RNA decapping is an important contributor to gene expression and is a critical determinant of mRNA decay. The recent demonstration that mammalian cells harbor at least two distinct decapping enzymes that preferentially modulate a subset of mRNAs raises the intriguing possibility of whether additional decapping enzymes exist. Because both known decapping proteins, Dcp2 and Nudt16, are members of the Nudix hydrolase family, the authors set out to determine whether other members of this family of proteins also contain intrinsic RNA decapping activity. Here they demonstrate that six additional mouse Nudix proteins—Nudt2, Nudt3, Nudt12, Nudt15, Nudt17, and Nudt19—have varying degrees of decapping activity in vitro on both monomethylated and unmethylated capped RNAs. These findings demonstrate that multiple Nudix family hydrolases may function in mRNA decapping and mRNA stability.
Keywords: decapping proteins, Nudix proteins, mRNA stability
Abstract
RNA decapping is an important contributor to gene expression and is a critical determinant of mRNA decay. The recent demonstration that mammalian cells harbor at least two distinct decapping enzymes that preferentially modulate a subset of mRNAs raises the intriguing possibility of whether additional decapping enzymes exist. Because both known decapping proteins, Dcp2 and Nudt16, are members of the Nudix hydrolase family, we set out to determine whether other members of this family of proteins also contain intrinsic RNA decapping activity. Here we demonstrate that six additional mouse Nudix proteins—Nudt2, Nudt3, Nudt12, Nudt15, Nudt17, and Nudt19—have varying degrees of decapping activity in vitro on both monomethylated and unmethylated capped RNAs. The decapping products from Nudt17 and Nudt19 were analogous to Dcp2 and predominantly generated m7GDP, while cleavage by Nudt2, Nudt3, Nudt12, and Nudt15 was more pleiotropic and generated both m7GMP and m7GDP. Interestingly, all six Nudix proteins as well as both Dcp2 and Nudt16 could hydrolyze the cap of an unmethylated capped RNA, indicating that decapping enzymes may be less constrained for the presence of the methyl moiety. Investigation of Saccharomyces cerevisiae Nudix proteins revealed that the yeast homolog of Nudt3, Ddp1p, also possesses decapping activity in vitro. Moreover, the bacterial Nudix pyrophosphohydrolase RppH displayed RNA decapping activity and released m7GDP product comparable to Dcp2, indicating that decapping is an evolutionarily conserved activity that preceded mammalian cap formation. These findings demonstrate that multiple Nudix family hydrolases may function in mRNA decapping and mRNA stability.
INTRODUCTION
Eukaryotic mRNAs are modified with a 5′-monomethyl guanosine cap structure and a 3′-poly(A) tail (Shatkin and Manley 2000), which are both important for mRNA translation and stability (Furuichi et al. 1977; Shimotohno et al. 1977; Shatkin 1985; Hentze 1997; Sachs et al. 1997; Gingras et al. 1999; Mangus et al. 2003). Bulk mRNA decay usually initiates with the removal of the 3′-poly(A) tail (Decker and Parker 1993), followed by decay of the RNA body from either the 5′ or 3′ ends. In the 3′ decay pathway, a multi-subunit exosome complex degrades the mRNA in a 3′-to-5′ direction (Mitchell et al. 1997; Anderson and Parker 1998), and the resulting cap dinucleotide is subsequently hydrolyzed by the scavenger decapping enzyme, DcpS (Wang and Kiledjian 2001; Liu et al. 2002). In the 5′ decay pathway, the monomethyl guanosine (m7G) mRNA cap is cleaved to generate a 5′-monophosphate RNA (Dunckley and Parker 1999; Lykke-Andersen 2002; van Dijk et al. 2002; Wang et al. 2002; Song et al. 2010) that is subsequently degraded by the 5′-to-3′ exoribonuclease Xrn1 (Decker and Parker 1993; Hsu and Stevens 1993).
Removal of 5′-cap structure is carried out by at least two mRNA decapping enzymes, Dcp2 (Dunckley and Parker 1999; Lykke-Andersen 2002; van Dijk et al. 2002; Wang et al. 2002) and Nudt16 (Song et al. 2010). Dcp2 activity is highly regulated, and numerous proteins have been identified to colocalize with Dcp2 in cytoplasmic foci termed P-bodies (Franks and Lykke-Andersen 2008). In yeast, Dcp1p forms a heterodimer with Dcp2p and enhances its decapping activity (Steiger et al. 2003; She et al. 2004). Its activity is also stimulated by various decapping enhancers, including Dcp1p, Edc1p, Edc2p, Edc3p, Dhh1p, and the Lsm1–7 protein complex (Coller and Parker 2004). Edc4 (also known as Hedls or Ge-1) was identified as a mammalian positive effector of Dcp2 (Fenger-Gron et al. 2005). Conversely, Dcp2 decapping activity is also negatively regulated. In both yeast and mammals, the cap-binding protein eIF4E inhibits Dcp2 decapping (Schwartz and Parker 1999, 2000; Ramirez et al. 2002; Khanna and Kiledjian 2004), possibly by preventing access of the Dcp2 to the cap (Grudzien et al. 2006) as well as the cytoplasmic poly(A)-binding protein PABPC1 (Khanna and Kiledjian 2004) and the 5′-capped RNA-binding protein VCX-A (Jiao et al. 2006, 2009).
In contrast to Dcp2, Nudt16 is less extensively studied. Nudt16 was initially identified in Xenopus as a nucleolar decapping enzyme that specifically binds the U8 snoRNA (Tomasevic and Peculis 1999; Ghosh et al. 2004). It was subsequently shown to be a cytoplasmic protein in mammalian cells and involved in mRNA decapping (Song et al. 2010). A comparison of the two decapping proteins revealed that they are both differentially used in specified mRNA decay processes: NMD preferentially uses Dcp2 rather than Nudt16; Dcp2 and Nudt16 are redundant in microRNA-mediated silencing; and Dcp2 and Nudt16 are differentially used for different ARE-mRNA decay (Li et al. 2011). Surprisingly, both Dcp2 and Nudt16 regulate the stability of only a subset of mRNA transcripts (Song et al. 2010; Li et al. 2011), indicating that there may be additional heretofore uncharacterized decapping enzyme(s).
Dcp2 and Nudt16 are members of the Nudix hydrolase superfamily that catalyzes the hydrolysis of a wide range of small nucleotide substrates composed of a nucleoside diphosphate linked to another moiety X (Nudix) (Bessman et al. 1996). The Nudix family is evolutionarily conserved and widely present in viruses, bacteria, archaea, and eukaryotes (McLennan 2006). The catalytic activity depends on a conserved Nudix motif consisting of the consensus sequence GX5EX7REUXEEXGU (where U represents a hydrophobic residue, and X represents any amino acid), which forms part of the versatile catalytic site for diphosphate hydrolysis (Bessman et al. 1996). The two glutamic acid residues in the core of the motif, REX2EE, are important for metal ion coordination (Abeygunawardana et al. 1995; Gabelli et al. 2001) and critical for pyrophosphatase activity of Nudix proteins (Lin et al. 1996; Safrany et al. 1998). To date, 22 Nudix hydrolase genes and at least five pseudogenes have been identified in mammals. Dcp2 and Nudt16 are the only known Nudix proteins with mRNA decapping activity. In this study, we analyzed the decapping capacity of all the Nudix proteins and identified six additional Nudix proteins with decapping activity in vitro. In addition, we demonstrate that the yeast Ddp1p Nudix protein and the bacterial RppH Nudix protein also possess decapping activity, indicating that decapping is an evolutionarily conserved activity.
RESULTS
Identification of mammalian Nudix proteins possessing decapping activity
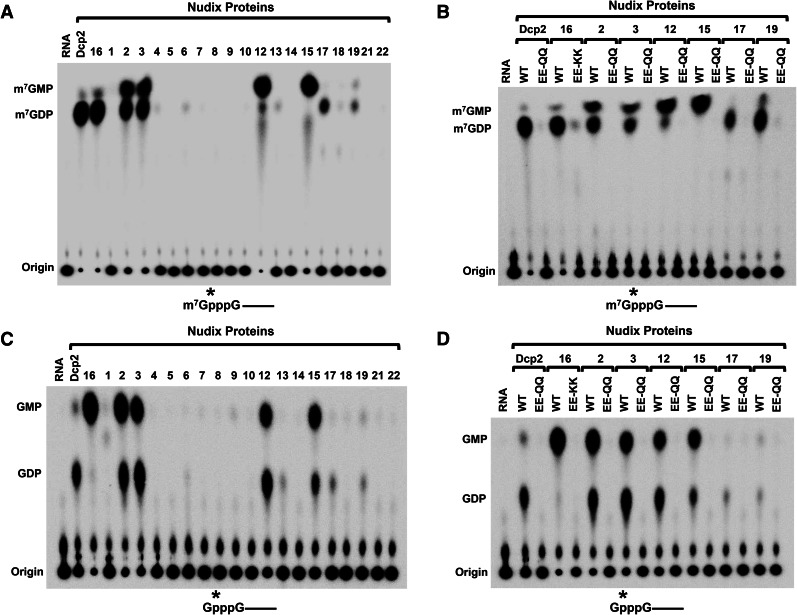
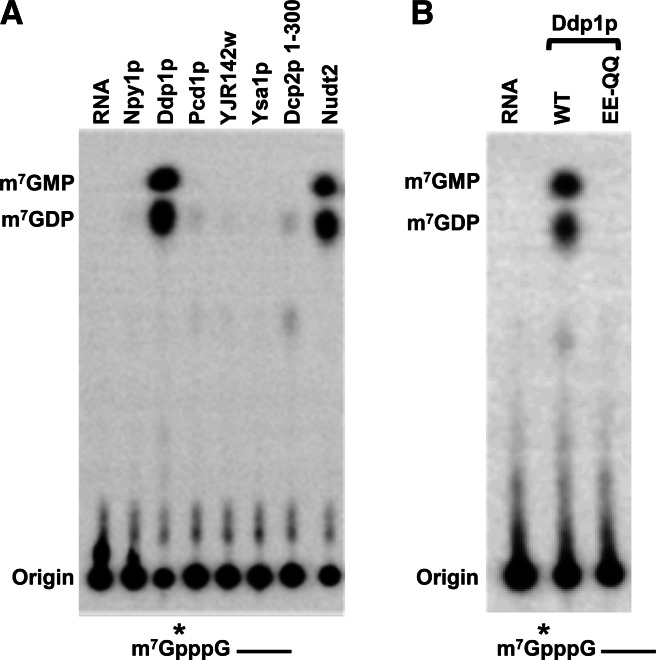
Mammalian cells contain 22 genes encoding proteins of the Nudix superfamily (McLennan 2006), two of which encode the Dcp2 and Nudt16 mRNA decapping enzymes. To assess whether any of the remaining Nudix proteins also possess decapping activity, cDNAs encoding the mouse Nudix family proteins were isolated and used to express His-tagged recombinant proteins purified from Escherichia coli. The Nudt10 and Nudt11 mRNAs encode identical proteins and are represented by Nudt10 below. The purified tagged-recombinant Nudix proteins were each individually tested for their capacity to hydrolyze cap-labeled RNAs. Surprisingly, decapping activity comparable to that of Dcp2 and Nudt16 could be detected with four of the proteins—Nudt2, Nudt3, Nudt12, and Nudt15—on monomethylated capped RNA substrate (Fig. 1A). Decapping activity was also detected with Nudt17 and Nudt19, albeit at lower efficiencies. Analysis of the decapping products revealed Nudt2 and Nudt3 generated both m7GMP and m7GDP, while the product of Nudt12 and Nudt15 was primarily m7GMP, and Nudt17 and Nudt19 released m7GDP (Fig. 1A). The decapping product of the latter two is analogous to the predominant product released by Dcp2 and Nudt16. The detected decapping activities with all six proteins were a function of the respective Nudix proteins as demonstrated by substitution mutations within the Nudix motif. Replacing the two glutamic acid residues within the Nudix motif necessary for metal ion coordination (Abeygunawardana et al. 1995; Gabelli et al. 2001) to two glutamines abolished their decapping activity (Fig. 1B), demonstrating that all six Nudix proteins contain intrinsic decapping activity.
FIGURE 1.
Identification of new Nudix proteins possessing RNA-decapping activity in mammals. In vitro decapping assays were carried out as described in Materials and Methods to screen all recombinant mouse Nudix proteins. Human Dcp2 and Nudt16 proteins were used as controls. Monomethylated (A) or unmethylated (C) capped RNAs, 32P-labeled at the γ-phosphate within the cap and containing a stretch of 16 guanosines at the 3′ end to minimize 3′-end decay, were used as substrates and decapping products resolved by TLC. Decapping activity for proteins containing substitutions of the two highly conserved glutamic acids within the Nudix motif are shown in B and D. The glutamic acids within the Nudt16 were substituted to lysines (EE-KK) and glutamines (EE-QQ) in the remaining Nudix proteins. The capped RNA substrates are shown schematically under each panel, where the line denotes the RNA and the asterisk represents the position of the 32P. Migration of the markers is shown on the left of each panel.
Having recently demonstrated that a subclass of decapping enzymes preferentially functions on incompletely capped mRNAs lacking the N7 methyl moiety (Jiao et al. 2010; Chang et al. 2012), we tested the capacity of the Nudix proteins to decap unmethylated capped RNA. Consistent with a previous report, Nudt16 hydrolyzed the GpppRNA unmethylated capped RNA and generated GMP decapping product (Fig. 1C; Ghosh et al. 2004). Unexpectedly, Dcp2 could also hydrolyze GpppRNA and released GDP. The apparent discrepancy between our previous data demonstrating Dcp2 specificity on the m7G-cap (Wang et al. 2002) and the current data appears to be due to the cation used. Minimal decapping activity is detected when Mg2+ was used and more robust activity when Mn2+ was used as the cation (Supplemental Fig. S1). Analysis of the remaining Nudix proteins revealed that all six proteins that could hydrolyze methylated capped RNA were also competent to decap unmethylated capped RNA. Nudt2, Nudt3, Nudt12, and Nudt15 all efficiently decapped unmethylated capped RNA, generating both GDP and GMP products (Fig. 1C). A low level of activity releasing GDP was also detected with Nudt17 and Nudt19. The detected decapping activities on unmethylated capped RNAs were an intrinsic property of the respective proteins since substitution of two critical glutamic acids within the Nudix motif ablated decapping activity in all six proteins (Fig. 1D). Collectively, our data demonstrate that in addition to Dcp2 and Nudt16, Nudt 2, 3, 12, 15, 17, and 19 also possess RNA decapping activity in vitro. However, the possibility remains that the use of different assay conditions may reveal additional Nudt proteins with decapping activity.
Cap cleavage specificity of the Nudix proteins
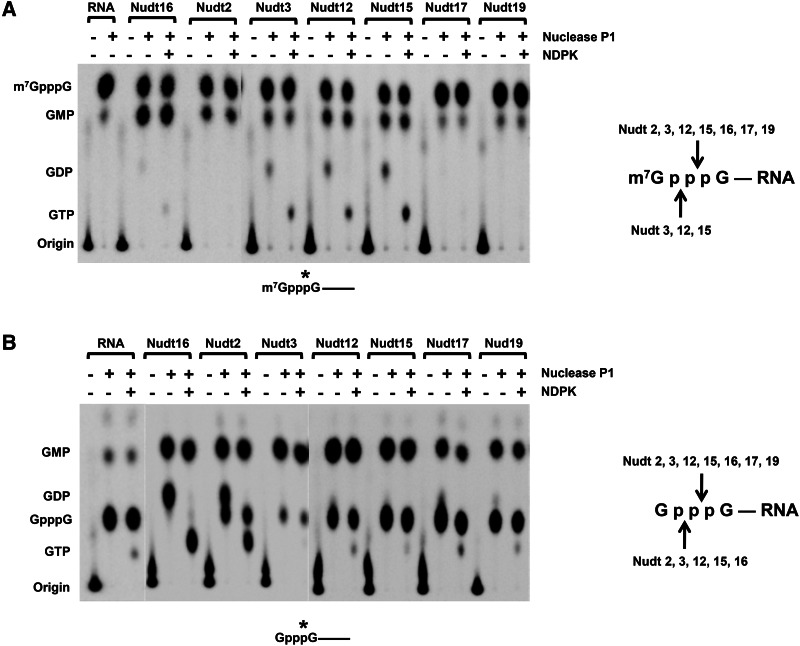
The above analysis was carried out with capped RNAs specifically 32P-labeled at the γ-phosphate of the cap (m7G32pppG-RNA), where decapping between the β-γ-phosphates would generate m7GMP while hydrolysis of the triphosphate between the α-β-phosphates would generate m7GDP, which could potentially be further processed to m7GMP by a phosphatase activity removing the terminal phosphate. To determine the cleavage specificity of the Nudix proteins, we expanded the above analysis with capped RNAs labeled at the α-phosphate of the cap triphosphate linkage (m7Gpp32pG-RNA) (see Materials and Methods) and followed the fate of the first nucleotide of the RNA rather than the m7G cap. The α-phosphate-labeled monomethylated capped RNA was subjected to decapping by the individual Nudix proteins followed by nuclease P1 treatment. Nuclease P1 hydrolyzes RNA into nucleotides containing a 3′-hydroxyl but is unable to cleave the pyrophosphate linkage of the cap (Cruz-Reyes et al. 1998) and would generate a cap structure with the α-phosphate labeled. If Nudix proteins can hydrolyze the cap pyrophosphate at both γ- and β-phosphates, both GDP (p32pG) and GMP (32pG) would be detected following nuclease P1 digestion. As shown in Figure 2A, both GMP and GDP are detected following nuclease P1 digestion of Nudt3, Nudt12, and Nudt15 decapping products. The identity of the GDP was further confirmed by treatment of the decapping and nuclease P1-digested products with NDP kinase (Fig. 2A), which converts GDP to GTP (Heidbuchel et al. 1992).
FIGURE 2.
Cleavage specificity of the Nudix proteins on capped RNA. Monomethylated (A) and unmethylated (B) α-phosphate-labeled capped RNAs were incubated with the indicated Nudix proteins for 1 h at 37°C. Reaction products were subjected to nuclease P1 and NDPK treatment as indicated before resolution by TLC. NDPK phosphorylates nucleoside diphosphates to nucleoside triphosphates. Labeling is as described in the legend to Figure 1. A summary of the likely cleavage site(s) within the cap triphosphate for each Nudix protein is summarized on the right.
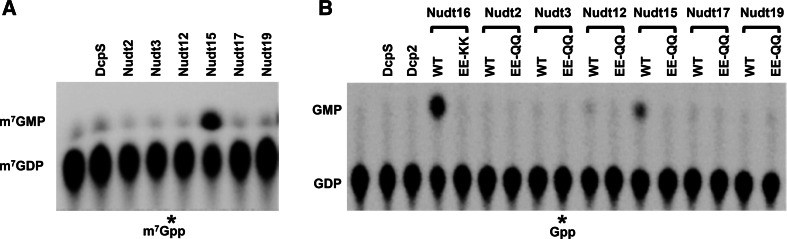
The above data suggest that Nudt3, Nudt12, and Nudt15 can decap between the α-β- and β-γ-phosphates. For Nudt17 and 19, only GMP can be detected, indicating that they hydrolyze the cap only between the α- and β-phosphates. As expected, Nudt16 also predominantly cleaves the cap between α- and β-phosphates and released 5′-monophosphorylated RNA. However, trace amounts of GDP can also be detected, suggesting a low level of activity between the β- and γ-phosphates. To assess whether any of the proteins possess nucleoside diphosphatase activity that might function on m7GDP decapping product, their activity on m7GDP was tested. Interestingly, Nudt15 contained nucleoside diphosphatase activity and could hydrolyze m7Gpp to m7Gp (Fig. 3), which may account for why only m7GMP was detected as the decapping product for Nudt15 in Figure 1A. Moreover, we were unable to detect a nucleoside diphosphatase activity for DcpS, which has been reported to convert m7Gpp to m7Gp (van Dijk et al. 2003) and is consistent with a more recent report that demonstrated although DcpS avidly binds m7Gpp, it does not hydrolyze this substrate (Wypijewska et al. 2012).
FIGURE 3.
m7GDP and GDP hydrolase activity of the Nudix proteins. Hydrolysis of m7GDP (A) and GDP (B) by the indicated Nudix proteins is shown. Labeling is as described in the legend to Figure 1.
The cleavage specificity of the Nudix proteins on unmethylated capped RNA was determined by a similar approach. As shown in Figure 2B, after Nudt2 decapping reaction followed by nuclease P1 digestion, both GMP and GDP can be detected, indicating that Nudt2 can cleave the unmethylated RNA following either the α- or β-phosphates. The remaining Nudix proteins—Nudt12, Nudt15, Nudt17, and Nudt19—all specifically generate GMP, indicating that they preferentially hydrolyze the cap between the α- and β-phosphates (Fig. 2B). However, considering that Nudt15 also contains GDPase activity (Fig. 3B), the possibility remains that the prominent GMP product detected from the Nudt15 decapping of unmethylated capped RNA in Figure 1B may be a result of an initial GDP converted to GMP. The cleavage sites within the pyrophosphate linkage of the cap for each Nudix protein are summarized in the schematics of Figure 2. Collectively, our data indicate that Nudt2, Nudt3, Nudt12, Nudt15, Nudt16, Nudt17, and Nudt19 cleave both methylated and unmethylated capped RNAs between the α- and β-phosphates to generate m7GDP and GDP, respectively. Cleavage within the β-γ-phosphates was detected with Nudt3, Nudt12, and Nudt15 with methyl-capped RNA, and Nudt2, Nudt3, Nudt12, and Nudt15 and Nudt16 with unmethylated capped RNA.
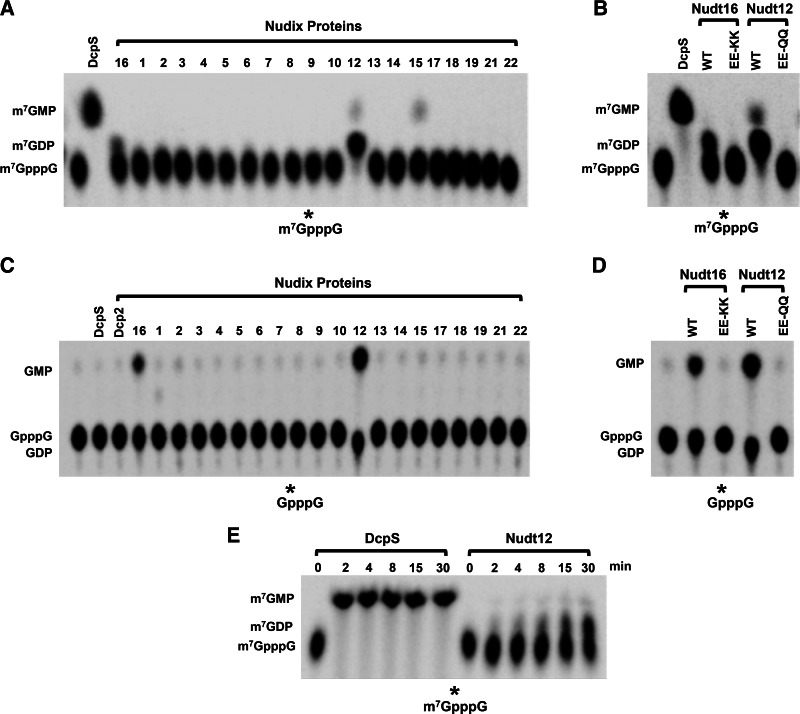
Decapping activity of Nudix proteins on cap structures
To date, the scavenger decapping enzyme DcpS is the only known protein that can specifically hydrolyze the m7GpppN cap dinucleotide (Liu et al. 2002). To assess whether any of the Nudix proteins can hydrolyze cap structure, we tested their ability to decap 32P-labeled cap structure. As shown in Figure 4A, Nudt12 was capable of hydrolyzing the cap dinucleotide and predominantly generated m7GDP product under these assay conditions and further confirmed it with NDPK treatment (Supplemental Fig. S2A). Low levels of activity could also be detected with Nudt16 generating m7GDP as well (Fig. 4A; Supplemental Fig. S2A). The glutamic acid substitution mutants within both, Nudt12 and Nudt16, disrupted the observed activities (Fig. 4B). Similarly, both Nudt12 and Nudt16 demonstrated pyrophosphohydrolase activity on unmethylated cap structure (Fig. 4C,D). As expected, decapping products of the symmetrical unmethylated GpppG cap structure were GMP and GDP as confirmed with NDPK treatment (Supplemental Fig. S2B). We conclude that Nudt12 and Nudt16 can hydrolyze methylated and unmethylated cap structures with Nudt16 displaying less robust activity (Fig. 4; Supplemental Fig. S2). In contrast, DcpS exclusively uses methylated cap structure as the substrate to generate m7GMP product (Fig. 4A,C; Liu et al. 2002).
FIGURE 4.
Cap structure decapping by Nudix proteins. The indicated Nudix proteins were incubated with 32P-labeled m7GpppG (A) and GpppG (C) cap structures for 30 min at 37°C, and the reaction products were resolved by TLC. Recombinant human DcpS protein was used as a control. The activity of the proteins with the indicated substitution mutations in the metal binding residues is denoted for m7GpppG (B) and GpppG (D). (E) A decapping time course with 50 nM DcpS or Nudt12 on m7GpppG. Labeling is as described in the legend to Figure 1.
The surprising finding that Nudt12 also appeared to possess robust activity on methylated cap structure prompted us to compare the decapping activities of DcpS and Nudt12 directly on this substrate. A time-course titration was carried out to assess the level of m7GpppG decapping activity of Nudt12 relative to the scavenger decapping activity of DcpS. As shown in Figure 4E, 50% of the m7GpppG cap structure was decapped by Nudt12 at 30 min under these assay conditions. In contrast, even at the first 2-min time point, DcpS already hydrolyzed 100% of the m7GpppG cap structure substrate. Although Nudt12 possesses scavenger decapping activity on m7GpppG cap structure, its level of activity, at least in vitro, is not comparable to that observed with DcpS.
Identification of potential decapping enzyme in yeast
Our recent demonstration that Nudt16 is a second decapping enzyme in mammals (Song et al. 2010) and the identification of six additional Nudix proteins with decapping activity in this study raised the intriguing possibility of whether Nudix proteins in other organisms also contain decapping activity. Search of the yeast Saccharomyces cerevisiae database revealed the presence of six Nudix proteins, including the Dcp2 decapping enzyme. To assess whether any of the remaining five yeast Nudix proteins (Npy1p, Ddp1p, Pcd1p, Ysa1p, and YJR142w) contain decapping activity, we undertook a similar analysis as above and generated His-tagged recombinant proteins for each and tested their decapping potential in vitro. Significantly, decapping activity was detected from one of the proteins, Ddp1p (Fig. 5A), and this activity was ablated by substitution mutations within the Nudix motif (Fig. 5B). Of note, the detected activity from Ddp1p is more robust than that observed with the first 300 amino acids of the Dcp2 protein, which contains the catalytically active Nudix fold. Interestingly, Ddp1, Npy1, and Pcd1p proteins appear to be homologs of Nudt3, Nudt12, and Nudt15, respectively (Supplemental Fig. S3), yet decapping activity was only detected from Ddp1p, suggesting that Nudt3 and Ddp1p may be evolutionarily conserved decapping enzymes.
FIGURE 5.
Yeast Ddp1p contains in vitro decapping activity. (A) Products of decapping assays with the indicated yeast Nudix proteins (50 nM) resolved by TLC are shown. The N-terminal 300 amino acids of yeast Dcp2p containing the catalytically active Nudix motif (Schwartz et al. 2003; Steiger et al. 2003) (Dcp2p 1–300) and recombinant mouse Nudt2 proteins were used as positive controls. Positions of decapping products are indicated at left. (B) Activity of the Ddp1p active-site EE-QQ mutant. Labeling is as described in the legend to Figure 1.
Decapping activity is not restricted to eukaryotic Nudix proteins
Having demonstrated a propensity for Nudix proteins beyond just Dcp2 and Nudt16 to decap mRNA, we asked whether this is a recent evolutionary acquisition in eukaryotes. Importantly, we had reported the presence of a bacterial decapping activity that copurified with tagged fusion proteins expressed in E. coli (Liu et al. 2008), suggesting that prokaryotic proteins may also possess decapping activity. Thirteen Nudix hydrolase genes have been reported in E. coli (McLennan 2006). To determine whether a bacterial protein may also have decapping activity, we tested the RppH Nudix protein. RppH is a pyrophosphohydrolase that hydrolyzes the 5′-end triphosphate linkage of prokaryotic RNAs to remove the terminal diphosphate, leaving a 5′-end monophosphate RNA (Deana et al. 2008) analogous to the RNA product generated by both Dcp2 and Nudt16 decapping enzymes. As shown in Figure 6, RppH decapped both methylated and unmethylated capped RNAs to generate m7GDP and GDP, respectively, and was dependent on the Nudix motif (Fig. 6). Importantly RppH demonstrated a requirement for the RNA body in conjunction with the cap and failed to hydrolyze cap structure in the absence of RNA (Fig. 6), analogous to the decapping properties of Dcp2 (Wang et al. 2002). These data demonstrate that RppH possesses the capacity to hydrolyze the 5′ cap of an mRNA and suggest that the decapping activity of Nudix proteins evolutionarily proceeded eukaryotic capping/decapping.
FIGURE 6.
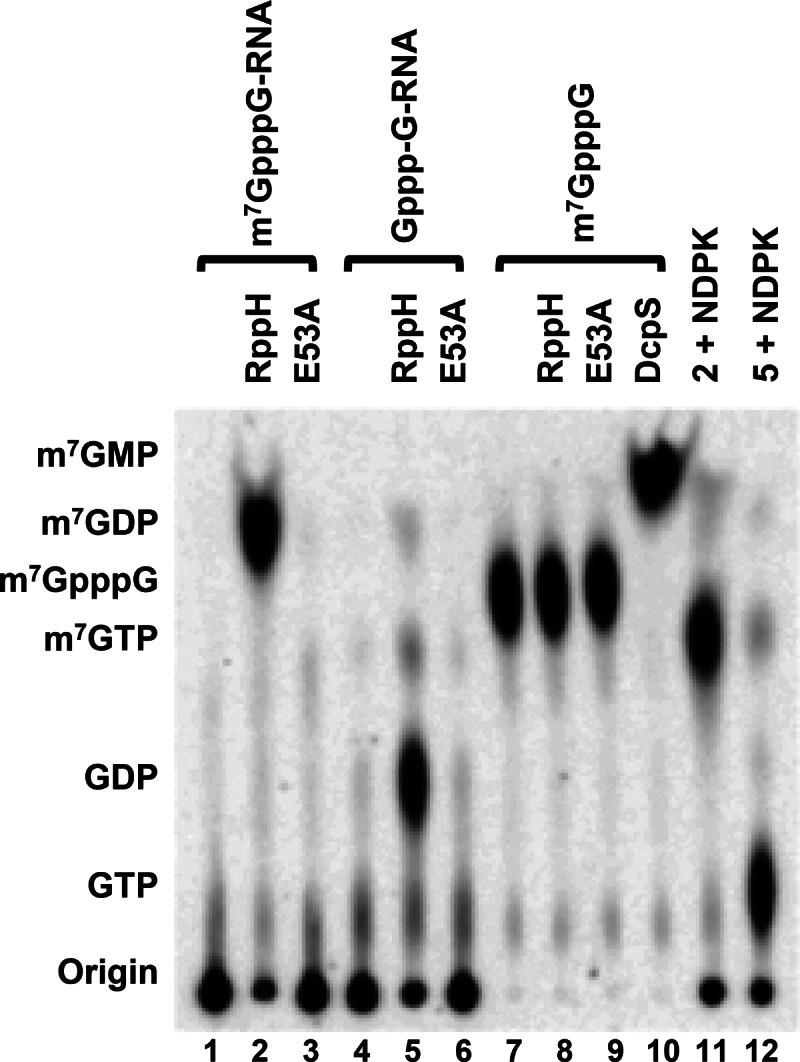
E. coli RppH protein has decapping activity on both monomethylated and unmethylated capped RNA but not on cap structure in vitro. Wild-type and the E53A active-site mutant bacterial RppH protein (50 nM) were used in decapping assays with methyl-capped RNA, unmethylated capped RNA, and methyl-cap structure as denoted at the top. Lanes 11 and 12 represent the products of lanes 2 and 5, respectively, treated with NDPK.
DISCUSSION
Our recent demonstration that Nudt16 is also a mammalian decapping protein and that both Dcp2 and Nudt16 each preferentially functions on a subset of mRNAs and pathways (Song et al. 2010; Li et al. 2011, 2012) raised the possibility that additional decapping enzymes are functional in mammalian cells. To evaluate whether proteins in addition to the two known Nudix decapping proteins possess decapping activity, we tested all known mouse Nudix proteins in vitro and identified six additional potential mammalian decapping enzymes—Nudt2, Nudt3, Nudt12, Nudt15, Nudt17, and Nudt19 (Fig. 1)—and the yeast Nudt3 homolog Ddp1p as a potential yeast decapping enzyme (Fig. 5). Similar to Dcp2 and Nudt16, Nudt17 and Nudt19 decap monomethylated capped RNA and release m7GDP, while Nudt12 and Nudt15 predominantly release m7GMP, and Nudt2 and Nudt3 are more pleiotropic and generate both m7GMP and m7GDP (Fig. 1A). Moreover, all the Nudix proteins that can decap m7G capped RNA, including Dcp2 and Nudt16, were also capable of hydrolyzing unmethylated capped RNA to various degrees (Fig. 1C). Interestingly, Nudt16 and Nudt12 were also capable of functioning on cap structure, suggesting that proteins other than DcpS may also hydrolyze cap structure generated by 3′-end decay; however, their relative levels of activity were substantially less than DcpS, at least in vitro (Fig. 4). Our data suggest that decapping is more complex than initially envisioned and may involve additional decapping enzymes other than Dcp2 and Nudt16. Lastly, we demonstrate that decapping is an evolutionarily conserved activity that preceded mammalian cap formation and is an intrinsic property of the E. coli RppH protein despite the lack of capped RNA in prokaryotes.
Many of the Nudix proteins had been extensively characterized in their capacity to hydrolyze small molecules, such as (d)NTPs (both canonical and oxidized derivatives), nucleotide sugars and alcohols, dinucleoside polyphosphates (NpnN), and dinucleotide coenzymes, as well as non-nucleotide substrates such as diphosphoinositol polyphosphates (DIPs), 5-phosphoribosyl 1-pyrophosphate (PRPP), thiamine pyrophosphate (TPP), and dihydroneopterin triphosphate (DHNTP) (McLennan 2006). Although a subset of mammalian Nudix proteins was shown to lack decapping activity (Nudt1, Nudt4, Nudt5, Nudt7) (Song et al. 2010), the more extensive analysis of all known mammalian Nudix proteins in this study revealed six additional mammalian Nudix proteins with decapping activity. Interestingly, similar to the canonical decapping enzymes that hydrolyze both methylated and unmethylated capped RNAs, all six proteins also function on both capped RNA substrates. Of the six proteins, Nudt17 and Nudt19 have modest activity, while the decapping activity of Nudt2, Nudt3, Nudt12, and Nudt15 is comparable to that of Dcp2 and Nudt16.
Our studies revealed that diverse decapping products were generated by the decapping-competent Nudix proteins. By using capped RNA labeled at either the γ- or α-phosphates within the cap triphosphate linkage, the cleavage specificity of the Nudix proteins was determined (schematically summarized in Fig. 2). Nudt2, Nudt3, Nudt12, and Nudt15 hydrolyze monomethylated capped RNA after both the α- and β-phosphates generating m7GMP + ppRNA and m7GDP + pRNA (Fig. 1A,B), while Nudt2, Nudt3, Nudt 12, Nudt15, and Nudt16 function similarly on unmethylated capped RNA (Figs. 1B, 2B). Demonstration of several Nudix proteins capable of potentially generating RNAs with a 5′ monophosphate or diphosphate raises an interesting question regarding the functional consequence of mRNAs with a 5′ diphosphate since these would not serve as a substrate for currently known 5′–3′ exoribonucleases. However, the recent demonstration that a cytoplasmic 5′-monophosphorylated mRNA can be recapped through a 5′-diphosphorylated intermediate (Otsuka et al. 2009; Mukherjee et al. 2012) indicates that 5′-diphosphorylated mRNAs may be more efficient substrates for recapping and entry of the mRNA into the translational pool. Whether decapping enzymes that can generate 5′-end diphosphorylated RNAs are involved in facilitating the cytoplasmic recapping pathway remains to be determined.
Nudt2 was reported to asymmetrically hydrolyze ApnA (n ≥ 4) and lead to the production of ATP as one of the products. Ap4A, a side product of protein synthesis catalyzed by amino-acyl-tRNA synthetases (Sillero and Sillero 2000), is assumed to be the principal substrate of Nudt2 in vivo (McLennan 2000). Although the biological significance of Ap4A is not fully understood, it has been proposed to control DNA replication and DNA repair, cell proliferation and apoptosis, and regulation of ATP-sensitive K+ channels (McLennan 2006). Nudt3 was identified as a diphosphoinositol-polyphosphate phosphohydrolase (DIPPase), which can hydrolyze both diphosphoinositol-polyphosphate and ApnA (Safrany et al. 1998). Expression of mouse Nudt3 attenuates signaling through the ERK1/2 pathway even if an active-site mutant is used (Chu et al. 2004). Both Nudt2 and Nudt3 can be detected in the cytoplasm (McLennan 2006), suggesting that they might be involved in cytoplasmic mRNA decay in cells. Human Nudt12 can hydrolyze NAD(P)H to NMNH and AMP (2′,5′-ADP) and is also active to a lesser extent toward NAD(P)+, ADP-ribose, and diadenosine triphosphate (Ap3A) (McLennan 2006). GFP-Nudt12 fusion protein localizes to peroxisomes and larger, unidentified cytoplasmic structures of unknown function (Abdelraheim et al. 2003). Peroxisomal localization is mediated through a C-terminal PNL tripeptide, which functions as a novel type 1 peroxisomal targeting signal (Abdelraheim et al. 2003). Nudt15 can hydrolyze 8-oxo-dGTP or dGTP to generate 8-oxo-dGMP or dGMP (Cai et al. 2003; Ishibashi et al. 2005). 8-Oxo-dGTP can be incorporated into DNA during DNA replication, which causes G:C to A:T or T:A to G:C transversion mutations. Expression of mouse Nudt15 in E. coli mutT− cells significantly reduced the elevated level of spontaneous mutation frequency, suggesting that it may function to minimize deleterious nucleotide incorporation into DNA (Cai et al. 2003). Nudt15 is also implicated in DNA replication and repair by directly interacting with and stabilizing the PCNA protein (Yu et al. 2009). Intriguingly, Nudt1 and Nudt5 are also 8-OH-dGTPases; however, they do not have RNA decapping activity (Fig. 1), demonstrating that decapping is not a general feature of 8-OH-dGTPases. Mouse Nudt19 (RP2p) was identified as a CoA diphosphatase (Ofman et al. 2006). Similar to Nudt12, Nudt19 also contains a C-terminus type 1 peroxisomal targeting signal and may also localize to peroxisomes. To date, Nudt17 is an uncharacterized protein with no known function. Whether any of these Nudix proteins are bifunctional and also possess mRNA decapping activity in cells in addition to their reported activities on nucleotide-containing molecules remains to be determined. However, it is interesting that the Nudt16 decapping enzyme is also a (deoxy)inosine diphosphatase that contributes to minimizing the deleterious incorporation of inosine nucleotides into DNA and RNA (Abolhassani et al. 2010; Iyama et al. 2010), and a dual activity on nucleotides and 5′ caps may be a general feature for these proteins.
The presence of mRNA decapping enzymes other than Dcp2 in mammalian cells was suggestive that the same may be true in nonmammalian cells. Toward that end, we have shown that one of the five yeast non-Dcp2 Nudix proteins also possesses robust mRNA decapping activity (Fig. 5). Ddp1p (YOR163w) is a member of the DIPP family of hydrolases with activity on the diphosphoinositol pentakisphosphate (PP-InsP5) and bisdiphosphoinositol tetrakisphosphate ([PP]2-InsP4) but also on highly phosphorylated (di)nucleotides such as Ap6A, Ap5A, Ap5A, and Ap4A (McLennan 2006). Importantly, other than Dcp2p, Ddp1p and its mammalian homolog Nudt3 are the only evolutionarily conserved Nudix proteins with detectable decapping activity. It will be interesting to determine whether Ddp1p functions as a decapping enzyme in yeast cells.
An unexpected finding from these studies was the demonstration that the bacterial pyrophosphohydrolase RppH (Deana et al. 2008) has potent decapping activity on a capped RNA but not cap structure (Fig. 6). The dependence of the cap being linked to an RNA moiety and the selective release of m7GDP as the decapping product is analogous to Dcp2 (Piccirillo et al. 2003) and suggests that RppH is likely an RNA-binding protein that binds the RNA at the 5′ end to hydrolyze within the triphosphate linkage, which would explain its requirement of the RNA body to be linked to the cap structure. The significance of a protein with decapping activity in prokaryotes is not evident. Nevertheless, this finding reveals that decapping activity predates eukaryotic capping and is an evolutionarily conserved function.
Identification of six Nudix family members in mammals and one in yeast with RNA decapping activity in vitro demonstrates that removal of the 5′ cap is more complex than originally envisioned. Multiple Nudix family members can decap mRNA in vitro and likely in cells, and experiments are under way to determine the latter. The recent demonstration that the yeast Rai1 (Xiang et al. 2009; Jiao et al. 2010) and Dxo1 (Chang et al. 2012) proteins, which are not members of the Nudix family of hydrolases, also contain decapping activity that preferentially functions on incompletely capped mRNAs, strongly suggests that there may yet be additional uncharacterized decapping proteins that modulate mRNA decay in cells. Furthermore, the surprising ability of all the decapping-competent Nudt proteins to also function on unmethylated capped RNAs to varying degrees raises an interesting question of whether these proteins may also function on incompletely capped mRNAs in cells and how they may be regulated. Future studies will provide new insights into the broad role of decapping enzymes in the regulation of mRNA decay and gene expression.
MATERIALS AND METHODS
Plasmid construction and protein purification
cDNAs encoding the various Nudix proteins were amplified from reverse-transcribed mouse embryonic fibroblast (MEF) cell mRNAs using the corresponding forward and reverse primer sets (primer sequences are listed in Supplemental Table S1). The cDNAs were inserted into the corresponding restriction enzyme sites shown in the primers within pET28a. Plasmids pET28a-Nudt1, 4, 5, and 7 were described previously (Song et al. 2010). The mutant constructs were generated by a one-step mutagenesis PCR approach. Briefly, the wild-type (WT) constructs were used as templates, and the mutant primer sets (listed in Supplemental Table S1) were used for PCR amplification. After digestion by Dpn1, PCR products were transformed into DH5α E. coli. All constructs were confirmed by sequencing. 6 × His-tagged proteins were purified using His Bind Resin (Novagen) according to the manufacturer’s instructions as modified in Liu et al. (2008).
RNA production and in vitro decapping assays
The pcDNA3 polylinker (pcP) RNA with a G tract at the 3′ end was transcribed by SP6 polymerase using PCR-generated DNA template with SP6 and C16T7 primers as previously described (Wang and Kiledjian 2001). To make DNA template containing only one guanosine as the first nucleotide, PCR amplification was carried out using the following three primers: 5′SP6-(CAT)n, (CAT)3template58 bp, and 3′(CAT)3-58 bp at a 200:1:200 ratio. The resolved DNA fragment was purified from an agarose gel and used for in vitro transcription in the presence of [α-32P]GTP and rATP, rCTP, and rUTP to generate an RNA with the first nucleotide labeled at the α-phosphate (pp32pG-RNA). Cap labeling of in vitro–transcribed RNAs were generated using the vaccinia virus capping enzyme (Shuman 1990) in the presence of [α-32P]GTP with or without S-adenosyl-dismethionine (SAM), as described (Wang and Kiledjian 2001). For RNAs with the α-phosphate containing the 32P-label, they were capped with unlabeled rGTP to generate m7Gpp32pG-RNA and Gpp32pG-RNA. Monomethylated and unmethylated cap structures were generated by nuclease P1 digestion of corresponding cap labeled RNAs. The nucleoside diphosphates, m7G32pp and G32pp, were generated as the hydrolysis product of the corresponding cap labeled RNAs with RppH.
In vitro decapping assays were carried out as previously described (Wang et al. 2002). In brief, 50 nM recombinant proteins was incubated with the indicated cap labeled RNAs in decapping buffer (10 mM Tris-HCl at pH 7.5, 100 mM KCl, 2 mM MgCl2, 2 mM DTT, 0.5 mM MnCl2, 40 units/mL recombinant RNase inhibitor). Decapping reactions were carried out for 30 min at 37°C and stopped by extracting once with phenol:chloroform (1:1). Decapping products were resolved by polyethyleneimine-cellulose TLC plates (Sigma-Aldrich) developed in 0.45 M (NH4)2SO4 in a TLC chamber at room temperature and exposed to a PhosphorImager.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Kiledjian laboratory for helpful discussions and critical reading of the manuscript. This work was supported by NIH grant GM67005 to M.K.
REFERENCES
- Abdelraheim SR, Spiller DG, McLennan AG 2003. Mammalian NADH diphosphatases of the Nudix family: Cloning and characterization of the human peroxisomal NUDT12 protein. Biochem J 374: 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeygunawardana C, Weber DJ, Gittis AG, Frick DN, Lin J, Miller AF, Bessman MJ, Mildvan AS 1995. Solution structure of the MutT enzyme, a nucleoside triphosphate pyrophosphohydrolase. Biochemistry 34: 14997–15005 [DOI] [PubMed] [Google Scholar]
- Abolhassani N, Iyama T, Tsuchimoto D, Sakumi K, Ohno M, Behmanesh M, Nakabeppu Y 2010. NUDT16 and ITPA play a dual protective role in maintaining chromosome stability and cell growth by eliminating dIDP/IDP and dITP/ITP from nucleotide pools in mammals. Nucleic Acids Res 38: 2891–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JSJ, Parker RP 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J 17: 1497–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessman MJ, Frick DN, O’Handley SF 1996. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem 271: 25059–25062 [DOI] [PubMed] [Google Scholar]
- Cai JP, Ishibashi T, Takagi Y, Hayakawa H, Sekiguchi M 2003. Mouse MTH2 protein which prevents mutations caused by 8-oxoguanine nucleotides. Biochem Biophys Res Commun 305: 1073–1077 [DOI] [PubMed] [Google Scholar]
- Chang JH, Jiao X, Chiba K, Oh C, Martin CE, Kiledjian M, Tong L 2012. Dxo1 is a new type of eukaryotic enzyme with both decapping and 5′–3′ exoribonuclease activity. Nat Struct Mol Biol 19: 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Alapat D, Wen X, Timo K, Burstein D, Lisanti M, Shears S, Kohtz DS 2004. Ectopic expression of murine diphosphoinositol polyphosphate phosphohydrolase 1 attenuates signaling through the ERK1/2 pathway. Cell Signal 16: 1045–1059 [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R 2004. Eukaryotic mRNA decapping. Annu Rev Biochem 73: 861–890 [DOI] [PubMed] [Google Scholar]
- Cruz-Reyes J, Piller KJ, Rusche LN, Mukherjee M, Sollner-Webb B 1998. Unexpected electrophoretic migration of RNA with different 3′ termini causes a RNA sizing ambiguity that can be resolved using nuclease P1-generated sequencing ladders. Biochemistry 37: 6059–6064 [DOI] [PubMed] [Google Scholar]
- Deana A, Celesnik H, Belasco JG 2008. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature 451: 355–358 [DOI] [PubMed] [Google Scholar]
- Decker CJ, Parker R 1993. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes Dev 7: 1632–1643 [DOI] [PubMed] [Google Scholar]
- Dunckley T, Parker R 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J 18: 5411–5422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger-Gron M, Fillman C, Norrild B, Lykke-Andersen J 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell 20: 905–915 [DOI] [PubMed] [Google Scholar]
- Franks TM, Lykke-Andersen J 2008. The control of mRNA decapping and P-body formation. Mol Cell 32: 605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuichi Y, LaFiandra A, Shatkin AJ 1977. 5′-Terminal structure and mRNA stability. Nature 266: 235–239 [DOI] [PubMed] [Google Scholar]
- Gabelli SB, Bianchet MA, Bessman MJ, Amzel LM 2001. The structure of ADP-ribose pyrophosphatase reveals the structural basis for the versatility of the Nudix family. Nat Struct Biol 8: 467–472 [DOI] [PubMed] [Google Scholar]
- Ghosh T, Peterson B, Tomasevic N, Peculis BA 2004. Xenopus U8 snoRNA binding protein is a conserved nuclear decapping enzyme. Mol Cell 13: 817–828 [DOI] [PubMed] [Google Scholar]
- Gingras AC, Raught B, Sonenberg N 1999. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem 68: 913–963 [DOI] [PubMed] [Google Scholar]
- Grudzien E, Kalek M, Jemielity J, Darzynkiewicz E, Rhoads RE 2006. Differential inhibition of mRNA degradation pathways by novel cap analogs. J Biol Chem 281: 1857–1867 [DOI] [PubMed] [Google Scholar]
- Heidbuchel H, Callewaert G, Vereecke J, Carmeliet E 1992. Membrane-bound nucleoside diphosphate kinase activity in atrial cells of frog, guinea pig, and human. Circ Res 71: 808–820 [DOI] [PubMed] [Google Scholar]
- Hentze MW 1997. eIF4G: A multipurpose ribosome adapter? Science 275: 500–501 [DOI] [PubMed] [Google Scholar]
- Hsu CL, Stevens A 1993. Yeast cells lacking 5′ → 3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol Cell Biol 13: 4826–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi T, Hayakawa H, Ito R, Miyazawa M, Yamagata Y, Sekiguchi M 2005. Mammalian enzymes for preventing transcriptional errors caused by oxidative damage. Nucleic Acids Res 33: 3779–3784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyama T, Abolhassani N, Tsuchimoto D, Nonaka M, Nakabeppu Y 2010. NUDT16 is a (deoxy)inosine diphosphatase, and its deficiency induces accumulation of single-strand breaks in nuclear DNA and growth arrest. Nucleic Acids Res 38: 4834–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Wang Z, Kiledjian M 2006. Identification of an mRNA-decapping regulator implicated in X-linked mental retardation. Mol Cell 24: 713–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Chen H, Chen J, Herrup K, Firestein BL, Kiledjian M 2009. Modulation of neuritogenesis by a protein implicated in X-linked mental retardation. J Neurosci 29: 12419–12427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao X, Xiang S, Oh C, Martin CE, Tong L, Kiledjian M 2010. Identification of a quality-control mechanism for mRNA 5′-end capping. Nature 467: 608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Kiledjian M 2004. Poly(A)-binding-protein-mediated regulation of hDcp2 decapping in vitro. EMBO J 23: 1968–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Song M, Kiledjian M 2011. Differential utilization of decapping enzymes in mammalian mRNA decay pathways. RNA 17: 419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Dai J, Song M, Fitzgerald-Bocarsly P, Kiledjian M 2012. Dcp2 decapping protein modulates mRNA stability of the critical interferon regulatory factor (IRF) IRF-7. Mol Cell Biol 32: 1164–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Abeygunawardana C, Frick DN, Bessman MJ, Mildvan AS 1996. The role of Glu 57 in the mechanism of the Escherichia coli MutT enzyme by mutagenesis and heteronuclear NMR. Biochemistry 35: 6715–6726 [DOI] [PubMed] [Google Scholar]
- Liu H, Rodgers ND, Jiao X, Kiledjian M 2002. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J 21: 4699–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SW, Jiao X, Welch S, Kiledjian M 2008. Analysis of mRNA decapping. Methods Enzymol 448: 3–21 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol Cell Biol 22: 8114–8121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A 2003. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLennan AG 2000. Dinucleoside polyphosphates—friend or foe? Pharmacol Ther 87: 73–89 [DOI] [PubMed] [Google Scholar]
- McLennan AG 2006. The Nudix hydrolase superfamily. Cell Mol Life Sci 63: 123–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D 1997. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell 91: 457–466 [DOI] [PubMed] [Google Scholar]
- Mukherjee C, Patil DP, Kennedy BA, Bakthavachalu B, Bundschuh R, Schoenberg DR 2012. Identification of cytoplasmic capping targets reveals a role for cap homeostasis in translation and mRNA stability. Cell Rep 2: 674–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofman R, Speijer D, Leen R, Wanders RJ 2006. Proteomic analysis of mouse kidney peroxisomes: Identification of RP2p as a peroxisomal nudix hydrolase with acyl-CoA diphosphatase activity. Biochem J 393: 537–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka Y, Kedersha NL, Schoenberg DR 2009. Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol Cell Biol 29: 2155–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo C, Khanna R, Kiledjian M 2003. Functional characterization of the mammalian mRNA decapping enzyme hDcp2. RNA 9: 1138–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez CV, Vilela C, Berthelot K, McCarthy JE 2002. Modulation of eukaryotic mRNA stability via the cap-binding translation complex eIF4F. J Mol Biol 318: 951–962 [DOI] [PubMed] [Google Scholar]
- Sachs AB, Sarnow P, Hentze MW 1997. Starting at the beginning, middle, and end: Translation initiation in eukaryotes. Cell 89: 831–838 [DOI] [PubMed] [Google Scholar]
- Safrany ST, Caffrey JJ, Yang X, Bembenek ME, Moyer MB, Burkhart WA, Shears SB 1998. A novel context for the ‘MutT’ module, a guardian of cell integrity, in a diphosphoinositol polyphosphate phosphohydrolase. EMBO J 17: 6599–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DC, Parker R 1999. Mutations in translation initiation factors lead to increased rates of deadenylation and decapping of mRNAs in Saccharomyces cerevisiae. Mol Cell Biol 19: 5247–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz DC, Parker R 2000. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol Cell Biol 20: 7933–7942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz D, Decker CJ, Parker R 2003. The enhancer of decapping proteins, Edc1p and Edc2p, bind RNA and stimulate the activity of the decapping enzyme. RNA 9: 239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shatkin AJ 1985. mRNA cap binding proteins: Essential factors for initiating translation. Cell 40: 223–224 [DOI] [PubMed] [Google Scholar]
- Shatkin AJ, Manley JL 2000. The ends of the affair: Capping and polyadenylation. Nat Struct Biol 7: 838–842 [DOI] [PubMed] [Google Scholar]
- She M, Decker CJ, Sundramurthy K, Liu Y, Chen N, Parker R, Song H 2004. Crystal structure of Dcp1p and its functional implications in mRNA decapping. Nat Struct Mol Biol 11: 249–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotohno K, Kodama Y, Hashimoto J, Miura KI 1977. Importance of 5′-terminal blocking structure to stabilize mRNA in eukaryotic protein synthesis. Proc Natl Acad Sci 74: 2734–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S 1990. Catalytic activity of vaccinia mRNA capping enzyme subunits coexpressed in Escherichia coli. J Biol Chem 265: 11960–11966 [PubMed] [Google Scholar]
- Sillero A, Sillero MA 2000. Synthesis of dinucleoside polyphosphates catalyzed by firefly luciferase and several ligases. Pharmacol Ther 87: 91–102 [DOI] [PubMed] [Google Scholar]
- Song MG, Li Y, Kiledjian M 2010. Multiple mRNA decapping enzymes in mammalian cells. Mol Cell 40: 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R 2003. Analysis of recombinant yeast decapping enzyme. RNA 9: 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasevic N, Peculis B 1999. Identification of a U8 snoRNA-specific binding protein. J Biol Chem 274: 35914–35920 [DOI] [PubMed] [Google Scholar]
- van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Seraphin B 2002. Human Dcp2: A catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J 21: 6915–6924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E, Le Hir H, Seraphin B 2003. DcpS can act in the 5′–3′ mRNA decay pathway in addition to the 3′–5′ pathway. Proc Natl Acad Sci 100: 12081–12086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kiledjian M 2001. Functional link between the mammalian exosome and mRNA decapping. Cell 107: 751–762 [DOI] [PubMed] [Google Scholar]
- Wang Z, Jiao X, Carr-Schmid A, Kiledjian M 2002. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc Natl Acad Sci 99: 12663–12668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wypijewska A, Bojarska E, Lukaszewicz M, Stepinski J, Jemielity J, Davis RE, Darzynkiewicz E 2012. 7-Methylguanosine diphosphate (m7GDP) is not hydrolyzed but strongly bound by decapping scavenger (DcpS) enzymes and potently inhibits their activity. Biochemistry 51: 8003–8013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S, Cooper-Morgan A, Jiao X, Kiledjian M, Manley JL, Tong L 2009. Structure and function of the 5′ → 3′ exoribonuclease Rat1 and its activating partner Rai1. Nature 458: 784–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Cai JP, Tu B, Wu L, Zhao Y, Liu X, Li L, McNutt MA, Feng J, He Q, et al. 2009. Proliferating cell nuclear antigen is protected from degradation by forming a complex with MutT Homolog2. J Biol Chem 284: 19310–19320 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.