In this study, Boeke and coworkers define the structure and significance of a pseudoknot in the retrotransposable element Ty1 RNA. Ty1 reverse transcription depends on pseudoknot structure, establishing a cis-acting signal of the Ty1 genome.
Keywords: RNA structure, retrotransposon, reverse transcription, pseudoknot, SHAPE
Abstract
Ty1 retrotransposon RNA has the potential to fold into a variety of distinct structures, mutation of which affects retrotransposition frequencies. We show here that one potential functional structure is located at the 5′ end of the genome and can assume a pseudoknot conformation. Chemoenzymatic probing of wild-type and mutant mini-Ty1 RNAs supports the existence of such a structure, while molecular genetic analyses show that mutations disrupting pseudoknot formation interfere with retrotransposition, indicating that it provides a critical biological function. These defects are enhanced at higher temperatures. When these mutants are combined with compensatory changes, retrotransposition is restored, consistent with pseudoknot architecture. Analyses of mutants suggest a defect in Ty1 reverse transcription. Collectively, our data allow modeling of a three-dimensional structure for this novel critical cis-acting signal of the Ty1 genome.
INTRODUCTION
Ty1 is the most abundant transposable element in the genome of the budding yeast Saccharomyces cerevisiae at about 30 copies in lab strains. Ty1 replicates via an RNA intermediate; the RNA molecule serves as a mRNA for the Gag and Gag-Pol proteins, as well as genomic RNA for reverse transcription. Here, we consider the role of novel cis-acting sequences in the genomic RNA. Earlier studies showed that sequences required for retrotransposition lie near the element termini, with a ∼600-nt sequence near the 5′ terminus playing an important role (Xu and Boeke 1990). Among other roles, this region sequesters the cellular tRNAi-Met, which serves as the primer of reverse transcription, and produces a short DNA molecule referred to as minus strand strong stop DNA (Chapman et al. 1992; Wilhelm et al. 1994; Keeney et al. 1995). Previous studies have predicted a variety of structures for this RNA based on modeling, phylogenetic analyses, and mutational studies (Friant et al. 1996, 1997, 1998; Cristofari et al. 2002; Bolton et al. 2005).
Since these studies were published, SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension), a novel chemoenzymatic probing strategy, has been applied to model the secondary structure of retroviral genomes (Wilkinson et al. 2006, 2008; Deigan et al. 2009; Weeks and Mauger 2011). Using SHAPE and retrotransposition analyses of wild-type (WT) and mutant mini-Ty1 RNAs, we investigate here the structure of the 5′ end of Ty1 RNA and provide evidence that pseudoknot formation is required for efficient retrotransposition. Loss of Watson-Crick base-pairing in the stems leads to decreased retrotransposition, and restoring it restores transposition frequency. The identity of the base at the pseudoknot junction does not have a major effect on the structure. The pseudoknot structure is conserved across Ty1, Ty1′, and Ty2, as well as Ty1s from different Saccharomyces species, providing independent biological evidence for its importance.
Analysis of the Ty1 RNA half-life, packaging of Ty1 RNAs into virus-like particles (VLPs) and cDNA accumulation suggests that the primary importance of the pseudoknot is during the reverse transcription step of the life cycle.
Three-dimensional (3-D) modeling of the pseudoknot suggests two coaxial stems. RNA pseudoknots were first discovered in plant viruses (Rietveld et al. 1982) and are present in many classes of RNA. The H-type pseudoknot corresponds to a hairpin where the single-stranded region of the loop is base-paired with a single-stranded region elsewhere in the RNA sequence (Brierley et al. 2007). Pseudoknots are present in ribozymes (Ferre-D'Amare et al. 1998; Baskerville and Bartel 2002) and telomerases (ten Dam et al. 1991; Bhattacharyya and Blackburn 1994; Comolli et al. 2002). They have been shown to be important for the internal ribosome entry site (IRES) and initiation of translation (Kanamori and Nakashima 2001; Berry et al. 2010) as well as in autoregulation (Benard et al. 1998) and frame-shifting (Giedroc et al. 2000; Theis et al. 2008; Giedroc and Cornish 2009). Kissing loops are specific examples of pseudoknots in which two loops form intermolecular interactions, for example, in HIV-1 replication (for review, see Lu et al. 2011). The Ty1 pseudoknot has two distinct structural features: (1) it is located precisely at the RNA 5′ end; and (2) it encompasses a large RNA segment (> 300 nt), making it a “long-range” pseudoknot.
RESULTS
A predicted long-range pseudoknot structure in Ty1 RNA; evidence from SHAPE
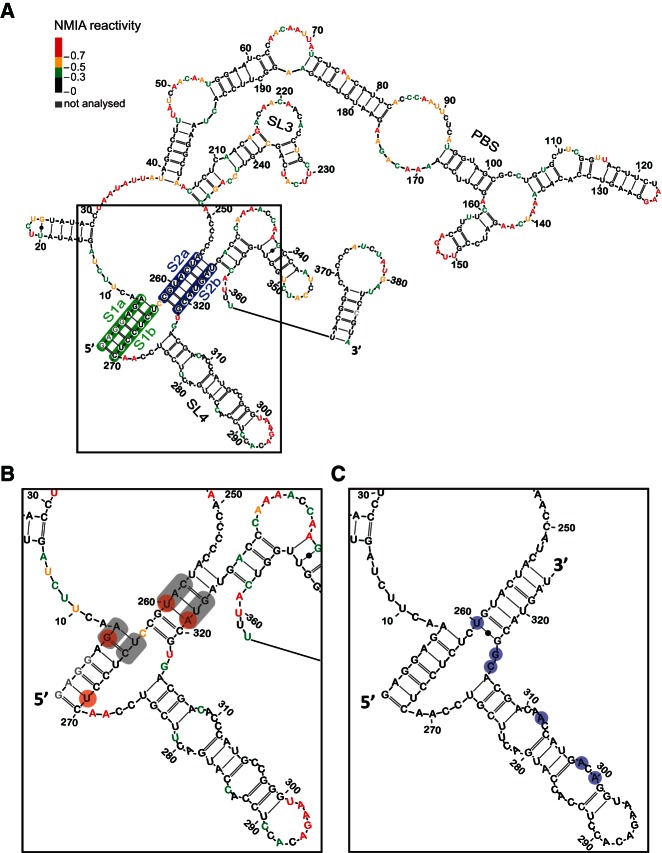
Initial evidence for a pseudoknot in the Ty1 RNA genome was based on the comparison between NMIA (N-methylisatoic anhydride) reactivities for the in vitro transcribed full-length Ty1 RNA and RNA in VLPs (Purzycka et al. 2013). No evidence supporting a pseudoknot was observed in full-length in vitro transcribed RNA. Since large RNAs rarely achieve functional structure without protein cofactors or chaperones (Gesteland et al. 2006), we elected to study this putative pseudoknot in vitro within a 560-nt “mini-Ty1” transcript (Fig. 1A). The length of this model transcript corresponds to a previously described functional fragment of Ty1 RNA (Bolton et al. 2005). SHAPE analysis of WT mini-Ty1 RNA revealed a reactivity pattern within the pseudoknot region comparable to that reported for RNA in VLPs (Purzycka et al. 2013). In particular, nucleotide region 256–262 was insensitive to NMIA modification (Fig. 1B). This allowed us to use this transcript as a model to characterize the structure of this region of Ty1 RNA in detail.
FIGURE 1.
In vitro RNA secondary structure model of (A) the first 388 nt of Ty1 genomic RNA and (B) the pseudoknot region. Each nucleotide is color-coded according to its normalized NMIA reactivity (color key, top left). Nonevaluated nucleotides are colored gray. The proposed pseudoknot region is boxed. The S1 stem is highlighted in green, S2 stem in blue. Sites of mutations in double/triple mutants GA6CU, UC264AG, CA258GC, UG322GC, CAU258GCC, and AUG321GGC are highlighted in gray. Sites of mutations in single mutants G6A, U269A, U260C, and A321G are highlighted by red circles. Neighboring stem–loop structure SL3 (nt 206–248) and SL4 (nt 275–316) are also designated. (C) Secondary structure model of the Ty2 pseudoknot region based on multiple sequence alignments of all known Saccharomyces cerevisiae Ty1 and Ty2 elements. Ty2 bases that differ from Ty1 are highlighted by light blue circles.
SHAPE analysis of pseudoknot mutants and suppressor mutants that restore complementarity
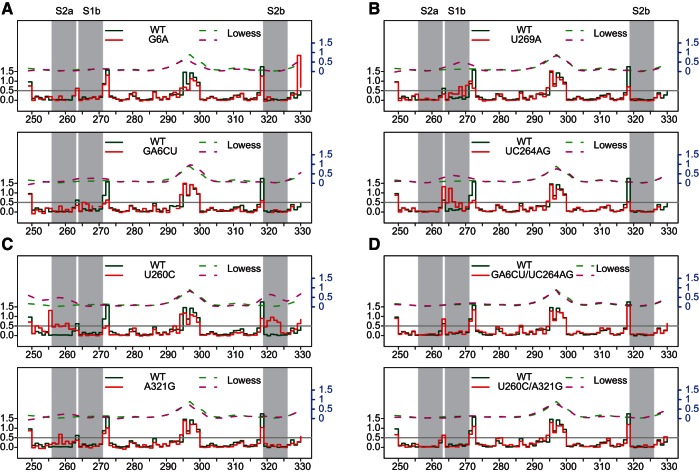
A previous mutagenesis screen identified a collection of transposition-deficient modular mini-Ty1-HIS3 elements with mutations in sequences required in cis for Ty1 replication and integration (Bolton et al. 2005). This genetic study showed an interaction between GAGGAGA and UCUCCUC sequences was essential for active retrotransposition. This stem constitutes part of the proposed pseudoknot. Our nomenclature for single and double mutants used here is as follows: The identity of nucleotide(s) in wild type RNA is given, followed by the corresponding position of the first base in question and the nucleotide identity(s) in mutant RNA. The single nucleotide substitution G6A, within GAGGAGA, did not result in a significant reactivity increase in the corresponding complementary region (Fig. 2A). Thus, a more dramatically destabilizing mutation was tested; in the double mutant GA6CU, we observed destabilization of the S1 stem; increased NMIA sensitivity was evident at positions C265, U266, and C268, confirming the involvement of these bases in the in vitro interaction. The strongest increase in reactivity corresponded to C265. This residue of WT RNA would normally base pair with the G6 that was substituted in the mutant. Interestingly, C258 from S2a also gained reactivity, suggesting an extensive network of interactions, as expected for a higher order structure.
FIGURE 2.
Step plots of NMIA reactivity (left y-axis) of the Ty1 pseudoknot region. (A) S1a mutants, (B) S1b mutants, (C) S2 mutants, and (D) S1/S2 compensatory mutants. Each mutant (solid red line) is plotted against the WT (solid green line) for comparison. Nucleotide positions corresponding to reverse transcriptase stops in the DMSO-control reaction appear as gaps. Lowess smoother fit curves (right y-axis, in blue) over a 10-nt window size were plotted in dashed lines. Nucleotides within the pseudoknot are indicated by gray stripes. The gray horizontal line at 0.5 represents an arbitrarily defined threshold for reactive residues.
When the structures of mutants with single U269A and double UC264AG substitutions in S1b were assessed, we observed localized reactivity changes mapping within the pseudoknot (Fig. 2B). Again, the highest increase was observed in residues closest to sites of mutation, suggesting that formation of the GAGGAGA/UCUCCUC stem is energetically favorable during RNA folding. Nucleotides 266–270 of the U269A mutant showed the greatest increase in reactivity, whereas the most pronounced changes mapped to 263–266 for the UC264AG mutant. Reactivity of only 2 nt from the corresponding S1a region could be detected in these mutants, due to the fact that 5′ end reactivity was masked by the abundant full-length product from the primer extension reaction (nucleotides that could not be evaluated are shown in gray in Fig. 1A,B), and the UC264AG mutant showed increased reactivity at those positions.
We also introduced mutations into the second pseudoknot helix (Fig. 2C). As predicted, the U260C mutation resulted in increased NMIA reactivity in the region 255–262 and, more importantly, a very strong effect was observed on complementary nucleotides 319–323. The strong destabilization introduced by single nucleotide substitution could be related to both the site of mutation, i.e., located closer to the middle of the stem, and the lower intrinsic stability of the S2 stem, which contains only three GC base pairs. As expected, predicted (RNAstructure) (Reuter and Mathews 2010) ΔG values for S1 and S2 were −10.1 and −7.6 kcal/mol, respectively. When the single mutation A321G in S2b substituted the original A-U base pair with a G-U wobble, we still observed increased reactivity in the corresponding S2a region. The observed change was small, indicating that some local destabilization had occurred but that the overall pseudoknot structure was preserved.
To thoroughly test the pseudoknot hypothesis, we analyzed RNAs with “suppressor” mutations restoring complementarity (Fig. 2D). GA6CU and U260C mutations were complemented with UC264AG and A321G, respectively. Both mutants showed restored WT-like NMIA reactivity patterns, consistent with a pseudoknot structure.
We performed an unsupervised clustering analysis (Fig. 3) on the pseudoknot mutants, which showed that WT and compensatory mutants are closely related structurally. Mutant A321G is similar to the compensatory mutant U260C/A321G, while U269A and UC264AG are also coclustered, suggesting the usefulness of this type of comparison. Interestingly, this analysis revealed that the global structure of mutant G6A resembled GA6CU despite the fact that only very modest destabilization within the pseudoknot region was observed for the G6A substitution. The observation that these mutants clustered distinctly from the WT suggests that localized changes within stem S1 had similar effects on global architecture.
FIGURE 3.
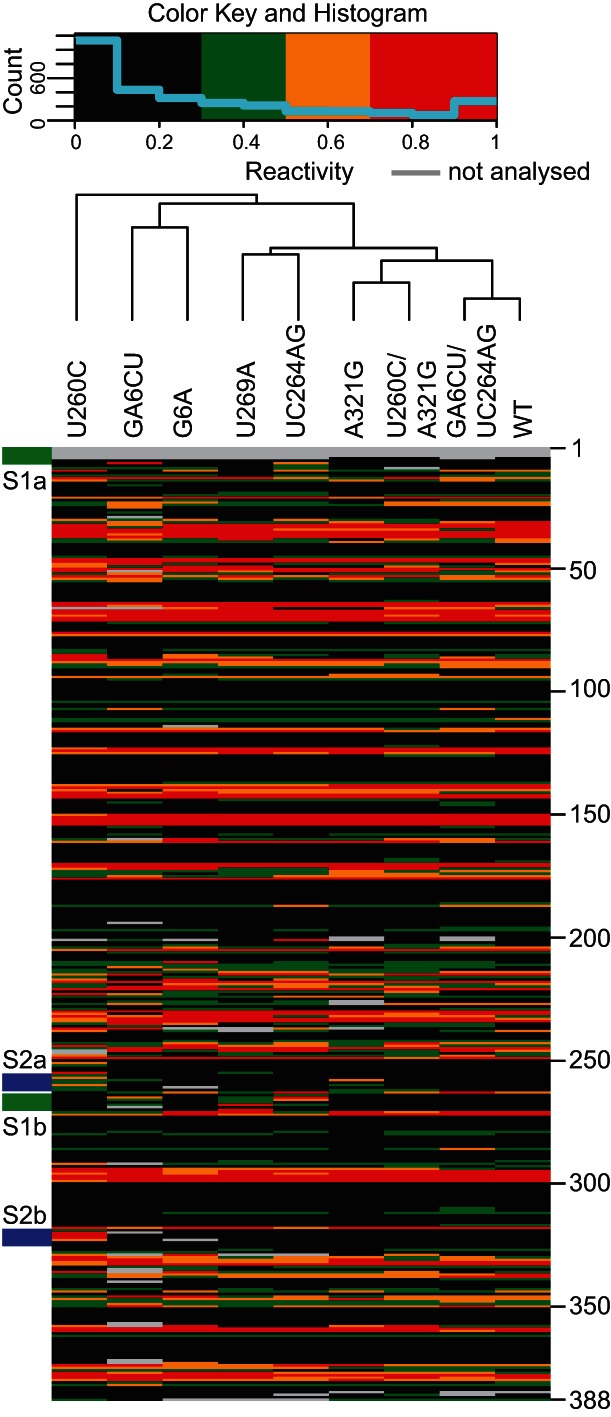
Clustering of Ty1 pseudoknot mutants. Each column consists of NMIA reactivity of the designated Ty1 RNA at single-nucleotide resolution (nt 1–388, color key at top). Green stripes on the left indicate the S1 stem. Blue stripes indicate the S2 stem. Nucleotide positions are marked on the right.
In the original mutagenesis screen by Bolton et al. (2005), several mutations within regions adjacent to the pseudoknot were shown to disrupt retrotransposition. We examined whether those defects were related to pseudoknot disruption by structurally characterizing mutants U207C, U209C, and C210U (Supplemental Table S1). The first two substitutions fall in stem SL3, while C210U is in a bulged region. We detected localized but extensive changes for U207C and U209C mutants that mapped proximal to the base substitution and in the complementary strand of the stem. Interestingly, single mutations within the internal loop caused a reactivity increase both at the substituted C and in the adjacent residues, U209 and A211. Also, C244 from this internal loop showed increased NMIA sensitivity. The above data provide experimental support for formation of stem SL3 in the structure. Residues within the pseudoknot did not show changes in reactivity pattern in these mutants, suggesting SL3 might function as a separate element and/or is necessary for the global architecture of the Ty1 RNA molecule. Interestingly, when G305 of SL4 was substituted by A (Supplemental Table S1), not only did reactivity of residues C286 and C304 increase but those of nucleotides 246–248 were also markedly reduced, suggesting that conformational changes in this region may influence global Ty1 RNA structure.
The pseudoknot is phylogenetically conserved
The pseudoknot lies within a highly constrained region in that it overlaps a coding region. Indeed, it is highly conserved within the family of Ty1 elements. In the emerging Ty1 subfamily designated Ty1′ (e.g., YBLWTy1-1; GenBank NC_001134.8), which is 91.7% (nucleotide) identical within the region encompassed by the pseudoknot sequences in Figure 1B, pseudoknot stem sequences are 100% identical. In the Saccharomces kluyveri Tsk1 element (GenBank AF492702.1), which is only 84% identical overall in the region, the pseudoknot stems are also identical. Even in Ty2, a sequence with only 82.9% identity (represented by Ty2-117; GenBank X03840.1), the pseudoknot stem sequences differ only in that Ty2 has a terminal G-U base pair in S2, where Ty1 has a G-C base pair (Fig. 1C).
Mutations in the pseudoknot interfere with retrotransposition
To directly address the impact of altered RNA structure on retrotransposition, mutant mini-Ty1-HIS3 elements were subcloned into a GAL1 promoter-driven full-length Ty1 element (pECB9C) (Bolton et al. 2005) carrying the retrotransposition reporter gene mhis3AI (Curcio and Garfinkel 1991) downstream from the GAG-POL stop codon. Retrotransposition was measured both in mini-Ty1 and full-length Ty1 contexts. Transposition frequencies of Ty1 elements were determined from the number of His+ colonies formed (relative to total colonies) following 24 h of galactose induction (Fig. 4A; Table 1). The single mutant G6A in S1a displayed an obvious defect, with a transposition frequency of 7% of WT. Transposition was further decreased to <5% of WT in mutant GA6CU, consistent with destabilization of stem S1 observed by SHAPE. Mutants U269A and UC264AG in S1b displayed similar trends in transposition defects, suggesting transposition frequency is directly correlated with the integrity of the S1 stem structure. Two additional single mutants in stem S2 were investigated. Mutant U260C showed ∼30% of the transposition activity of WT, despite the fact that the majority of the S2a-S2b interaction was suggested by SHAPE to be disrupted. A parallel study involving ex virio and in virio Ty1 genome analysis (Purzycka et al. 2013) identified stem S2 as a potential protein binding site on Ty1 RNA. The nucleic acid chaperone activity of the Gag polyprotein may facilitate pseudoknot folding and stabilization (Cristofari et al. 2000), potentially explaining the discrepancy observed between in vitro structural analysis and in vivo transposition data. The other mutant A321G in S2b, predicted to form a G-U base pair, had a close-to-normal transposition phenotype, suggesting that pseudoknot structure was not affected in vivo in this mutant. Transposition assays performed in a modular mini-Ty1-HIS3 system (Bolton et al. 2005) yielded similar results.
FIGURE 4.
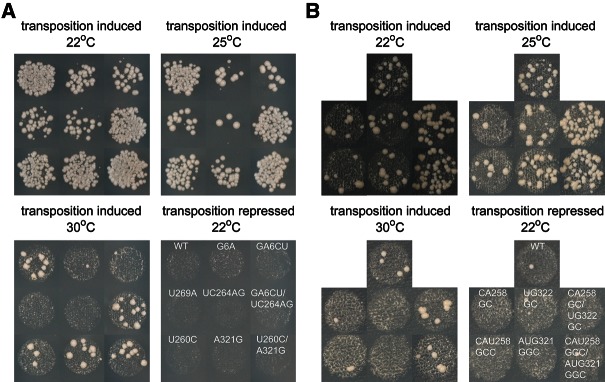
Ty1 pseudoknot formation is required for active transposition. (A) Transposition of full-length Ty1-mhis3AI elements containing designated mutations in the JB970 strain background was assayed at 22°C, 25°C, and 30°C. (B) Transposition of mini-Ty1-HIS3 elements containing designated mutations in the YQH055 strain background was assayed at 22°C, 25°C, and 30°C. Transposition under repressed conditions (glucose) is shown as a control. Quantitative retrotransposition frequency data are presented in Table 1.
TABLE 1.
Ty1 retrotransposition is affected by mutations in pseudoknot region
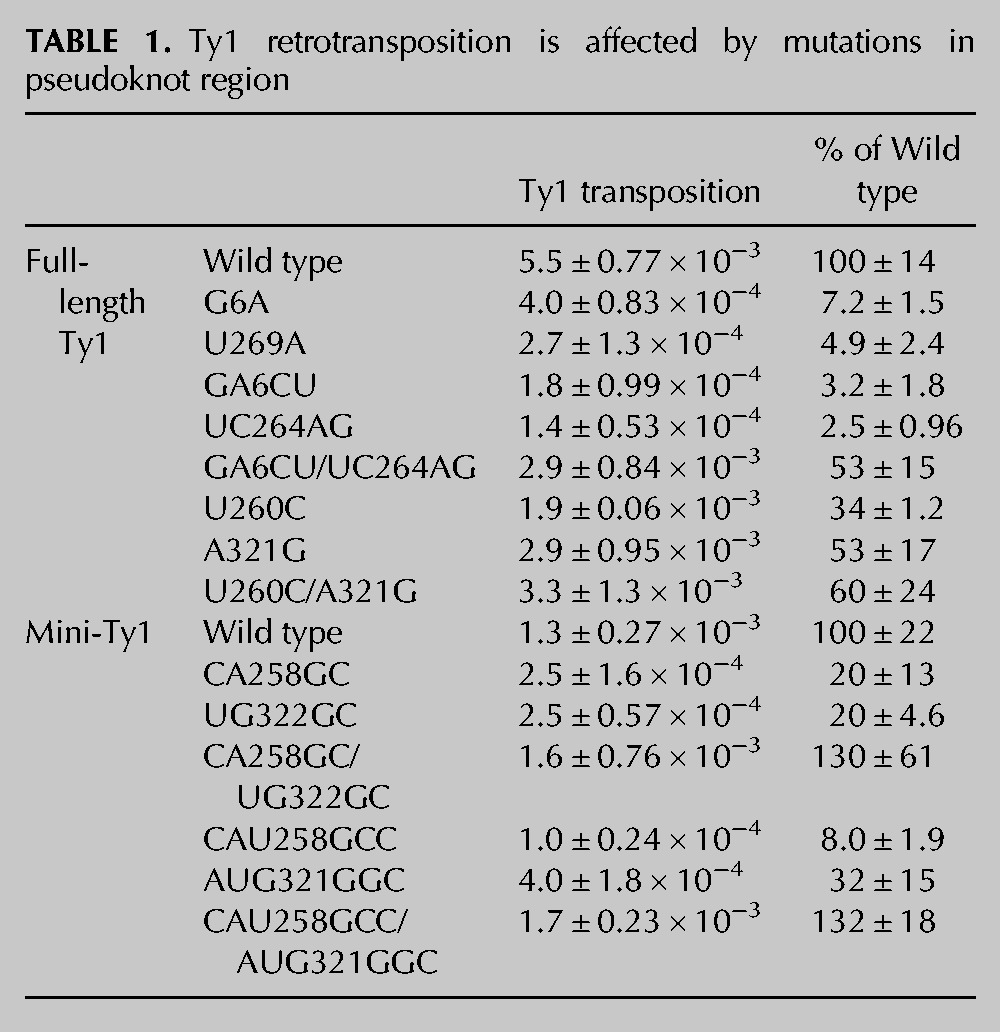
Defects observed for mutants were more prominent when the galactose induction temperature was increased (Fig. 4), which may reflect further pseudoknot destabilization at higher temperatures, in addition to previous findings that Ty1 transposition is a temperature-sensitive process with reduced protease and reverse transcriptase (RT) activities at high temperature (Garfinkel et al. 1985; Wilhelm et al. 2000; Lawler et al. 2002). Stronger pseudoknot destabilization was also negatively correlated with the transposition efficiency. The GA6CU and UC264AG mutants showed more profound transposition defects in comparison to G6A and U269A mutants, respectively. This effect became even more pronounced when the temperature was increased during galactose induction.
To further validate the existence of S2 duplex structure as part of the pseudoknot, we generated mini-Ty1-HIS3 mutants in S2a and S2b with more extensive mutations (Fig. 4B; Table 1). With double mutations in S2a or S2b, mutants CA258GC and UG322GC exhibit a fivefold decrease in transposition activity compared to WT. When one more mutation was introduced in S2a, the triple mutant CAU258GCC displayed an even more severe transposition defect (∼8% of WT). Interestingly, with the A321G mutation replacing the original A-U base pair with a G-U wobble base pair, triple mutant AUG321GGC in S2b resulted in a transposition frequency similar to the double mutants, suggesting that transposition level is negatively correlated with the extent of S2 destabilization.
Suppressor mutations restore retrotransposition
To recover pseudoknot functionality in mutants with disrupted base-pairing and decreased transposition frequency, suppressor mutations were introduced to restore stem complementarity and tested for their ability to relieve retrotransposition defects (Fig. 4; Table 1). A suppressor mutation in the S1 stem combining both GA6CU and UC264AG mutations restored transposition activity from <4% to >50% of WT. Similarly, a suppressor mutant in the S2 stem with U260C and A321G mutations restored transposition from 30% to 60% of WT. In this experiment, it was unavoidable to change one amino acid of the GAG open reading frame; in spite of this, useful experimental data were obtained. The close-to-normal transposition level in the new S2 suppressor mutant suggests that there is no deleterious effect of the M90V amino acid substitution in Gag.
With more extensive mutations introduced into S2, the mini-Ty1-HIS3 system had to be used to supply wild-type Ty1 proteins from a helper plasmid during transposition (Bolton et al. 2005). When CA258GC mutations were combined with UG322GC mutations, transposition levels increased from 20% to 130% of WT. Similarly, a combination of triple mutations on S2a and S2b (CAU258GCC/AUG321GGC) increased transposition from <10% to >130% of WT. Interestingly, these S2 suppressor mutants transpose even better than WT and contain only G-C base pairs at mutation sites, resulting in more stable base-pairing in S2 compared to WT sequences. This observation suggests that elevated transposition activity may be facilitated by a more stabilized pseudoknot structure.
The above results on suppressor mutants revealed a strong correlation between restored stem complementarity and recovery of retrotransposition frequency, providing further evidence supporting the proposed pseudoknot structure.
Identity of the junction nucleotide is not critical for pseudoknot formation
It was previously demonstrated for pseudoknots that induce frame-shifting in various viruses that the single nucleotide at the junction between the two helices can affect both structure and function (Chen et al. 1995). We tested whether the identity of the intervening nucleotide at the junction affected the structure of the Ty1 pseudoknot by evaluating all possible mutants with synonymous mutation in the intervening nucleotide (Fig. 5A). Substituting C263 at the junction with any other nucleotide caused only modest reactivity changes at C262, adjacent to the substituted residue. A modest increase in NMIA sensitivity at this position was also observed for the compensatory U260C/A321G mutant, which did not show other indications of pseudoknot disruption. All three mutants displayed normal levels of transposition (Fig. 5B), suggesting that pseudoknot structure was preserved in the junction mutants.
FIGURE 5.
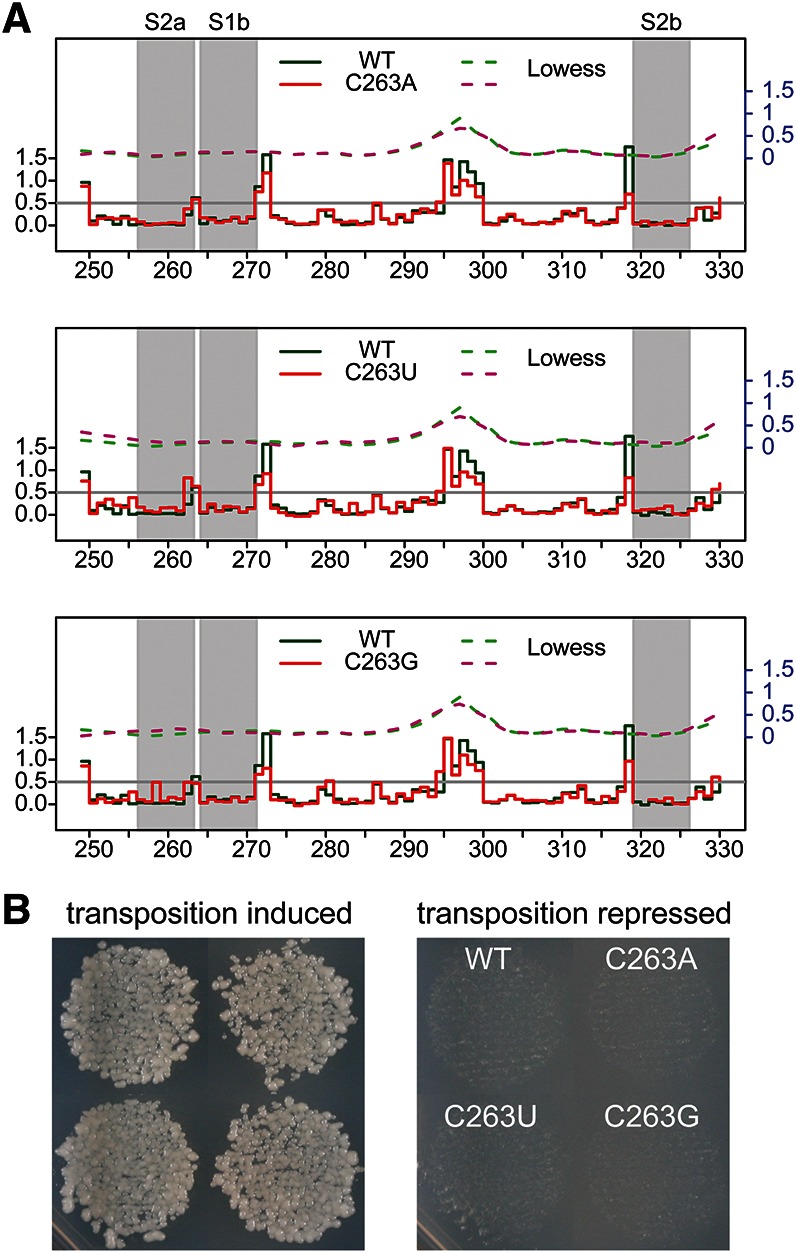
Mutations in the Ty1 pseudoknot junction do not affect pseudoknot formation or transposition efficiency. (A) Step plots of NMIA reactivity of the pseudoknot region in junction mutants C263A, C263U, and C263G. (B) Transposition of junction mutants compared with WT Ty1-mhis3AI in the JB740 strain background at 22°C. Transposition under repressed conditions (glucose) is shown as a control.
Pseudoknot mutations have no obvious effects on Ty1 RNA transcription, stability, and packaging
Ty1 RNA transcription/turnover and packaging into VLPs are two key processes preceding reverse transcription. We, therefore, examined whether any defects were associated with these steps in the Ty1 life cycle. To monitor Ty1 RNA transcription and stability, Ty1 RNA levels were assessed after adding glucose to shut off steady state Ty1 transcription. No obvious decrease in absolute RNA level was observed for S1 stem mutant GA6CU, whereas mutant UC264AG showed slightly increased RNA expression compared to WT at steady state (Fig. 6A). Furthermore, WT and mutant Ty1 RNAs have almost identical half-lives (Fig. 6B), suggesting that RNA transcription/turnover is normal in pseudoknot mutants. As a further test for subtle effects on expression, we examined production of Ty1 Gag in cells expressing WT and mutant elements (Fig. 6E). No expression defect was detected in the mutants, even though, in an earlier study, we could readily detect differences as small as threefold by this method (Yarrington et al. 2012). If anything, we see modestly increased expression in the mutants, but this is also seen in the suppressor mutants; thus, we do not believe it is responsible for the decreased retrotransposition seen in the mutants.
FIGURE 6.
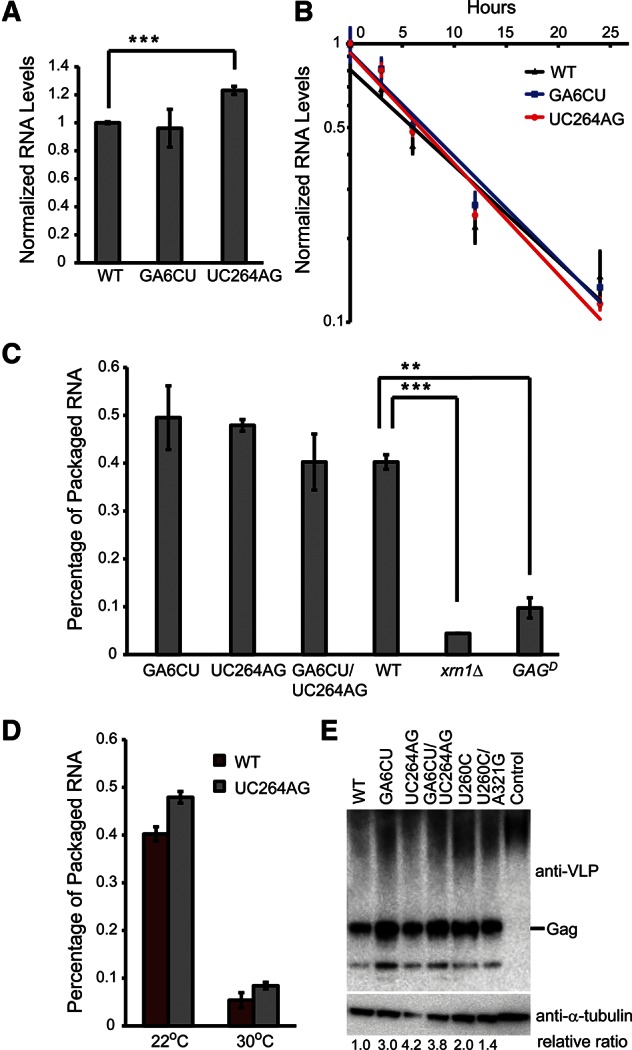
Ty1 RNA half-life and packaging into VLPs are not affected in pseudoknot mutants. (A) Steady state Ty1 RNA levels were measured by QRT-PCR in WT and S1 mutants. RNA levels were normalized to WT, and error bars represent the standard deviation of three biological replicates. (B) Ty1 RNA levels were measured by QRT-PCR at the indicated time points post-glucose exposure. RNA levels in each strain were normalized to the RNA level at the 0-h time point and plotted on a log scale. Solid lines are fitted using an exponential decay function. The slopes of the fitted lines reflect the decay rates of Ty1 RNA. Error bars represent the standard deviation of three replicates. (C) Ty1 RNA levels from samples with and without benzonase treatment were measured by QRT-PCR, and their ratios were plotted as a percentage of packaged Ty1 RNA. Error bars represent the standard deviation of two independently treated lysates. (D) Percentage of Ty1 RNA packaging is shown for WT and the UC264AG mutant at normal (22°C) and high (30°C) temperatures. (**) P < 0.01, (***) P < 0.001. P-values were determined by Student's t-test. (E) Immunoblot analysis of Ty1 Gag from JB970 strains expressing WT or designated mutant Ty1 element. Control lane is the JB970 strain with no plasmid transformed. Protein levels were normalized to α-tubulin levels.
To determine whether pseudoknot architecture is required for packaging of Ty1 RNA into VLPs, benzonase digestion of total RNA was performed to detect the amount of Ty1 RNA packaged/protected by Gag protein surrounding the mutant VLPs (Lin and Levin 1998). We have extensively modified this assay to make it more quantitative by combining benzonase digestion with a QRT-PCR-based readout and a “spike-in” of synthetic Ty1 RNA as an internal control for RNA recovery (see Materials and Methods). A packaging defect was observed in a dominant-negative Ty1 GAG mutant (Monokian et al. 1994) known to form defective VLPs (Fig. 6C). Similarly, XRN1 encodes a P-body 5′-3′ exonuclease essential for efficient Ty1 RNA packaging (Dutko et al. 2010). WT Ty1 RNA expressed in an xrn1Δ strain showed greatly reduced packaging, validating the assay. In contrast, S1 stem mutants did not show enhanced benzonase sensitivity, suggesting either that the pseudoknot is not involved in Ty1 RNA packaging or that packaging is signaled by an extensive network of RNA tertiary interactions as well as nucleic acid-protein interactions that extend beyond the pseudoknot region, thus disrupting the pseudoknot alone is insufficient to impair packaging. It is noteworthy that Ty1 RNA packaging is significantly affected by temperature (Fig. 6D). At 30°C, both WT and the UC264AG mutant have diminished levels of RNA packaging. This could be a consequence of decreased protease activity, which reduces the efficiency of VLP maturation (Curcio and Garfinkel 1992; Lawler et al. 2002).
Decreased Ty1 cDNA is detected in pseudoknot mutants
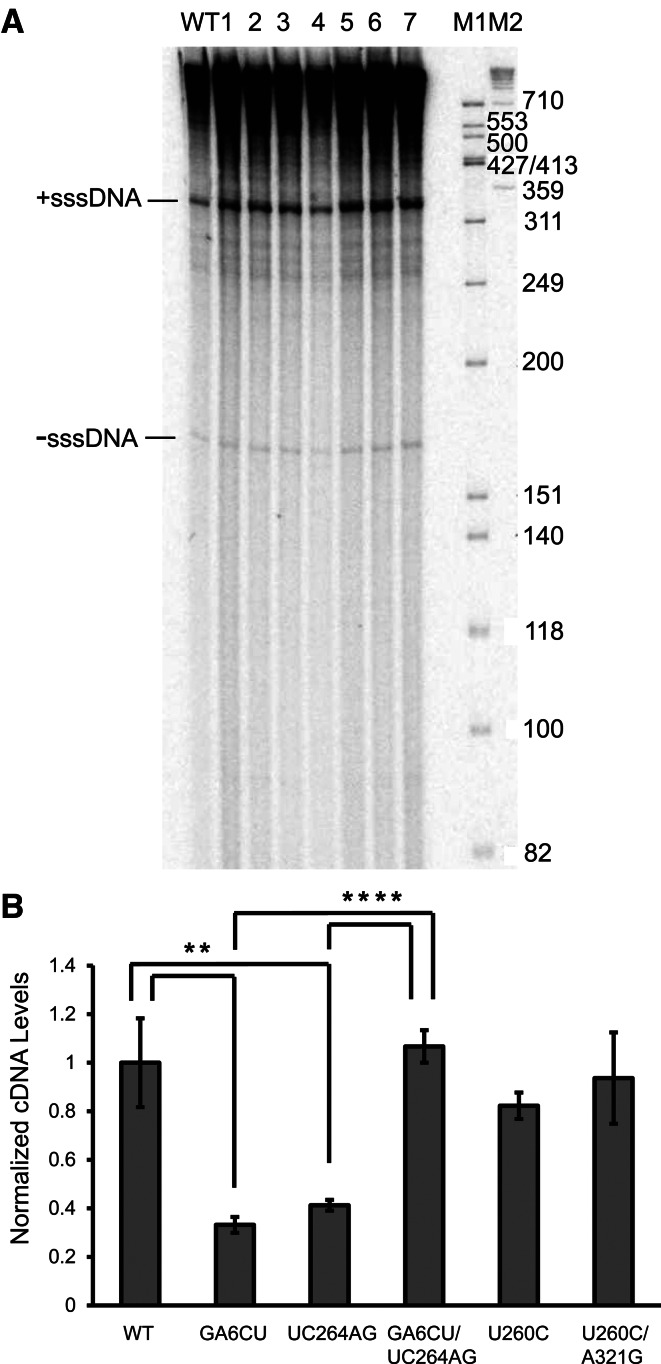
To further probe transposition defects observed in pseudoknot mutants, reverse transcription of Ty1 RNA was examined. In pseudoknot mutants, both minus strand strong stop DNA (−sssDNA) and plus strand strong stop DNA (+sssDNA) were produced at similar levels as WT in vitro (Fig. 7A). However, when we analyzed the amount of Ty1 cDNA in cell lysates with Q-PCR (Fig. 7B), greater than twofold reductions in cDNA levels were detected in S1 mutants GA6CU and UC264AG, and a 1.2-fold reduction was detected for S2 mutant U260C, in agreement with the mild transposition defect observed for this mutant. The cDNA level was clearly restored in the compensatory mutant GA6CU/UC264AG. Our results are consistent with earlier studies by Bolton et al. (2005), who tested a subset of these mutants and measured cDNA production by Southern blotting, indicating that the Ty1 pseudoknot is involved in mediating efficient cDNA synthesis during retrotransposition. Because the reduction in retrotransposition frequency is larger than the cDNA synthesis defect, it is likely that the quality as well as the quantity of Ty1 cDNA is adversely affected in the mutants.
FIGURE 7.
Decreased cDNA level in pseudoknot mutants. (A) Endogenous RT reactions for WT and pseudoknot mutants. (1) G6A, (2) U269A, (3) U260C, (4) GA6CU/UC264AG, (5) UC264AG, (6) U260C/A321G, (7) A321G, (M1, M2) DNA markers. Positions of −sss- and +sssDNA are indicated. (B) Ty1 cDNA levels were measured by Q-PCR and normalized to WT cDNA level; error bars represent standard deviation of three biological replicates. (**) P < 0.01, (****) P < 0.0001. P-values were determined by Student's t-test.
DISCUSSION
The pseudoknot structure
The Ty1 pseudoknot, analogous to more compact pseudoknots in retroviruses and other viruses that mediate translational frame-shifting, is imbedded in the GAG coding region, greatly constraining its evolution. However, the Ty1 pseudoknot is noteworthy structurally in that (1) it is the only example where the pseudoknot forms at the extreme 5′ end of the RNA, and (2) the region encompassed by the structure is >320 nt in length and contains at least three other potential stem-forming regions, one of which is the primer binding site (PBS). Most pseudoknots are far more compact and can be <35 nt long, many functioning to promote ribosomal frame-shifting, and their potency in controlling a translating ribosome to effect frame-shifting depends on their structural stability, which is presumably enhanced by their more compact structure (Hansen et al. 2007). Thus, the Ty1 pseudoknot more closely resembles the long-range pseudoknots found in the Varkud satellite (VS) ribozyme (∼90 nt) (Rastogi et al. 1996), human telomerase RNA (∼95 nt) (Chen et al. 2000), a plasmid copy number control system (∼130 nt) (Asano and Mizobuchi 1998), a proposed ribosomal operon translational control region (∼300 nt) (Chiaruttini et al. 1996), and the Escherichia coli 16S ribosomal RNA (∼900 nt) (Poot et al. 1998).
Phylogenetic conservation of the Ty1 pseudoknot suggests it evolved long ago. This sequence is under a dual constraint, being part of an important structure, while part of the sequence is also embedded within the Gag coding region. The Ty1 packaging signal has not yet been clearly identified. RNA kissing loops (“intermolecular” pseudoknots) and the NC domain of Gag are the determinants of HIV dimerization and packaging (for review, see Lu et al. 2011). Since the Ty1 pseudoknot may mediate protein binding (Purzycka et al. 2013) and is somewhat similar structurally to the HIV kissing-loop motif, we investigated its potential involvement in RNA packaging. However, the Ty1 pseudoknot does not seem to play a role in this process but rather to have a role later in the life cycle.
A model for the 3-D structure of the pseudoknot
Pseudoknots perform a variety of functions depending on their structure (Pleij 1990; Westhof and Jaeger 1992). Such motifs are also used to organize global RNA folds, as exemplified by the ribosome. Here, we attempted to predict the structure of the Ty1 pseudoknot in terms of its 3-D fold. This structural model of Figure 8 was developed on the context of the first 362 nt of Ty1 RNA using RNAComposer (Popenda et al. 2012). The most energetically favorable structure predicts that helices S1 and S2 are coaxial with C263 stacked between them (Fig. 8B). Stacking between C263 and the C262-G319 pair may explain increased NMIA reactivity of C262 when C263 was mutated in the junction mutants. It was demonstrated for mouse mammary tumor virus (MMTV) that an unpaired A creates a bent structure, preventing coaxial stacking of helical stems (Shen and Tinoco 1995). Substituting C263 with A, G, or U did not interfere with pseudoknot function in our experimental system. Indeed, for several viral pseudoknots, as long as their conformation and overall stability is maintained, the exact nucleotide sequence may not be important (Giedroc and Cornish 2009). On the other hand, it was recently demonstrated that the murine leukemia virus (MLV) pseudoknot regulates translational recoding by a protonation-dependent conformational switch that involves interactions of the stem with the loop (Houck-Loomis et al. 2011). Although the Ty1 G6A mutant did not show substantial reactivity changes in the pseudoknot region, our clustering analysis demonstrated that this mutant is distinct from the WT in global structure. At this stage, the details are hard to characterize using secondary structure probing, but a base pair involving nucleotides 6 and 265 might be important for contacts regulating pseudoknot function. This idea is supported by the relatively severe transposition defect of this single-nucleotide substitution mutant.
FIGURE 8.
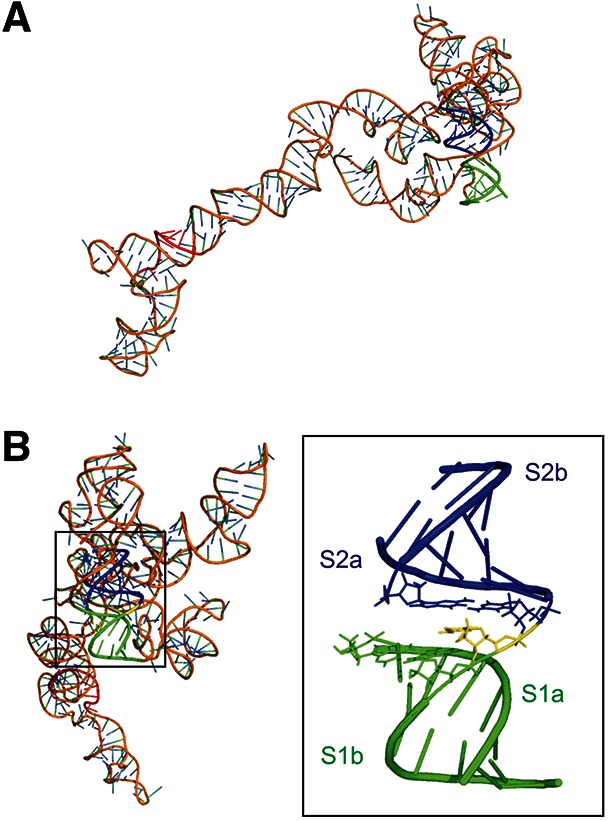
3D structure model of Ty1 RNA (nt 1–362). (A) Relative orientation of the PBS and the pseudoknot. S1 is indicated in green, S2 in blue, C263 in the junction is shown in yellow, Ty1 PBS is shown in red. (B) View rotated 90° along y-axis and pseudoknot with the central segment (nt 7, 262–264, 319) shown as sticks.
Effects on retrotransposition
In general, mutations affecting pseudoknot structure have the predicted effects, i.e., disrupting either stem reduces retrotransposition and correcting base-pairing restores retrotransposition frequency. The early stages of the Ty1 life cycle, including RNA expression and stability, translation (as measured by steady state Ty1 protein expression), packaging, and VLP assembly appear mostly unaffected, although we cannot rule out a subtle qualitative defect in the assembled structure. The one exception is that the abundance of Ty1 Gag protein is somewhat increased in the pseudoknot mutants, consistent with a modest increase in translation rate. However, this increase is also observed in the “suppressor” mutants and thus is unlikely to explain the observed transposition defect. It is remarkable that the ribosome can enter a mRNA which has such a stable fold precisely at its 5′ end. The 5′ cap, of course, would protrude from the structure and be available for recognition by the cap-binding complex and initiation factors.
Collectively, these results suggest a defect later in the life cycle, and we observe reduced Ty1 cDNA production in the mutants that is restored in “suppressor mutant” constructs. This reverse transcription defect could have many consequences, such as a qualitative difference in the folded structure of the Ty1 RNA, which might affect proper priming of –sssDNA, a process that occurs within the RNA loop that is “closed” by the pseudoknot structure, albeit at least 100 nt away in the linear sequence. However, we observed normal levels of −sss- and +sssDNA from VLPs in vitro, suggesting the defect might be at a later stage in reverse transcription. Elongation of the reverse transcript of Ty1 has been shown to be a relatively inefficient process in Ty1, and a potential point of regulation and control by host factors (Lee et al. 1998). Multiple regulators of Ty1 transposition in S. cerevisiae have conserved roles in genome maintenance (Scholes et al. 2001). Our data suggest that the Ty1 pseudoknot may play a role in regulating the efficiency of formation of the complete Ty1 reverse transcript. Also, since the cDNA accumulation defect is quantitatively subtler than the decrease in retrotransposition frequency, it suggests that the quality as well as the quantity of the Ty1 cDNA may be adversely affected in the pseudoknot mutants. Further studies will be required to dissect the detailed mechanism of the defect.
MATERIALS AND METHODS
Media, plasmids, and strains
Yeast strains used in this study were JB970 (MATa, lys2 his3Δ200 trp1-289 ura3-52 spt3-101), YQH055 (MATα his3Δ200 leu2Δ1 ura3-167 rad52Δ::NATMX4 [pECB2B2, Ty1-helper-2 µ-LEU2]), JB740 (MATα his3Δ200 leu2Δ1 ura3-167), JB879 (MATa ura3-52 his3Δ200 ade2-101 lys2-801 Met− reg1-501), and JB503 (MATa his3Δ200 lys2 ura3-52 trp1-1). Media were prepared as described (Sherman et al. 1986).
Mini-Ty1-HIS3 mutant plasmids, Ty1 helper plasmid pECB2B2, and the full-length wild-type pGAL-Ty1-mhis3AI plasmid, pECB9C, were previously described (Bolton et al. 2005). pECB9C derivatives were constructed by subcloning XhoI-HpaI or XhoI-BbvCI fragments of mini-Ty1-HIS3 plasmids into the pECB9C backbone. Compensatory pseudoknot mutants and junction mutants were generated by quick-change mutagenesis.
YQH136, a control strain used in Ty1 packaging analysis, was generated by transforming pECB9C into the yeast xrn1Δ knockout strain 14540 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 xrn1Δ::KanMX). The dominant-negative GAG mutant was expressed from a galactose-inducible neo-marked Ty1 plasmid, pGM335 (Monokian et al. 1994), in the JB503 strain background. pQH170 was generated by cloning the SP6 promoter directly upstream of pRY098 (pGALCO760-2882Ty1mhis3AI) (Yarrington 2009).
SHAPE
Templates for in vitro transcription were obtained by PCR amplification of mini-Ty1 fragments (+1–560 nt) from corresponding plasmids using a forward primer containing an SP6 promoter followed by WT or mutant 5′ Ty1 RNA sequence [WT: 5′-GATTTAGGTGACACTATAGAGGAGAACTTCTAGT-3′] and a reverse primer [5′-GAACCGGACCGAAGCCCGATTTGGATCCGGCGAACCGGATCGAAACATTGGTGGTGGTCTGAC-3′]. Corresponding mutations were incorporated into the forward primer when amplifying S1a mutant sequences. SP6-MEGAscript (Life Technologies) was used for in vitro transcription following the manufacturer's protocol. RNA was DNase I-treated and recovered by LiCl precipitation. For NMIA modification, 8 pmol RNA were refolded in 20 µL of buffer (10 mM Tris-HCl [pH 8.0], 100 mM KCl, and 0.1 mM EDTA) by heating for 1 min at 85°C, slow cooling to 4°C, adding 130 µL of folding buffer (final concentration: 40 mM Tris-HCl [pH 8.0], 200 mM KCl, 0.5 mM EDTA, and 5 mM MgCl2), and incubating for 20 min at 37°C. The RNA was divided into two tubes and treated with 8 µL of NMIA in DMSO ([+], final concentration 3 mM) or DMSO alone (−), and the modification reaction was carried for 45 min at 37°C. After recovering RNA by ethanol precipitation, it was resuspended in 10 µL of 5 mM Tris-HCl [pH 7.0] and 0.5 mM EDTA.
For detection of 2′-O-adducts, 12 µL of primer-template solution containing fluorescently labeled primer (TCAGGTGATGGAGTGCTCAG; Cy5 [+] and Cy5.5 [−]; 8 µM and 6 µM, respectively] and 1.5 pmol of RNA were incubated at 85°C for 1 min, 60°C for 5 min, 35°C for 5 min, and 50° for 2 min. Reverse transcription was carried out at 50°C for 50 min (Invitrogen SuperScript III, Life Technologies). Subsequently, 1 µL of 4 M NaOH was added, and RNA was hydrolyzed by heating 3 min at 95°C. Reactions were neutralized with 2N HCl and combined. Thermo Sequenase Cycle Sequencing kit (Affymetrix) and primers labeled with WellRed D2 and Licor IR-800 were used to generate sequencing ladders. Combined samples and sequencing ladders were ethanol-precipitated, washed twice with 70% ethanol, dried, and resuspended in deionized formamide. A CEQ8000 Genetic Analysis System (Beckman-Coulter) was used to analyze primer extension products.
SHAPE data analysis and clustering
Electropherograms were processed using ShapeFinder software (Vasa et al. 2008). NMIA reactivity was normalized using ShapeNorm, a custom data-processing script developed in the statistical language R. Peak intensity (peak area differences between reaction and background) for each nucleotide is divided by the average intensity of the top 8% most reactive peaks excluding outliers, which are defined as peaks greater than 1.5 times the interquartile difference above the 3rd quartile (as described in Low and Weeks 2010). Reverse transcription stops in the DMSO-control reaction were identified as outlying high peaks in the plotted background area, and the corresponding nucleotide positions were marked as nonevaluated residues (gray colored nucleotides in Figs. 1 and 3). All reactivity data reported were averaged from at least two independent experiments.
Euclidean distances, i.e., dissimilarities, among different pseudoknot mutants were calculated by regarding each mutant as a point in a 388-dimensional space, where its Cartesian coordinates are NMIA reactivity at the corresponding 388 nt (nt 1 to 388). Mutants (columns) were then reordered and clustered according to the computed dissimilarities. All statistical computing was conducted using open-source statistical software R version 2.10.1; scripts ShapeNorm, ShapePlots, and ShapeCluster are available on request.
Transposition assays
Transformants containing WT and mutant GAL-Ty1-mhis3AI URA3-marked plasmids were streaked out on SC–Ura plates with 2% glucose. Approximately a match head-sized patch of cells was scraped from plates and resuspended in 150 µL SC–Ura medium. A600 was measured by a Tecan Safire 2 microplate reader. Cells were diluted to equal A600 values, and 50 µL diluted cells were spotted onto SC–Ura plates and incubated at 30°C for 2 d. Cells were then replicated onto both 2% galactose and 2% glucose SC–Ura plates and grown at various target induction temperatures (22°C, 25°C, or 30°C). After 24 h galactose induction, plates were replica-plated to YPD plates for 1 d at 30°C before being replica-plated to SC–His plates. Formation of His+ colonies was assessed after 1–2 d. Three independent transformants were analyzed for each WT/mutant Ty1 yeast strain. Transposition frequencies were calculated as the ratio of His+ CFU/total CFU after 24 h galactose induction.
Transformants containing WT and mutant GAL-mini-Ty1-HIS3 URA3-marked plasmids in a YQH055 strain background were streaked out on SC–Ura–Leu plates with 2% glucose. Fifty microliters of cells at equivalent A600 values were spotted onto SC–Ura–Leu plates and incubated at 30°C for 2 d. Cells were then replicated onto both 2% galactose and 2% glucose SC–Ura–Leu plates and grown at various target induction temperatures (22°C, 25°C, or 30°C). After 48 h of galactose induction, plates were replica-plated to YPD plates for 1 d at 30°C before being replica-plated to SC–His + 5-FOA plates. Formation of His+ colonies was assessed after 2–3 d. Three independent transformants were analyzed for each WT/mutant Ty1 yeast strain. Transposition frequencies were calculated as the ratio of His+Ura− CFU/Ura− CFU after 48 h of galactose induction.
RNA half-life measurement
Yeast cells (JB970) harboring pECB9C or derivatives were grown in 20 mL SC–Ura medium with 1% raffinose overnight at 30°C. A600 was measured the next day, and 4 A600 of cells were subcultured in 40 mL YNB medium containing 2% casamino acids and 2% galactose. Induced cultures were incubated at 22°C for 45 h, then diluted by adding an equal volume of YNB/CAA medium containing 4% glucose to inhibit Ty1 expression. Time points were taken at 0, 3, 6, 12, and 24 h post-glucose exposure. For each time point, 10 mL of yeast culture was harvested by centrifugation at 1500g for 5 min, washed once with ice-cold DEPC-treated water, and frozen in liquid nitrogen. Total RNA was extracted with hot acid phenol (Schmitt et al. 1990; Collart and Oliviero 2001), turbo DNase (Ambion) treated, and reverse transcribed into cDNA using an Oligo(dT)20 primer and SuperScript III reverse transcriptase (Invitrogen) for quantitative real-time PCR (Q-PCR) analysis.
Packaging assay
Lawns of yeast cells (strain JB970) harboring pECB9C or derivatives were scraped from SC–Ura plates and resuspended in 40 mL YNB containing 2% casamino acids and 1% raffinose. Cells were grown for ∼6 h at 30°C to A600 = 0.4–0.6. Galactose was added to 2% [w/v], and cultures were incubated at 22°C for an additional 40 h to A600 = 4–5. Cells were harvested by centrifugation at 1500g for 10 min and washed once with ice-cold DEPC-treated water. Eighty A600 units of cells were lysed using 0.5-mm glass beads in 1 mL buffer B/EDTA (15 mM KCl, 10 mM HEPES-KOH [pH 7.8], 5 mM EDTA) containing 0.2% Triton X-100, 3 mM dithiothreitol, 2 mM phenylmethylsulfonyl fluoride, 1× protease inhibitor cocktail (complete EDTA-free cocktail tablet, Roche), and 60 U of RNase inhibitor (RNasin; Promega). Glass beads were washed once with 1 mL buffer B/EDTA plus additives and pooled with cell lysates. Cellular extracts containing VLPs were centrifuged at 1000g at 4°C for 5 min, and supernatants were transferred to ice-cold tubes. Aliquots of 133 µL of cell extract were added to 15 µL 10× buffer M (100 mM Tris-HCl [pH 7.5], 100 mM MgCl2, 500 mM NaCl) and 2 µL benzonase (250 U/μL, Sigma E-8263) or enzyme dilution buffer (20 mM Tris-HCl [pH8.0], 2 mM MgCl2, 20 mM NaCl, 50% glycerol). Reactions were incubated at room temperature for 15 min and stopped by addition of 5 µL 0.5 M EDTA, 300 µL nucleic acid extraction buffer (50 mM Tris-HCl [pH7.5], 150 mM NaCl, 10 mM EDTA, 5% SDS), and 450 µL phenol:chloroform:isoamylalcohol (PCIA; 25:24:1). After vigorous vortexing, 5 µL of 2.15 nM synthetic Ty1 in vitro transcript synthesized by SP6 polymerase using pQH170 as a template was added to each reaction as an internal control to monitor RNA recovery through the extraction and precipitation steps. Nucleic acids were extracted twice with PCIA and once with chloroform, precipitated by isopropanol and ammonium acetate, and resuspended in 100 µL DEPC-treated water. DNA was removed by turbo DNase digestion. Purified RNA was then reverse transcribed into cDNA using a gene-specific primer QH217 (5′-TCCTTGCGTTTCAGCTTCC-3′) and SuperScript III reverse transcriptase for Q-PCR analysis.
Q-PCR
Q-PCR was performed with fast SYBR green reagent as previously described (Livak and Schmittgen 2001). Primer sequences used for DNA amplification are listed in Table 2. In the packaging assay, native Ty1 RNA levels post-benzonase treatment were calculated by the 2−ΔΔCT method, using synthetic Ty1 as the internal control for RNA recovery, and normalized to the amount of native Ty1 RNA in samples without benzonase treatment. For determining the RNA half-life, Ty1 RNA levels were calculated by the 2−ΔΔCT method, using ACT1 RNA as the endogenous control, and normalized to the amount of WT Ty1 RNA at the 0-h time point. The 0-h time point nucleic acid samples, without turbo DNase treatment, were used for cDNA analysis. Full-length Ty1s were expressed from pGAL-Ty1-mhis3AI plasmids. Transcribed Ty1 RNA underwent a splicing event, in which the artificial intron (AI) inside the HIS3 gene was spliced out. As a result, Ty1 cDNA reverse transcribed from mature RNA lacked the AI sequence. Ty1 cDNAs were then specifically amplified by intron-spanning HIS3 primers; cDNA levels were calculated by the 2−ΔΔCT method, using ACT1 gDNA as the endogenous control, and normalized to the amount of WT Ty1 cDNA. Either the minus or plus strand cDNA will give a signal in this assay.
TABLE 2.
List of Q-PCR primers used in this study
Endogenous reverse transcription
Cellular extracts containing VLPs were prepared as described for the packaging assay. A cell extract equivalent to 35 µg of total protein was used for each endogenous RT reaction. Reactions were initiated by addition of a RT cocktail (final concentrations: 100 µM each of dATP, dCTP, and dGTP, 0.1 µM dTTP, 0.3 µM α-32P-dTTP, 10 mM MgCl2, 50 mM Tris-HCl [pH 8.0], and 2% β-mecaptoethanol) and incubated 1 h at room temperature. Subsequently, reactions were incubated with proteinase K (5 µg) in a buffer of 50 mM Tris-HCl [pH 7.0], 10 mM EDTA, 100 mM NaCl, and 1% SDS for 30 min at 37°C. Samples were then extracted twice with PCIA, twice with chloroform, ethanol-precipitated, and fractionated on a 5% 7 M urea polyacrylamide gel to visualize −sssDNA and +sssDNA.
RNA 3-D structure prediction
Ty1 RNA 3-D structure was generated using RNAComposer (http://rnacomposer.cs.put.poznan.pl) and following secondary structure dot-bracket notation: (((((((.......((((((...))))))......(..(((((((.........(((((.((......(..((((.((((((............(((.(((...((((....((..(((((....))))))).))))......((..(((...)))..)).))).)))........))))))))))))).)))))..))))))).((((.((((...........(((......))).))))..)))))......[[[[[[[.)))))))....((((...((((..(((.........)))..))))....))))..]]]]]]]((((........(((((.......)))))))))
Eight hundred ninety-four distance constraints (Supplemental Table S2) were introduced for nt 256–262 and 319–321 to increase accuracy. Those constraints were subtracted from the element of 1YHQ (PDB ID) found using RNA FRABASE 2.0 (Popenda et al. 2010). The stereochemical quality of the resulting structure was validated using the PDB validation tool http://deposit.pdb.org/adit/.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joan Curcio, David Garfinkel, Eric Bolton, Robert Yarrington, and Candice Coombes for helpful discussions, Lixin Dai and Zheng Kuang for advice on Q-PCR, Mariusz Popenda for advice on 3-D structure prediction, and Marty Taylor for help with immunoblots. The work was supported in part by NIH grant GM36481 to J.D.B.
REFERENCES
- Asano K, Mizobuchi K 1998. Copy number control of IncIα plasmid ColIb-P9 by competition between pseudoknot formation and antisense RNA binding at a specific RNA site. EMBO J 17: 5201–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP 2002. A ribozyme that ligates RNA to protein. Proc Natl Acad Sci 99: 9154–9159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benard L, Mathy N, Grunberg-Manago M, Ehresmann B, Ehresmann C, Portier C 1998. Identification in a pseudoknot of a U·G motif essential for the regulation of the expression of ribosomal protein S15. Proc Natl Acad Sci 95: 2564–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry KE, Waghray S, Doudna JA 2010. The HCV IRES pseudoknot positions the initiation codon on the 40S ribosomal subunit. RNA 16: 1559–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A, Blackburn EH 1994. Architecture of telomerase RNA. EMBO J 13: 5721–5731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton E, Coombes C, Eby Y, Cardell M, Boeke J 2005. Identification and characterization of critical cis-acting sequences within the yeast Ty1 retrotransposon. RNA 11: 308–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I, Pennell S, Gilbert RJ 2007. Viral RNA pseudoknots: Versatile motifs in gene expression and replication. Nat Rev Microbiol 5: 598–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman K, Bystrom A, Boeke J 1992. Initiator methionine tRNA is essential for Ty1 transposition in yeast. Proc Natl Acad Sci 89: 3236–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Chamorro M, Lee SI, Shen LX, Hines JV, Tinoco I Jr, Varmus HE 1995. Structural and functional studies of retroviral RNA pseudoknots involved in ribosomal frameshifting: Nucleotides at the junction of the two stems are important for efficient ribosomal frameshifting. EMBO J 14: 842–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Blasco MA, Greider CW 2000. Secondary structure of vertebrate telomerase RNA. Cell 100: 503–514 [DOI] [PubMed] [Google Scholar]
- Chiaruttini C, Milet M, Springer M 1996. A long-range RNA-RNA interaction forms a pseudoknot required for translational control of the IF3-L35-L20 ribosomal protein operon in Escherichia coli. EMBO J 15: 4402–4413 [PMC free article] [PubMed] [Google Scholar]
- Collart MA, Oliviero S 2001. Preparation of yeast RNA. In Current protocols in molecular biology (ed. Ausubel FM et al. ), Unit13 12 Wiley and Sons, Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- Comolli LR, Smirnov I, Xu L, Blackburn EH, James TL 2002. A molecular switch underlies a human telomerase disease. Proc Natl Acad Sci 99: 16998–17003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristofari G, Ficheux D, Darlix JL 2000. The GAG-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J Biol Chem 275: 19210–19217 [DOI] [PubMed] [Google Scholar]
- Cristofari G, Bampi C, Wilhelm M, Wilhelm FX, Darlix JL 2002. A 5′–3′ long-range interaction in Ty1 RNA controls its reverse transcription and retrotransposition. EMBO J 21: 4368–4379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio MJ, Garfinkel DJ 1991. Single-step selection for Ty1 element retrotransposition. Proc Natl Acad Sci 88: 936–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio MJ, Garfinkel DJ 1992. Posttranslational control of Ty1 retrotransposition occurs at the level of protein processing. Mol Cell Biol 12: 2813–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deigan KE, Li TW, Mathews DH, Weeks KM 2009. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci 106: 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutko JA, Kenny AE, Gamache ER, Curcio MJ 2010. 5′ to 3′ mRNA decay factors colocalize with Ty1 gag and human APOBEC3G and promote Ty1 retrotransposition. J Virol 84: 5052–5066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre-D'Amare AR, Zhou K, Doudna JA 1998. Crystal structure of a hepatitis Δ virus ribozyme. Nature 395: 567–574 [DOI] [PubMed] [Google Scholar]
- Friant S, Heyman T, Wilhelm ML, Wilhelm FX 1996. Extended interactions between the primer tRNAi(Met) and genomic RNA of the yeast Ty1 retrotransposon. Nucleic Acids Res 24: 441–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friant S, Heyman T, Poch O, Wilhelm M, Wilhelm FX 1997. Sequence comparison of the Ty1 and Ty2 elements of the yeast genome supports the structural model of the tRNAiMet-Ty1 RNA reverse transcription initiation complex. Yeast 13: 639–645 [DOI] [PubMed] [Google Scholar]
- Friant S, Heyman T, Bystrom AS, Wilhelm M, Wilhelm FX 1998. Interactions between Ty1 retrotransposon RNA and the T and D regions of the tRNAiMet primer are required for initiation of reverse transcription in vivo. Mol Cell Biol 18: 799–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfinkel DJ, Boeke JD, Fink GR 1985. Ty element transposition: Reverse transcriptase and virus-like particles. Cell 42: 507–517 [DOI] [PubMed] [Google Scholar]
- Gesteland RF, Cech T, Atkins JF 2006. The RNA world: The nature of modern RNA suggests a prebiotic RNA world. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Giedroc DP, Cornish PV 2009. Frameshifting RNA pseudoknots: Structure and mechanism. Virus Res 139: 193–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedroc DP, Theimer CA, Nixon PL 2000. Structure, stability and function of RNA pseudoknots involved in stimulating ribosomal frameshifting. J Mol Biol 298: 167–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TM, Reihani SN, Oddershede LB, Sorensen MA 2007. Correlation between mechanical strength of messenger RNA pseudoknots and ribosomal frameshifting. Proc Natl Acad Sci 104: 5830–5835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houck-Loomis B, Durney MA, Salguero C, Shankar N, Nagle JM, Goff SP, D'Souza VM 2011. An equilibrium-dependent retroviral mRNA switch regulates translational recoding. Nature 480: 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanamori Y, Nakashima N 2001. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA 7: 266–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney J, Chapman K, Lauermann V, Voytas D, Astrom S, von Pawel-Rammingen U, Bystrom A, Boeke J 1995. Multiple molecular determinants for retrotransposition in a primer tRNA. Mol Cell Biol 15: 217–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler JF Jr, Haeusser DP, Dull A, Boeke JD, Keeney JB 2002. Ty1 defect in proteolysis at high temperature. J Virol 76: 4233–4240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BS, Lichtenstein CP, Faiola B, Rinckel LA, Wysock W, Curcio MJ, Garfinkel DJ 1998. Posttranslational inhibition of Ty1 retrotransposition by nucleotide excision repair/transcription factor TFIIH subunits Ssl2p and Rad3p. Genetics 148: 1743–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Levin HL 1998. Reverse transcription of a self-primed retrotransposon requires an RNA structure similar to the U5-IR stem-loop of retroviruses. Mol Cell Biol 18: 6859–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Low JT, Weeks KM 2010. SHAPE-directed RNA secondary structure prediction. Methods 52: 150–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Heng X, Summers MF 2011. Structural determinants and mechanism of HIV-1 genome packaging. J Mol Biol 410: 609–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monokian GM, Braiterman LT, Boeke JD 1994. In-frame linker insertion mutagenesis of yeast transposon Ty1: Mutations, transposition and dominance. Gene 139: 9–18 [DOI] [PubMed] [Google Scholar]
- Pleij CW 1990. Pseudoknots: A new motif in the RNA game. Trends Biochem Sci 15: 143–147 [DOI] [PubMed] [Google Scholar]
- Poot RA, van den Worm SH, Pleij CW, van Duin J 1998. Base complementarity in helix 2 of the central pseudoknot in 16S rRNA is essential for ribosome functioning. Nucleic Acids Res 26: 549–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popenda M, Szachniuk M, Blazewicz M, Wasik S, Burke EK, Blazewicz J, Adamiak RW 2010. RNA FRABASE 2.0: An advanced web-accessible database with the capacity to search the three-dimensional fragments within RNA structures. BMC Bioinformatics 11: 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popenda M, Szachniuk M, Antczak M, Purzycka KJ, Lukasiak P, Bartol N, Blazewicz J, Adamiak RW 2012. Automated 3D structure composition for large RNAs. Nucleic Acids Res 40: e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purzycka KJ, Legiewicz M, Matsuda E, Eizentstat LD, Lusvarghi S, Saha A, Le Grice SFJ, Garfinkel DJ 2013. Exploring Ty1 retrotransposon RNA structure within virus-like particles. Nucleic Acids Res 41: 463–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi T, Beattie TL, Olive JE, Collins RA 1996. A long-range pseudoknot is required for activity of the Neurospora VS ribozyme. EMBO J 15: 2820–2825 [PMC free article] [PubMed] [Google Scholar]
- Reuter JS, Mathews DH 2010. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinformatics 11: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld K, Van Poelgeest R, Pleij CW, Van Boom JH, Bosch L 1982. The tRNA-like structure at the 3′ terminus of turnip yellow mosaic virus RNA. Differences and similarities with canonical tRNA. Nucleic Acids Res 10: 1929–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18: 3091–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes DT, Banerjee M, Bowen B, Curcio MJ 2001. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159: 1449–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen LX, Tinoco I Jr 1995. The structure of an RNA pseudoknot that causes efficient frameshifting in mouse mammary tumor virus. J Mol Biol 247: 963–978 [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB 1986. Methods in yeast genetics: A laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- ten Dam E, van Belkum A, Pleij K 1991. A conserved pseudoknot in telomerase RNA. Nucleic Acids Res 19: 6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theis C, Reeder J, Giegerich R 2008. KnotInFrame: Prediction of −1 ribosomal frameshift events. Nucleic Acids Res 36: 6013–6020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC 2008. ShapeFinder: A software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA 14: 1979–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks KM, Mauger DM 2011. Exploring RNA structural codes with SHAPE chemistry. Acc Chem Res 44: 1280–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E, Jaeger L 1992. RNA pseudoknots. Curr Opin Struct Biol 2: 327–333 [Google Scholar]
- Wilhelm M, Wilhelm F-X, Keith G, Agoutin B, Heyman T 1994. Yeast Ty1 retrotransposon: The minus-strand primer binding site and a cis-acting domain of the Ty1 RNA are both important for packaging of primer tRNA inside virus-like particles. Nucleic Acids Res 22: 4560–4565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Boutabout M, Wilhelm FX 2000. Expression of an active form of recombinant Ty1 reverse transcriptase in Escherichia coli: A fusion protein containing the C-terminal region of the Ty1 integrase linked to the reverse transcriptase-RNase H domain exhibits polymerase and RNase H activities. Biochem J 348 (Pt 2): 337–342 [PMC free article] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 1: 1610–1616 [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM 2008. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol 6: e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Boeke JD 1990. Localization of sequences required in cis for yeast Ty1 element transposition near the long terminal repeats: Analysis of mini-Ty1 elements. Mol Cell Biol 10: 2695–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarrington RM 2009. “Synthetic and biochemical studies of retrotransposon Ty1.” PhD thesis, Johns Hopkins University School of Medicine, Baltimore, MD [Google Scholar]
- Yarrington RM, Richardson SM, Lisa Huang CR, Boeke JD 2012. Novel transcript truncating function of Rap1p revealed by synthetic codon-optimized Ty1 retrotransposon. Genetics 190: 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.