc6c (right) on their interactions with PABPC1 or other partners. GAPDH served as a control for nonspecific co-IP. (B) Functional effects of NP and PM mutations of Pan3 (left) and Tob2 (right). Poly(A) size-distribution profile analysis for Pan3 and Tob2 was performed as described in Materials and Methods. Cells were transiently transfected with the plasmids encoding the proteins indicated. Western blot analysis assesses protein expression levels. (C) Results of dual luciferase assays showing the gene silencing actions of WT Tnrc6c and its NP and PM mutants. Histograms (left) show the relative changes of Renilla luciferase (RL) activity detected in extracts from cells that were transiently cotransfected with a plasmid coding for firefly luciferase (FL) mRNA and RL mRNA that contain four boxB sites in the 3′ UTR, and a plasmid coding for λN-HA-Tnrc6c (WT or mutant). The RL/FL activity observed in the extract expressing λN-HA-lacZ was set at 100%. All data represent the normalized mean ± standard errors (n = 3). Western blot analysis (right) was used to assess the expression levels of the recombinant proteins.
Phosphomimetic mutations alter Tob2, Pan3, and Tnrc6c functions
Pan3 and Tob2 act as positive regulators of Pan2 and Caf1 deadenylases, respectively, in a PABPC1-dependent manner. To test the functional significance of the effect of PM mutations on the interactions of Pan3 and Tob2 with PABPC1 (Fig. 4A, left and middle), we carried out a cell-based poly(A) size-distribution assay to determine whether the PM mutations alter the deadenylation-enhancing function of the proteins (Fig. 4B). Total cytoplasmic mRNA was extracted, and the poly(A) tails were labeled at the 3′ end with α-[32P] 3′-deoxyadenosine (cordycepin) (Rose et al. 1977), followed by digestion with RNases T1 and A to remove the mRNA bodies. The resulting poly(A) tail samples were then analyzed by denaturing gel electrophoresis. The results showed that WT Pan3 and Tob2 enhance the Pan2 and Caf1 activities, respectively, resulting in accumulation of mRNAs with shorter poly(A) tails (Fig. 4B). The NP mutants of Pan3 and Tob2 remained capable of enhancing deadenylation mediated by Pan2 and Caf1, respectively, resulting in poly(A) size-distribution patterns similar to those caused by WT Pan3 or Tob2 (Fig. 4B). In contrast, Pan3 PM and Tob2 PM mutants, which exhibit diminished interactions with PABPC1, did not enhance Pan2- or Caf1-mediated deadenylation; the poly(A) tail size distributions were similar to those observed in the absence of Pan3 or Tob2 (Fig. 4B). Together, results from this functional assay showed that PM mutations near the PAM2 motif in IDRs of Pan3 and Tob2 have a functional consequence, namely, decreasing the ability of Pan3 and Tob2 to promote deadenylation mediated by Pan2 and Caf1, respectively.
To test whether PM or NP mutations in the silencing domain of Tnrc6c affect its function in miRNA-mediated gene silencing, we used an RNA-tethering approach combined with a dual luciferase assay. λ N-peptide (λN), a high-affinity ligand for the boxB RNA stem–loop, was fused to HA-Tnrc6c and its PM and NP mutants. Each fusion protein vector was cotransfected with the vector encoding Renilla luciferase (RL) mRNA that carried four boxB sites in the 3′ UTR (Pillai et al. 2004; Chen et al. 2009). The results of the dual luciferase assay showed that WT Tnrc6c effectively repressed RL activity (Fig. 4C; compare to negative control that coexpressed with λN-HA-lacZ). The overall RL activity with ectopic expression of the Tnrc6c PM mutant was higher than that with the Tnrc6c WT or the NP mutant (Fig. 4C, left), indicating that the PM mutation most impaired the gene silencing function of Tnrc6c. These results led us to conclude that both NP and PM mutations in the IDR around the PAM2 motif in Tnrc6c compromise its silencing function, with the PM mutation having a larger impact than the NP mutation. The observation that both PM and NP mutations of Tnrc6c compromise silencing suggests that this silencing function requires reversible Tnrc6c phosphorylation.
DISCUSSION
This study focuses on the PAM2-motif containing partners of PABPC1 in mammalian cells. Bioinformatics and structural analyses revealed that thirteen human PAM2-motif containing proteins exhibit similar structural features: (1) They contain IDRs; (2) PAM2 motifs are embedded within the IDRs; and (3) the PAM2 motifs are near one or more clusters of potential phosphorylation sites (Fig. 1; Supplemental Table S1). Based on these observations, we hypothesized that protein phosphorylation at these Ser/Thr clusters may modulate the interactions between PABPC1 and the PAM2-containing proteins. The results of experiments with four of these PAM2-containing proteins (Tob2, Pan3, Tnrc6c, and eRF3b) support our hypothesis. We thus propose that interaction between PABPC1 and a PAM2 motif-containing partner is brought about by a two-step mechanism (Fig. 5). First, the PAM2 motif makes the initial contact with the MLLE domain in PABPC1. This induces folding in the IDRs, promoting further contacts with the MLLE domain and strengthening the interaction. In this process, reversible phosphorylation at the IDRs near the PAM2 motifs serves to modulate the interaction. This mechanism may represent a general means to regulate the functions of the various PAM2-containing proteins in eukaryotes.
FIGURE 5.
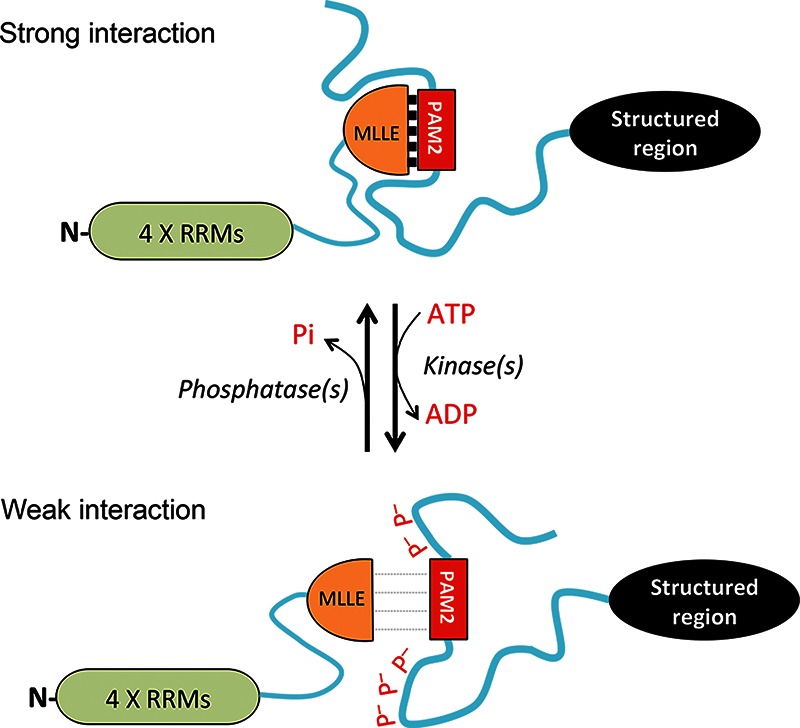
A model for regulation of interaction between eukaryotic PABPC1 and a PAM2-containing protein by reversible phosphorylation. (P– [in red font]) negatively charged phosphate group, (RRM) RNA-Recognition Motif, (blue wavy lines) intrinsically disordered regions, (black ovals) structured regions.
The results of the present study support our model (Fig. 5). We show that Tob2, Pan3, and Tnrc6c all can undergo appreciable degrees of reversible phosphorylation. Among the PAM2-containing proteins tested here, eRF3b is the least phosphorylated and Tnrc6c is the most phosphorylated, while Tob2 and Pan3 appear moderately phosphorylated. Using two complementary approaches, we showed that the degree of phosphorylation of these PAM2-containing proteins correlates inversely with their ability to interact with PABPC1. First, dephosphorylation by alkaline phosphatase treatment greatly enhances the ability of Tob2, Pan3, and Tnrc6c proteins to bind to PABPC1 (Fig. 2). The PABPC1 binding of eRF3b, whose electrophoretic mobility is not affected by phosphatase treatment, is not altered by the phosphatase treatment (Fig. 2B, right). It is worth noting that eRF3b is the least phosphorylated of the four proteins and binds more PBAPC1 than the other three proteins (Fig. 2). Second, calyculin A treatment, which greatly increased phosphorylation of Tob2 and Pan3, decreased their association with PABPC1 (Fig. 3A,B, upper left). On the other hand, calyculin A did not change the apparent phosphorylation state of eRF3b or alter its interaction with PABPC1 (Fig. 3B, lower left). Calyculin A treatment also did not greatly increase the phosphorylation or decrease the PABPC1 binding of Tnrc6 (which is predicted to be already highly phosphorylated) (Fig. 3B, upper right).
The amounts of PABPC1 pulled down by Tnrc6 in our co-IP experiments seem lower than those reported in previous studies (Fabian et al. 2009; Zekri et al. 2009; Huntzinger et al. 2010). It is worth noting that the earlier studies did not use full-length Tnrc6 paralogs for the co-IP experiments. Instead, the earlier experiments used nonphosphorylated recombinant Tnrc6, the Tnrc6 silencing domain alone (Fabian et al. 2009; Zekri et al. 2009), or a GFP-Tnrc6 fusion (Huntzinger et al. 2010) that could affect phosphorylation and expression of Tnrc6. It is plausible that, in the present study, Tnrc6 has more phosphorylation and, thus, lower PABPC1 binding. Our finding that Tnrc6c pulled down more PABPC1 after phosphatase treatment (Fig. 2B, left) supports this notion.
Functional assays showed that PM mutations of Tob2, Pan3, and Tnrc6c that diminish their interactions with PABPC1 (Fig. 4A) impair their biological functions to a greater degree than do NP mutants in the same proteins (Fig. 4B,C). Pan3 PM and Tob2 PM mutants were less effective in enhancing the Pan2 and Caf1 deadenylases' activities in mammalian cells, respectively (Fig. 4B). Similarly, the PM mutant of Tnrc6c exhibited a decreased gene silencing effect (Fig. 4C). It should be noted that the functional impairment seen in PM mutants could have other explanations, e.g., fortuitous alteration of protein conformation or folding as a result of introducing a number of Asp/Glu residues. Moreover, as some NP mutants showed moderately decreased functionality, overly persistent binding of a PAM2 motif-containing protein to PABPC1 could also hinder the cellular function. Thus, the results here suggest that the cellular functions of PAM2-containing proteins require their binding to PABPC1 to be both specific and reversible.
Tnrc6c, Pan3, and Tob2 are all positive regulators for mRNA decay, with Tnrc6c being specific for miRNA targets (Mangus et al. 2004; Liu et al. 2005; Ezzeddine et al. 2007; Funakoshi et al. 2007). Moreover, all three proteins participate in mRNA decay by promoting deadenylation (for review, see, e.g., Shyu et al. 2008; Chen and Shyu 2011). Previously, we showed that mRNA deadenylation is biphasic, with Pan2-Pan3 and Ccr4-Caf1 involved in the first and second phases, respectively (Yamashita et al. 2005). Through simultaneous interactions with PABPC1 and Pan2, Pan3 recruits Pan2 deadenylase directly to the 3′ poly(A)-PABPC1 complex to initiate the first phase of deadenylation (Yamashita et al. 2005; Funakoshi et al. 2007). Tob2 enhances the second phase of deadenylation by recruiting the Ccr4-Caf1 complex directly to the 3′ poly(A)-PABPC1 complex (Ezzeddine et al. 2007; Funakoshi et al. 2007; Mauxion et al. 2008). Interestingly, Tnrc6c is an integral part of a miRNA-RISC complex on miRNA-targeted mRNAs, a complex that recruits Pan2-Pan3 and Caf1-Ccr4 deadenylases to the mRNAs without involving PABPC1 (Braun et al. 2011; Chekulaeva et al. 2011; Fabian et al. 2011). This raises the question of the functional significance of the interaction between Tnrc6 and PABPC1. A recent study (Moretti et al. 2012) shed new light on this issue by showing that PABPC1, through its interaction with Tnrc6, helps binding of the miRISC complex on the target mRNA during the early phase of silencing. Subsequently, during the silencing phase, when deadenylases are recruited, the interaction between PABPC1 and Tnrc6 weakens, allowing deadenylation to proceed. In light of this observation, we proposed that during anchoring of miRISC, a less-phosphorylated state of Tnrc6 allows stronger interaction with PABPC1, whereas during the silencing phase, increasing phosphorylation of Tnrc6 lessens its interaction with PABPC1 and allows deadenylation. One important implication of the results in the present study is that a global change in miRNA-mediated gene silencing can be accomplished through signal-dependent, reversible phosphorylation of Tnrc6 proteins.
In summary, our finding that PAM2-containing proteins generally contain IDRs and are subject to dynamic phosphorylation provides a framework for understanding how interactions of various PAM2-containing proteins with PABPC1 are regulated to coordinate PABPC1’s multiple roles in mRNA turnover and translation. It is worth noting that the linker between the four RNA-recognition motifs (RRMs) and the MLLE domain in PABPC1 is also intrinsically disordered according to our analyses using PONDR and Globplot. Moreover, a recent study (Brook et al. 2012) reported that PABPC1 is subjected to extensive post-translational methylation and acetylation, some of which was suggested to impact the interactions between PABPC1 and PAM2-containing proteins. It will be interesting to learn if reversible phosphorylation at the IDR in PABPC1 has a role in its interactions with PAM2-containing proteins. Our findings open up new avenues to decipher the kinase/phosphatase signaling pathways that modulate the interactions between PABPC1 and PAM2-containing proteins. Given that most of the RNA-binding proteins in the mRNA interactome have IDRs (Castello et al. 2012), the mechanism we report here may represent a general new paradigm in which reversible phosphorylation of intrinsically disordered mRNA-binding proteins helps remodel mRNPs from their biogenesis in the nucleus to their turnover in the cytoplasm.
MATERIALS AND METHODS
Plasmid construction
The plasmids for myc-PABPC1, HA-Pan3, HA-Tnrc6c, λN-HA-lacZ, HA-Caf1, Tob2-V5, and the Tob2-V5 (FF: F140A/F260A) mutants were created as described previously (Yamashita et al. 2005; Ezzeddine et al. 2007; Zheng et al. 2008; Chen et al. 2009). To generate pcDNA6-eRF3b-V5, mRNA from human testis was reverse-transcribed into cDNA. The gene was then amplified using specific primers (5′-TGGAATTCTGCAGATACCATGGATTCGGGCAGCAGC-3′ and 5′-GCCACTGTGCTGGATTGTCCTTCTCTGGGACCAAT-3′) and subcloned into the EcoRV-linearized pcDNA6-V5 vector. To construct phosphomimetic (PM, Ser/Thr → Asp/Glu) mutants and nonphospho (NP, Ser/Thr → Ala) mutants of Tob2, Pan3, and Tnrc6c, minigene fragments spanning the targeted area of proteins with multiple mutations (Supplemental Fig. S3) were synthesized by IDT and Invitrogene. The In-fusion kit (Clontech) was used to replace the targeted regions in wild-type expression constructs with the minigenes, following the manufacturer’s manual. DNA sequencing was performed to confirm all the desired mutations and in-frame fusions.
Cell culture and transfection
NIH 3T3 B2A2 cells or U2OS cells were transfected using Lipofectamine 2000 (Invitrogen). Briefly, NIH3T3 B2A2 cells were maintained in DMEM (Invitrogen) medium containing 10% calf serum and 500 ng/mL tetracycline in an 8% CO2 atmosphere. For U2OS cells, 10% FBS (Invitrogen) was used in DMEM medium. On the day of transfection, 4.5 × 106 suspended cells were seeded onto a 10-cm dish immediately before adding the DNA-lipid (1:3) mixture, which was prepared by blending DNA-OptiMEM solution with Lipofetamine2000-OptiMEM solution as described in the manufacturer’s manual. Each 10-cm dish of cells was transfected with 18 μg of total plasmid DNA, including carrier plasmid and 6 μg of each expression plasmid, and incubated for 24 h. In experiments where increased protein phosphorylation was desired, calyculin A (Cell Signaling) was added to the cultured cells 1 h before the cells were harvested and lysed. DMSO was used as a solvent control.
Coimmunoprecipitation and Western blot analysis
Cytoplasmic lysate preparation and co-IP experiments were as described previously (Ezzeddine et al. 2007). Briefly, 24 h after transfection, cells expressing transfected plasmid DNA were harvested and lysed at 4°C in 600 μL lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM Na-orthovanadate, 1 mM Na-pyrophosphate, and 1 mM NaF, supplemented with a protease inhibitor cocktail [Roche]). Fifty microliters of cell lysate were saved as an “input” control, and the rest was incubated with anti-V5 or -HA epitope antibody-conjugated agarose beads (Sigma) in the presence of 0.1 mg/mL RNase A at 4°C for 2 h. The beads were subjected to five washes with the lysis buffer and one wash with PBS. The coprecipitated proteins were separated by SDS-PAGE and visualized by Western blotting with the cognate antibodies. For phosphatase treatment, the washed immunoprecipitation beads were incubated with alkaline phosphatase (Fermentase) after for 1 h at 37°C, with gentle agitation every 10 min. The lysate from cells ectopically expressing PABPC1 was incubated with beads treated with either phosphatase or vehicle before a second round of immunoprecipitation. For Western blot analysis, the protein samples were resolved on a 7% or 10% SDS-PAGE and transferred to Immobilon-P membranes (Millipore). To detect V5-tagged or HA-tagged proteins, membranes were incubated with horseradish peroxidase (HRP)-conjugated monoclonal anti-V5 antibody (1/5000; Invitrogen) or HRP-conjugated monoclonal anti-HA antibody (1/1000; Roche), respectively. For detecting endogenous proteins, membranes were first incubated with rabbit anti-Caf1-B peptide antibody (1/1000) (Yamashita et al. 2005), rabbit anti-Ago2 (1/1000; Biosensis), rabbit anti-eRF3b (1/2000; Proteintech), rabbit anti-Pan2-B peptide (1/2000) (Yamashita et al. 2005), rabbit anti-PABPC1 (1/4000; a gift from R. Lloyd, Baylor College of Medicine), or mouse anti-β-actin-HRP monoclonal antibody (1/10,000; Santa Cruz). Secondary HRP-conjugated goat anti-rabbit antibodies (1/4000; Bethyl) or goat anti-mouse antibodies (1/5000; Bio-Rad) were used for detection with chemiluminescence reagent (peroxide/luminol enhancer; Pierce). Images were acquired by GeneGnome (Syngene) and analyzed by GeneSnap software.
Poly(A) tail distribution
Total RNA was isolated from U2OS cells with the RNeasy Mini kit (Qiagen). Poly(A) tail size-distribution profile analysis was performed as described previously (Lingner and Keller 1993). Briefly, total RNA was end-labeled with [α-32P]3′-Cordycepin and yeast poly(A) polymerase (Affimatrix). Labeled RNA was then digested with RNase T1 (Sigma) and RNase A (Sigma). Radioactivity was measured after precipitation with trichloroacetic acid, and 20,000 CPM of each sample was loaded and separated on an 8% denaturing polyacrylamide-urea gel. Autoradiograms were scanned and analyzed by ImageJ software (NIH) to create poly(A) length-distribution profiles.
Renilla and firefly dual luciferase assay
U2OS cells were transiently transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Briefly, 0.6 × 106 of U2OS cells were seeded in six-well plates before addition of 20 ng of the reporter plasmid (psi-check2 or psi-check2-4xboxB) and 3 ug of pcDNA-λN-HA-lacZ, pcDNA-λN-HA-TNRC6C (WT or mutant). Cells were harvested 42 h after transfection, and samples were analyzed simultaneously for firefly and Renilla luciferase activities using the Dual-Glo Luciferase Assay System (Promega) according to the manufacturer’s instructions. The relative levels of Renilla luciferase activity were normalized by dividing each reporter’s Renilla luciferase activity by its firefly luciferase control.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Richard Kulmacz (University of Texas Medical School at Houston) for critical reading of the manuscript, Richard Lloyd (Baylor College of Medicine) for anti-PABP antibodies, Lingzhi Liu and Kristen Rogers for technical assistance, and Dinghai Zheng for initiating the Pan3 mutants test. This work was supported by the National Institutes of Health (RO1 GM046454 to A.-B.S.) and by the Houston Endowment, Inc. to A.-B.S.
REFERENCES
- Albrecht M, Lengauer T 2004. Survey on the PABC recognition motif PAM2. Biochem Biophys Res Commun 316: 129–138 [DOI] [PubMed] [Google Scholar]
- Braun JE, Huntzinger E, Fauser M, Izaurralde E 2011. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell 44: 120–133 [DOI] [PubMed] [Google Scholar]
- Brautigan DL 2012. Protein Ser/Thr phosphatases—the ugly ducklings of cell signalling. FEBS J 10.1111/j.1742-4658.2012.08609.x [DOI] [PubMed] [Google Scholar]
- Brook M, Gray NK 2012. The role of mammalian poly(A)-binding proteins in co-ordinating mRNA turnover. Biochem Soc Trans 40: 856–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook M, McCracken L, Reddington JP, Lu ZÄ, Morrice NA, Gray NK 2012. The multifunctional poly(A)-binding protein (PABP) 1 is subject to extensive dynamic post-translational modification, which molecular modelling suggests plays an important role in co-ordinating its activities. Biochem J 441: 803–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Johnson AK, Dunker AK, Daughdrill GW 2011. Evolution and disorder. Curr Opin Struct Biol 21: 441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. 2012. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149: 1393–1406 [DOI] [PubMed] [Google Scholar]
- Chekulaeva M, Mathys H, Zipprich JT, Attig J, Colic M, Parker R, Filipowicz W 2011. miRNA repression involves GW182-mediated recruitment of CCR4-NOT through conserved W-containing motifs. Nat Struct Mol Biol 18: 1218–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CYA, Shyu AB 2011. Mechanisms of deadenylation-dependent decay. Wiley Interdiscip Rev RNA 2: 167–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-YA, Zheng D, Xia Z, Shyu A-B 2009. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat Struct Mol Biol 16: 1160–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R 2004. Eukaryotic mRNA decapping. Annu Rev Biochem 74: 861–890 [DOI] [PubMed] [Google Scholar]
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z 2002. Intrinsic disorder and protein function. Biochemistry 41: 6573–6582 [DOI] [PubMed] [Google Scholar]
- Dyson HJ, Wright PE 2005. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 6: 197–208 [DOI] [PubMed] [Google Scholar]
- Eystathioy T, Jakymiw A, Chan EKL, Seraphin B, Cougot N, Fritzler MJ 2003. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA 9: 1171–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezzeddine N, Chang T-C, Zhu W, Yamashita A, Chen C-YA, Zhong Z, Yamashita Y, Zheng D, Shyu A-B 2007. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol Cell Biol 27: 7791–7801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Mathonnet G, Sundermeier T, Mathys H, Zipprich JT, Svitkin YV, Rivas F, Jinek M, Wohlschlegel J, Doudna JA, et al. 2009. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol Cell 35: 868–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, Cieplak MK, Frank F, Morita M, Green J, Srikumar T, Nagar B, Yamamoto T, Raught B, Duchaine TF, et al. 2011. miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR4-NOT. Nat Struct Mol Biol 18: 1211–1217 [DOI] [PubMed] [Google Scholar]
- Funakoshi Y, Doi Y, Hosoda N, Uchida N, Osawa M, Shimada I, Tsujimoto M, Suzuki T, Katada T, Hoshino S 2007. Mechanism of mRNA deadenylation: Evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev 21: 3135–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnad F, Gunawardena J, Mann M 2011. PHOSIDA 2011: The posttranslational modification database. Nucleic Acids Res 39: D253–D260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T 1999. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-Poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J Biol Chem 274: 16677–16680 [DOI] [PubMed] [Google Scholar]
- Hunter T 2007. The age of crosstalk: Phosphorylation, ubiquitination, and beyond. Mol Cell 28: 730–738 [DOI] [PubMed] [Google Scholar]
- Huntzinger E, Braun JE, Heimstadt S, Zekri L, Izaurralde E 2010. Two PABPC1-binding sites in GW182 proteins promote miRNA-mediated gene silencing. EMBO J 29: 4146–4160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK 2004. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res 32: 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikematsu N, Yoshida Y, Kawamura-Tsuzuku J, Ohsugi M, Onda M, Hirai M, Fujimoto J, Yamamoto T 1999. Tob2, a novel anti-proliferative Tob/BTG1 family member, associates with a component of the CCR4 transcriptional regulatory complex capable of binding cyclin-dependent kinases. Oncogene 18: 7432–7441 [DOI] [PubMed] [Google Scholar]
- Jacobson A 1996. Poly(A) metabolism and translation: The closed-loop model. In Translational control (ed. Hershey JWB et al. ), pp. 451–480 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Jinek M, Fabian MR, Coyle SM, Sonenberg N, Doudna JA 2010. Structural insights into the human GW182-PABC interaction in microRNA-mediated deadenylation. Nat Struct Mol Biol 17: 238–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LN, Lewis RJ 2001. Structural basis for control by phosphorylation. Chem Rev 101: 2209–2242 [DOI] [PubMed] [Google Scholar]
- Khaleghpour K, Kahvejian A, De Crescenzo G, Roy G, Svitkin YV, Imataka H, O’Connor-McCourt M, Sonenberg N 2001. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol Cell Biol 21: 5200–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, Gehring K 2010. Molecular basis of eRF3 recognition by the MLLE domain of Poly(A)-binding protein. PLoS One 5: e10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, De Crescenzo G, Lim N, Siddiqui N, Fantus D, Kahvejian A, Trempe J, Elias D, Ekiel I, Sonenberg N, et al. 2004. Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase. EMBO J 23: 272–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn U, Wahle E 2004. Structure and function of poly(A) binding proteins. Biochim Biophys Acta 1678: 67–84 [DOI] [PubMed] [Google Scholar]
- Linding R, Russell RB, Neduva V, Gibson TJ 2003. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res 31: 3701–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Keller W 1993. 3′-end labeling of RNA with recombinant yeast poly(A) polymerase. Nucleic Acids Res 21: 2917–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Rivas FV, Wohlschlegel J, Yates JR III, Parker R, Hannon GJ 2005. A role for the P-body component GW182 in microRNA function. Nat Cell Biol 7: 1261–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A 2003. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4: 233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Agrin NS, Smith M, Gongidi P, Jacobson A 2004. Positive and negative regulation of poly(A) nuclease. Mol Cell Biol 24: 5521–5533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauxion F, Faux C, Seraphin B 2008. The BTG2 protein is a general activator of mRNA deadenylation. EMBO J 27: 1039–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasaka T, Morita M, Ito K, Suzuki T, Fukuda H, Takeda S, Inoue J-I, Semba K, Yamamoto T 2008. Interaction of antiproliferative protein Tob with the CCR4-NOT deadenylase complex. Cancer Sci 99: 755–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti F, Kaiser C, Zdanowicz-Specht A, Hentze MW 2012. PABP and the poly(A) tail augment microRNA repression by facilitated miRISC binding. Nat Struct Mol Biol 19: 603–608 [DOI] [PubMed] [Google Scholar]
- Okochi K, Suzuki T, Inoue J, Matsuda S, Yamamoto T 2005. Interaction of anti-proliferative protein Tob with poly(A)-binding protein and inducible poly(A)-binding protein: Implication of Tob in translational control. Genes Cells 10: 151–163 [DOI] [PubMed] [Google Scholar]
- Pillai RS, Artus CG, Filipowicz W 2004. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA 10: 1518–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK 2007. Intrinsic disorder and functional proteomics. Biophys J 92: 1439–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiland S, Messerli G, Baerenfaller K, Gerrits B, Endler A, Grossmann J, Gruissem W, Baginsky S 2009. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol 150: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P, Obradovic Z, Dunker AK 2004. Natively disordered proteins: Functions and predictions. Appl Bioinformatics 3: 105–113 [DOI] [PubMed] [Google Scholar]
- Rose KM, Bell LE, Jacob ST 1977. Specific inhibition of chromatin-associated poly(A) synthesis in vitro by cordycepin 5′-triphosphate. Nature 267: 178–180 [DOI] [PubMed] [Google Scholar]
- Roy G, De Crescenzo G, Khaleghpour K, Kahvejian A, O’Connor-McCourt M, Sonenberg N 2002. Paip1 interacts with poly(A) binding protein through two independent binding motifs. Mol Cell Biol 22: 3769–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB 2000. Physical and functional interactions between the mRNA cap structure and the poly(A) tail. In Translational control of gene expression (ed. Sonenberg N et al. ). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sanduja S, Blanco FF, Dixon DA 2011. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdiscip Rev RNA 2: 42–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu AB, Wilkinson MF, van Hoof A 2008. Messenger RNA regulation: To translate or to degrade. EMBO J 27: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG 2009. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma M, Fujiki H, Furuya-Suguri H, Yoshizawa S, Yasumoto S, Kato Y, Fusetani N, Sugimura T 1990. Calyculin A, an inhibitor of protein phosphatases, a potent tumor promoter on CD-1 mouse skin. Cancer Res 50: 3521–3525 [PubMed] [Google Scholar]
- Sugiyama N, Nakagami H, Mochida K, Daudi A, Tomita M, Shirasu K, Ishihama Y 2008. Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol Syst Biol 4: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temporini C, Calleri E, Massolini G, Caccialanza G 2008. Integrated analytical strategies for the study of phosphorylation and glycosylation in proteins. Mass Spectrom Rev 27: 207–236 [DOI] [PubMed] [Google Scholar]
- Tiedje C, Kotlyarov A, Gaestel M 2010. Molecular mechanisms of phosphorylation-regulated TTP (tristetraprolin) action and screening for further TTP-interacting proteins. Biochem Soc Trans 38: 1632–1637 [DOI] [PubMed] [Google Scholar]
- Tompa P, Csermely P 2004. The role of structural disorder in the function of RNA and protein chaperones. FASEB J 18: 1169–1175 [DOI] [PubMed] [Google Scholar]
- Tritschler F, Huntzinger E, Izaurralde E 2010. Role of GW182 proteins and PABPC1 in the miRNA pathway: A sense of déjà vu. Nat Rev Mol Cell Biol 11: 379–384 [DOI] [PubMed] [Google Scholar]
- Uchida N, Hoshino S, Katada T 2004. Identification of a human cytoplasmic poly(A) nuclease complex stimulated by poly(A)-binding protein. J Biol Chem 279: 1383–1391 [DOI] [PubMed] [Google Scholar]
- Uversky VN 2011. Intrinsically disordered proteins from A to Z. Int J Biochem Cell Biol 43: 1090–1103 [DOI] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, Sachs AB 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 2: 135–140 [DOI] [PubMed] [Google Scholar]
- Wilusz CW, Wormington M, Peltz SW 2001. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol 2: 237–246 [DOI] [PubMed] [Google Scholar]
- Xue B, Dunbrack RL, Williams RW, Dunker AK, Uversky VN 2010. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta 1804: 996–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB 2005. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 12: 1054–1063 [DOI] [PubMed] [Google Scholar]
- Zekri L, Huntzinger E, Heimstadt S, Izaurralde E 2009. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol Cell Biol 29: 6220–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Ezzeddine N, Chen C-YA, Zhu W, He X, Shyu A-B 2008. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol 182: 89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.