A plethora of noncoding (nc) RNAs have been revealed through the application of high-throughput analysis of the transcriptome, and this has led to an intensive search for possible biological functions attributable to these transcripts. A major category of functional ncRNAs that has emerged concerns those that are implicated in coordinate gene silencing, either in cis or in trans. Xist and other ncRNAs have been shown to bind to PRC2, a repressive chromatin modifying complex. Silencing has been proposed to involve direct interaction between PRC2 proteins and the different ncRNAs. This review discusses the evidence for and against this hypothesis, in the process highlighting alternative models and discussing experiments that in the future will help to resolve existing discrepancies.
Keywords: noncoding RNA, lincRNA, Xist RNA, polycomb, chromatin
Abstract
A plethora of noncoding (nc) RNAs has been revealed through the application of high-throughput analysis of the transcriptome, and this has led to an intensive search for possible biological functions attributable to these transcripts. A major category of functional ncRNAs that has emerged is for those that are implicated in coordinate gene silencing, either in cis or in trans. The archetype for this class is the well-studied long ncRNA Xist which functions in cis to bring about transcriptional silencing of an entire X chromosome in female mammals. An important step in X chromosome inactivation is the recruitment of the Polycomb repressive complex PRC2 that mediates histone H3 lysine 27 methylation, a hallmark of the inactive X chromosome, and recent studies have suggested that this occurs as a consequence of PRC2 interacting directly with Xist RNA. Accordingly, other ncRNAs have been linked to PRC2 targeting either in cis or in trans, and here also the mechanism has been proposed to involve direct interaction between PRC2 proteins and the different ncRNAs. In this review, I discuss the evidence for and against this hypothesis, in the process highlighting alternative models and discussing experiments that, in the future, will help to resolve existing discrepancies.
INTRODUCTION
The advent of oligonucleotide microarrays and advanced sequencing technologies has revealed an unexpected plethora of previously unrecognized noncoding RNAs (ncRNAs) in the genomes of species from budding yeast to man (for recent reviews, see Wang and Chang 2011; Rinn and Chang 2012). In mouse, ncRNAs identified by cDNA sequencing (macro ncRNAs) and long intergenic noncoding RNAs (lincRNAs) predicted on the basis of histone modification signatures that are hallmarks of transcription collectively comprise ∼4–5 × 103 individual loci (Kapranov et al. 2002; Bertone et al. 2004; Carninci et al. 2005). A more recent analysis estimated that there are ∼8 × 103 human lincRNAs (Cabili et al. 2011). The ncRNA loci are often rapidly evolving, and the transcripts are generally very low abundance. This may indicate a lack of biological function or, alternatively, a limited necessity to constrain sequence. However, a subset of these loci do show sequence conservation at ncRNA exons relative to neighboring neutral sequences, consistent with functional constraints on at least a proportion of these genes (Ponjavic et al. 2007; Marques and Ponting 2009).
Consistent with apparent functional constraints, specific roles have been attributed to a growing list of the novel ncRNAs. In several cases, these functions are analogous to those ascribed to known ncRNAs discovered in prior hypothesis-driven experiments. Notable examples are piwi-interacting RNAs (piRNAs) and microRNAs discovered through studies on transgene silencing phenomena in higher plants (Hamilton and Baulcombe 1999), and nematodes (Fire et al. 1998), and the noncoding RNAs, Xist and Tsix, discovered through studies on the mechanism of X chromosome inactivation in mammals (Brockdorff et al. 1992; Brown et al. 1992; Lee et al. 1999). Although well-studied ncRNAs have served as archetypes guiding the functional analysis of novel ncRNAs, the latter have also been implicated in biological functions that had not been previously anticipated, acting as molecular signals, decoys, guides, and scaffolds (for review, see Wang and Chang 2011; Rinn and Chang 2012).
Although it is probable that many ncRNAs do not have a discernible function, they, nevertheless, constitute a pool from which natural selection can evolve functional ncRNAs, conferring a selective advantage to the organism. In support of this view, recent evidence has demonstrated a remarkable example of convergent evolution whereby eutherian (placental) and metatherian (marsupial) mammals independently evolved a ncRNA, Xist and Rsx, respectively, to regulate dosage compensation (inactivation) of the X chromosome (Duret et al. 2006; Grant et al. 2012).
It is clear that the continually expanding list of ncRNA loci should not be considered as a single class, and indeed, a number of subdivisions have been defined to facilitate further analyses. An important distinction discriminates ncRNAs >200 nt in length, referred to as l(long)ncRNAs. Within this class, there are further subdivisions; thus, NATs (natural antisense RNAs) are produced in various configurations antisense to other, usually protein-coding transcripts and can function either in repressing or activating paired loci. Such functions may be ascribed to the lncRNAs or to the act of transcription, for example, where antisense ncRNAs overlap with sense promoters, leading to transcriptional interference at the sense locus, as has been demonstrated for ncRNA transcription in the Drosophila melanogaster bithorax complex (Petruk et al. 2006).
A number of lncRNAs function in coordinate gene silencing, either in cis or in trans, and in cases documented to date, the RNA is apparently required for function. Xist RNA, a prototypical example of this class, has been extensively studied and, as such, provides a paradigm underpinning functional analysis of other potentially related lncRNAs. This is exemplified by studies on recruitment of Polycomb repressive complex 2 (PRC2) by lncRNA, first demonstrated for Xist RNA (Mak et al. 2002; Plath et al. 2003; Silva et al. 2003; Zhao et al. 2008) and then extrapolated to Kcnqtlot1, a lncRNA required for silencing a cluster of imprinted genes on mouse chromosome 7 (Fitzpatrick et al. 2002; Pandey et al. 2008), to the lncRNA HOTAIR transcribed from the HOXD locus on human chromosome 12, required to direct PRC2 in trans to the HOXC locus on chromosome 2 (Rinn et al. 2007), and further, to a class of short ncRNAs produced at CpG island loci in mammalian cells and implicated more widely in PRC2 recruitment to target genes throughout the genome (Kanhere et al. 2010). Finally, a novel ncRNA COLDAIR transcribed from an intron in the Arabidopsis thaliana flowering control locus FLC has recently been implicated in the control of vernalization (regulation of flowering time by periods of cold) through direct recruitment of the A. thaliana PRC2 complex (Heo and Sung 2011). A key concept to emerge from these studies is that of a direct biochemical interaction between specific ncRNAs and proteins of the PRC2 complex (Zhao et al. 2008), an idea that has generated considerable excitement in the field. However, the picture is not entirely clear-cut, and there are some observations, notably regarding the link between Xist RNA and PRC2, which are difficult to reconcile with this emerging consensus. In this review, I provide an overview of the evidence in favor of direct recruitment of the PRC2 complex by ncRNA and then a discussion of confounding observations and alternative interpretations of existing data. Finally, I discuss future experiments that could shed further light on this important idea.
Polycomb recruitment by Xist RNA
X inactivation is the process that in mammals ensures equalization of X-linked gene dosage in XX females relative to XY males (Lyon 1961). Classical genetic studies demonstrated that X inactivation is regulated by a single cis-acting master switch locus, the X inactivation center (Xic), which was later identified as the Xist gene locus (Brown et al. 1991a,b). The Xist gene is transcribed exclusively from the inactive X chromosome (Xi), producing a ncRNA ∼17 kb in length (Fig. 1A; Brockdorff et al. 1992; Brown et al. 1992). Although Xist RNA is capped, spliced, and polyadenylated, the transcript somehow bypasses nuclear export pathways and, instead, accumulates and spreads in cis, covering the territory of the chromosome from which it is transcribed. This unique and unusual characteristic led to the proposal that Xist RNA is the cis-acting inactivation signal emanating from the Xic, an idea that has since been rigorously verified in a number of gene knockout and transgenic experiments (Lee et al. 1996; Penny et al. 1996; Herzing et al. 1997; Lee and Jaenisch 1997; Wutz and Jaenisch 2000).
FIGURE 1.
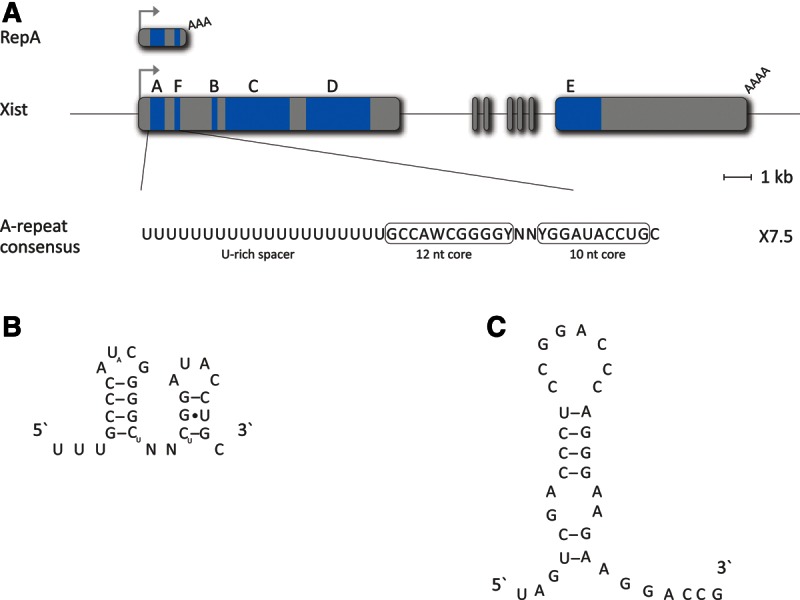
The A-repeat of Xist RNA. (A) Schematic representing the Xist gene locus and indicating the location of different tandemly repeated regions, labeled A–F (blue shading), and a short RNA, RepA, proposed to arise from within Xist exon 1 and to be important in PRC recruitment in X inactivation (see text). The A-repeat is essential for Xist-mediated chromosome silencing and is present in both full-length Xist RNA and RepA RNA. The consensus sequence of the A-repeat monomer with GC rich core (boxed) and U-rich spacer is shown below. (B) Proposed secondary structure of an A-repeat monomer, as determined by thermodynamic folding predictions and biophysical assays (see text). (C) Proposed secondary structure of a monomer of a tandem repeat located at the 5′ end of the marsupial ncRNA Rsx thought to function equivalently to Xist RNA A-repeat. This hypothetical structure is based solely on thermodynamic folding predictions.
Our understanding of the molecular mechanisms by which Xist RNA brings about chromosome-wide silencing is relatively superficial and likely incomplete. The predominating view is that Xist RNA directly recruits chromatin-/chromosome-modifying factors that then establish Xi heterochromatin. Identification of these factors has, however, been challenging, not least because Xist RNA is relatively intractable to biochemical analysis. The field has, instead, relied on candidate-based studies and serendipitous observations. A key breakthrough was the finding that early embryos homozygous for a mutation in the gene eed, encoding a protein belonging to the Polycomb group (PcG) of repressors, show a female-specific phenotype attributable to failure of X inactivation in a specific population of cells in the developing placenta (Wang et al. 2001). PcG proteins were originally identified in D. melanogaster as key mediators of heritable gene silencing, constituting a memory system for stable propagation of gene silencing through multiple cell generations (for a recent review, see Simon and Kingston 2009). Given that the inactive X chromosome is a classical model for developmentally regulated heritable gene silencing, an important role for PcG proteins in this process makes good sense.
Subsequent analysis substantiated that PcG proteins have an important role in the X inactivation pathway. Immunofluorescence studies demonstrated that the protein EED is highly enriched within the interphase Xi territory and, moreover, that EED localizes to the Xi on metaphase chromosomes, exhibiting a banded localization that closely resembles that previously described for Xist RNA (Duthie et al. 1999; Mak et al. 2002). This provided the first indication that PcG proteins may be targeted directly by Xist RNA. Additionally, it was found that the SET domain containing protein EZH2, previously shown to interact with EED, is also enriched within Xi territories. Further advances came with the recognition that EED/EZH2 recruitment to Xi occurs coincident with the onset of Xist RNA expression and in all cells of the developing embryo (Plath et al. 2003; Silva et al. 2003). In independent studies, it was determined that EED and EZH2 are core components of a multi-subunit histone methyltransferase complex, PRC2 (see Fig. 2), with specificity for lysine 27 (H3K27) of histone H3 (Cao et al. 2002; Czermin et al. 2002; Muller et al. 2002). Consistent with this H3K27, methylation (H3K27me2/3) was found to be highly enriched on Xi (Plath et al. 2003; Silva et al. 2003) and to be dependent on PRC2 function (Silva et al. 2003), indicating that this chromatin modification may be important in Xi silencing. Additionally, analysis of PRC2 recruitment in early mouse embryos (Mak et al. 2004) and in response to Xist transgene expression (Plath et al. 2003; Kohlmaier et al. 2004) demonstrated that recruitment is dependent on ongoing Xist RNA expression, further reinforcing the idea that PRC2 interacts physically with Xist RNA.
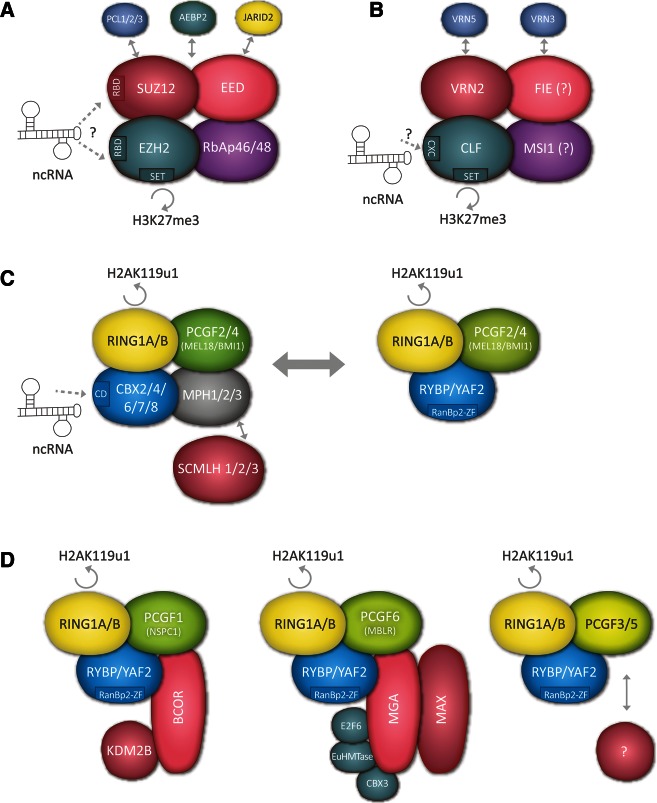
FIGURE 2.
Major Polycomb complexes in mammals and proposed ncRNA interactions. (A) Representation of Polycomb repressive complex 2 (PRC2) illustrating core components with putative RNA binding domains (RBD) and catalytic histone methyltransferase domain (SET). Several ncRNAs, including Xist, Kcnq1ot1, and HOTAIR, and short RNAs transcribed from CpG island promoters of PcG target genes, have been proposed to interact with the putative RBDs of either EZH2 or SUZ12 (see text). Additional subunits that are either weakly associated or substoichiometric PRC2 components are linked by double-headed arrows. (B) Representation of the Arabidopsis thaliana PRC2 complex linked to regulation of vernalization. Color-coding indicates subunit homology with mammalian PRC2. The CXC domain in the CLF subunit is a putative RBD interacting with the ncRNA COLDAIR (see text). VRN5 and VIN3 are weakly associated or substoichiometric components potentially homologous to PCL1/2/3 in mammals and required to fully activate PRC2 at the FLC locus. (C) The classical PRC1 complex exists in two forms in which either CBX and MPH or RYBP subunits associate with the catalytic core subunits RING1A/B and PCGF2/4. The weakly associated/substoichiometric PRC1 component SCMLH1/2/3 is linked with a double-headed arrow. The chromodomain (CD) of the CBX7 protein has been shown to bind the ncRNA ANRIL (see text). The RanBP-ZF domain present in RYBP/YAF2 has an RNA binding function in some related proteins but apparently not in RYBP/YAF2. (D) Representation of three additional PRC1-related complexes in which the subunit composition results from incorporation of different homologs of the PCGF subunit, specifically PCGF1, PCGF6, and PCGF3/5. These complexes include the RYBP/YAF2 subunit but not PcG CBX or MPH proteins. A non-PcG CBX homolog, CBX3, that binds H3K9me3 as opposed to H3K27me3, is found in PCGF6 complexes. To date, there is no evidence that these complexes bind to ncRNA.
Although PRC2 proteins emerged as central candidates for Xist RNA-mediated silencing, analysis of PRC2 mutant embryos suggested a more complex picture. Thus, although X reactivation was observed in extraembryonic trophectoderm cells of eed mutant embryos, re-expression of X-linked genes in cells of the embryo proper was observed only sporadically, if at all (Silva et al. 2003; Kalantry and Magnuson 2006). Moreover, analysis of other Xi markers, for example, histone hypoacetylation, indicated that X inactivation proceeds normally in the absence of core PRC2 proteins (Silva et al. 2003; Schoeftner et al. 2006). Thus, it was considered that PRC2 function on Xi is redundant, most likely due to the contribution of parallel silencing pathways.
Analysis of differentiating XX embryonic stem (ES) cells, an important cell culture model for X inactivation, and of embryos at different stages of development, demonstrated that enrichment of core PRC2 proteins diminishes as differentiation and development proceeds, likely attributable, at least in part, to falling levels of core PRC2 proteins (Plath et al. 2003; Silva et al. 2003). H3K27me3 enrichment on Xi, however, remained throughout development, indicating that relatively low levels of the PRC2 complex are sufficient to establish enhanced levels of H3K27me3.
At this point in time, it had been established that the second major PcG complex, PRC1 (Fig. 2C), is recruited to target sites via interaction of a chromodomain within the core PRC1 protein Polycomb (mammalian homologs CBX2, CBX4, CBX6, CBX7, and CBX8) with the H3K27me3 modification deposited by PRC2 (Cao et al. 2002; Fischle et al. 2003). Consistent with this, PRC1 core components were also found to be enriched over the Xi territory (de Napoles et al. 2004; Plath et al. 2004). Moreover, PRC1 was, at this time, shown to have a distinct histone modification activity, monoubiquitylation of histone H2A at lysine 119 (H2AK119u1), mediated by the core subunits Ring1 and homologs of the Drosophila protein Posterior sex combs (PSC) (de Napoles et al. 2004; Wang et al. 2004), and H2AK119u1 was found to be enriched on Xi at interphase and at metaphase, the latter again resembling the banded pattern observed previously for Xist RNA (de Napoles et al. 2004). Thus, the picture that began to emerge was that Xist RNA recruits PRC2 to Xi, leading to deposition of H3K27me3 with subsequent indirect recruitment of PRC1 and associated H2AK119u1. More recently, it has been shown that an H3K27me3-independent pathway also recruits PRC1 to Xi (Schoeftner et al. 2006) and that this is attributable to complexes that are related to PRC1 but in which CBX2/4/6/7/8 is substituted by the protein RYBP (see Fig. 2C,D; Tavares et al. 2012).
The overlapping localization patterns seen for Xist RNA and PcG-mediated histone modifications, coupled to rapid recruitment of PRC2 and PRC1 at the onset of Xist RNA expression and strict dependence of PRC2/PRC1 recruitment on ongoing Xist RNA expression, collectively supported a view that the Polycomb system is directly targeted by Xist RNA. However, it was formally possible that recruitment of PcG complexes was indirect, occurring in response to other Xist RNA-mediated changes in Xi heterochromatin. A key observation that lent strong support to the direct recruitment model was that PRC2 localization on Xi occurs in response to induced expression of an Xist transgene lacking a domain critical for Xist RNA-mediated silencing, the A-repeat (Kohlmaier et al. 2004). That PcG recruitment occurred in the absence of chromosome silencing seemed to affirm a direct interaction between Xist RNA and PcG proteins, as the indirect model could not be invoked in this circumstance. These experiments also indicated that PcG recruitment to Xi is not sufficient for chromosome silencing, although a caveat to this conclusion is that the chromosomal distribution of PcG proteins and associated histone modifications may differ following expression of Xist RNA lacking the A-repeat relative to wild-type Xist RNA.
Up to this point, our understanding of the link between Xist RNA and PcG recruitment relied heavily on immunofluorescence studies and, as such, was limited in terms of defining the molecular interactions that occur. For example, the resolution of light microscopy is no better than 200 nm in X- and Y-axes and ∼600 nm in the Z-axis, and this needs to be factored into conclusions regarding the apparent colocalization of Xist RNA and PcG protein immunofluorescence signals. Attention, therefore, turned to biochemical analysis of the proposed interaction of PcG proteins with Xist RNA. A key study identified RepA, a short and rare transcript embedded within Xist intron 1 and encompassing the A-repeats (see Fig. 2), and provided evidence, first, that RepA is important for Xist-mediated recruitment of PcG proteins and secondly, that this interaction is mediated by direct interaction of the PRC2 core component EZH2 with a stem–loop structure present in A-repeat monomers (Zhao et al. 2008). Key experiments in this study were RNA immunoprecipitation (RIP), in which Xist RNA was found to co-IP with core PRC2 proteins, and EMSA assays in which recombinant EZH2 subunit was found to bind to A-repeat RNA. An unstructured A-repeat mutant served as a negative control in the EMSA experiment, although a second control, A-repeat antisense RNA, which is predicted to form a different structure, did interact with EZH2, indicating that RNA binding is not entirely specific. A further indication that the reported interaction is relatively nonspecific is that E(Z), the D. melanogaster homolog of EZH2, also bound A-repeat RNA.
The A-repeat is a tandemly arranged repeat comprising 7.5 copies of a 26-nt GC-rich core sequence separated by AT-rich spacers of variable length (see Fig. 1A; Brockdorff et al. 1992; Brown et al. 1992). It has been proposed that each monomer folds into a structure comprising two stem–loops, one of which forms a stable AUCG tetraloop, and that monomers multimerize via inter-repeat duplex formation (Fig. 1B). Xist transgenes with A-repeat mutations predicted to disrupt these structures fail to silence (Wutz et al. 2002; Duszczyk et al. 2011). An alternative model proposes that multiple monomers interact to form two large structures involving inter-repeat base-pairing (see Fig. 2; Maenner et al. 2010). This latter study examined the assembly of PRC2 components following incubation of A-repeat probes with ES cell nuclear extracts and found that each PRC2 core component interacts with A-repeat probes corresponding to one of the two predicted structures, although efficient interaction of the SUZ12 subunit required the full-length A-repeat multimer.
Further analysis of the interaction between EZH2 and lncRNAs demonstrated a requirement for a domain within EZH2 comprising a tract of basic amino acids (Kaneko et al. 2010). The observed interaction was enhanced by phosphorylation at a specific residue, tyrosine 345, that occurs in mitosis through activity of CDK kinase. As such, phosphorylated EZH2 is present at very low abundance, and it is unclear if this is sufficient to entirely account for RNA-mediated recruitment of PRC2.
It should be noted that interaction of PRC2 subunits with the A-repeat element cannot be the whole story because, as discussed above, Xist RNA lacking A-repeats does recruit PRC2, albeit less robustly (Kohlmaier et al. 2004). One possibility is that other elements within the Xist transcript also bind directly to core PRC2 components. A further study demonstrated that Polycomblike2 (PCL2), a substoichiometric component of PRC2, is important for PRC2 recruitment by Xist RNA (Casanova et al. 2011). However, depletion of PCL2 also affected PRC2 recruitment at target sites other than Xi, suggesting a more general function in PRC2 binding to chromatin.
PRC2-mediated H3K27me3 has also been observed on Xi in marsupial mammals (Mahadevaiah et al. 2009; Chaumeil et al. 2011), and as discussed above, X inactivation in marsupials has recently been shown to be linked to expression of a novel cis-acting ncRNA, Rsx. Although the mechanism by which Rsx RNA targets H3K27me3 has not been studied in any detail, it is interesting to note that the 5′ end of the Rsx transcript is comprised of a tandemly repeated sequence similar to the A-repeats in Xist RNA (Grant et al. 2012). RNA folding algorithms predict that this repeat should form stem–loop structures that are possibly analogous to Xist A-repeats (see Fig. 1C) and, as such, may be candidates for recruitment of PRC2 complexes in marsupials.
Polycomb recruitment by lncRNAs at imprinted loci
Genes subject to parental imprinting in mammals are often found in clusters, which in several instances include lncRNA loci that are reciprocally expressed relative to the imprinted protein-coding genes within the same cluster. This has led to the idea that some imprinted ncRNAs may function in a similar manner to Xist RNA, silencing genes located in cis, albeit within a relatively limited range. Studies to date indicate that this could be the case for some, but not all, imprinted clusters. Thus, for example, the imprinted ncRNA gene H19 located on mouse chromosome 7 regulates imprinting of the neighboring Igf2 gene in cis, but this is attributable to enhancer competition and does not require the ncRNA per se (Ripoche et al. 1997; Hark et al. 2000). Interestingly, recent studies have revealed that H19 ncRNA does play a role in trans, repressing several imprinted genes located at other sites in the genome (Gabory et al. 2009), and, in addition, that H19 RNA is a microRNA precursor (Cai and Cullen 2007).
Conversely, the imprinted ncRNA Kcnqt1ot1, also located on mouse chromosome 7, does appear to function in a similar manner to Xist RNA. Kcnqt1ot1 RNA is expressed from the paternal allele, reciprocal to the expression of several maternally expressed imprinted genes located within a domain of ∼1 Mb. The extent of the Kcnqt1ot1 domain varies in different tissues. Thus, genes within a 200-kb domain centered on Kcnqt1ot1 are repressed in placental and embryo tissues in a manner dependent on CpG island DNA methylation, whereas genes located farther away are repressed only in placental tissues, and in these cases, silencing occurs independent of DNA methylation (Lewis et al. 2004; Umlauf et al. 2004). Deletion of the Kcnq1ot1 promoter on the paternal allele results in loss of imprinting of most genes within the cluster (Fitzpatrick et al. 2002). Maternally repressed genes within the Kcnq1ot1 domain have several hallmarks of silent heterochromatin, notably DNA methylation of associated CpG islands and enrichment of H3K9me2/3 and H3K27me2/3 histone modifications (Lewis et al. 2004; Umlauf et al. 2004), resembling the chromatin status of silenced genes on Xi. Mutation of G9a, required for H3K9 methylation, and of EED, required for H3K27 methylation, results in partial derepression of some genes within the cluster (Mager et al. 2003; Wagschal et al. 2008).
The Kcnqtlot1 ncRNA is, like Xist RNA, localized primarily within the nucleus, but the transcript is unspliced and is contiguous with genomic DNA spanning ∼50 kb (Engemann et al. 2000; Pandey et al. 2008). Distribution of Kcnqt1ot1 RNA within the imprinted domain has been investigated using a method termed ChRIP involving immunoprecipitation of chromatin coupled to analysis of associated RNA by RT-PCR (Pandey et al. 2008). These studies indicated that Kcnqtlot1 RNA localizes to chromatin within the domain encompassing repressed genes in placenta but to only a limited extent in fetal liver, a conclusion that was subsequently reinforced by RNA FISH analysis (Redrup et al. 2009). The lineage-specific chromatin interaction of Kcnqt1ot1 RNA was found to correlate broadly with lineage-specific chromatin structure, suggesting that the RNA may recruit histone-modifying factors. Evidence in support of this was provided by RIP assays in which both PRC2 core components (EZH2 and SUZ12) and G9a coimmunoprecipitated Kcnqt1ot1 RNA in placenta but not in fetal liver. Thus, silencing by Kcnqt1ot1 RNA has clear similarities with Xist RNA-mediated silencing, including the recruitment of PRC2 proteins and other chromatin-modifying factors.
The link between PcG proteins and Kcnq1ot1 RNA was further reinforced in studies analyzing imprinted domains in early mouse embryos that were either wild type or PcG mutant (Terranova et al. 2008). These studies confirmed that Kcnq1ot1 RNA, and also another imprinted ncRNA, Air, describe distinct nuclear domains in interphase nuclei, similar to Xist RNA. The domains showed exclusion of RNA polymerase II, consistent with the ncRNAs defining repressive compartments, again similar to Xist RNA. In both PRC2 and PRC1 mutant embryos, the compaction of the repressive compartments was disrupted, suggesting a role for PcG proteins in organizing their architecture.
As indicated above, the paternally expressed ncRNA Air also has parallels with Xist RNA, being required to repress the paternal allele of three imprinted protein-coding genes, Igf2r, Slc22a2, and Slc22a3, located within the same cluster (Sleutels et al. 2002). However, in this case, PcG-associated chromatin modifications have not been observed (Regha et al. 2007), and expression of genes in the cluster is unaffected in PcG mutants (Mager et al. 2003). There is, however, evidence that Air ncRNA directly recruits the G9a HMTase (Nagano et al. 2008).
In summary, studies on Kcnq1ot1 and Air ncRNAs reinforce the idea that ncRNA plays a role in recruitment of PcG proteins and other chromatin-modifying enzymes to defined targets. Evidence to date favors a direct recruitment mechanism, although further studies are needed to establish this unequivocally (see Discussion below).
PRC2 recruitment and lncRNAs in other organisms
Reports linking ncRNAs to PcG recruitment in D. melanogaster, where the PcG system has been extensively analyzed, are limited. Thus, it has been reported that ncRNAs transcribed at regulatory elements within the homeotic gene clusters recruit trithorax factors that, in turn, oppose activity of PcG proteins (Sanchez-Elsner et al. 2006). Thus, in this context, the ncRNAs appear to function as anti-PcG factors. An analysis of the properties of D. melanogaster lincRNA loci revealed enrichment in PcG-associated chromatin (Young et al. 2012). This may indicate a role for PcG recruitment in cis but could also be interpreted as preferential association of lincRNA loci with developmentally regulated genes.
A recent study in the higher plant A. thaliana, does point to a role for a specific ncRNA, designated COLDAIR, in PRC2 recruitment at the flowering control (FLC) locus (Heo and Sung 2011). Prior work demonstrated a role for A. thaliana PRC2 in repression of the FLC locus by vernalization, a process whereby flowering is accelerated by prolonged cold (for review, see Letswaart et al. 2012). FLC functions as a negative regulator of flowering and PRC2-mediated repression of FLC, therefore, promotes flowering. The mechanism whereby PRC2 is targeted to FLC is not fully understood, but it has been shown that conventional PRC2 covers the entire locus at the onset of vernalization but is only converted to an active complex through recruitment of a PHD domain protein (forming PHD-PRC2), initially at a single nucleation site, close to the FLC promoter. Active PHD-PRC2 then spreads across the entire locus, in the process depositing H3K27me3 (De Lucia et al. 2008). The recently described ncRNA, COLDAIR, transcribed in a sense direction from within FLC intron 1, has been found to play a role in PRC2 targeting (Heo and Sung 2011). Depletion of COLDAIR RNA by RNAi attenuated PRC2-mediated repression of FLC, and enrichment of COLDAIR RNA was observed in RIP experiments performed by IP of the A. thaliana EZH2 homolog, curly leaf (CLF) (see Fig. 2B). This interaction was mapped to a specific domain in CLF, the CXC domain, found in a number of other plant proteins. Although the function of this domain has not been extensively characterized, a CXC protein within the maize protein CBBP, that has a role in paramutation, was found to mediate DNA binding (Brzeska et al. 2010).
A second ncRNA, COOLAIR, has also been implicated in regulating the FLC locus (Swiezewski et al. 2009). COOLAIR is transcribed antisense to FLC and produces two transcripts through alternate use of polyadenylation signals. COOLAIR transcription is not required for vernalization, but it has been implicated in FLC repression early during cold treatment, possibly mediated by direct effects on the FLC promoter.
lncRNAs linked to PRC2 targeting in trans—HOTAIR
The examples discussed above established a precedent for lncRNAs functioning in the targeting of PRC2 and/or other chromatin-modifying complexes in cis. This concept was significantly expanded with the description of HOTAIR, a 2.2-kb lncRNA implicated in targeting chromatin-modifying complexes in trans (Rinn et al. 2007). HOTAIR lncRNA is transcribed from a site in the HOXC locus located on human chromosome 12. RNAi-mediated depletion of HOTAIR, surprisingly, led to activation of HOX genes within the HOXD locus on human chromosome 2. Activation of HOXD genes was accompanied by loss of PcG-mediated repression, and it was suggested that HOTAIR may function by targeting PcG repressors to sites in trans. Consistent with this idea, RIP assays identified HOTAIR RNA following IP of PRC2 core components SUZ12 or EZH2. Additionally, biotinylated HOTAIR RNA was used to pull down PRC2 from nuclear extracts. Phylogenetic analysis has demonstrated that HOTAIR is conserved in a number of mammalian species, although, perhaps surprisingly, gene knockout of the mouse HOTAIR ortholog was found not to affect HoxD gene silencing (Schorderet and Duboule 2011).
A new method, chromatin isolation by RNA purification (ChIRP) was developed to map HOTAIR binding sites genome-wide. Here, biotinylated oligonucleotides antisense to the RNA of interest are used to pull out chromatin fragments obtained by sonication following glutaraldehyde cross-linking (Chu et al. 2011). This approach identified >800 sites where HOTAIR binding occurred. Metanalysis indicated enrichment for promoters and gene bodies and an overlap with peaks of PRC2 protein occupancy. Although it is not clear how HOTAIR RNA is directed to target loci, this was seen to occur normally in EZH2-deficient cells.
A subsequent analysis demonstrated that the 5′ end of the HOTAIR transcript is required for interaction with PRC2. Surprisingly, the 3′ end of the transcript was reported to interact with a different chromatin-modifying factor, LSD1, a histone demethylase enzyme that is a component of the REST corepressor complex (Tsai et al. 2010). This observation, again based on data from RIP assays, led to the suggestion that HOTAIR functions as a molecular scaffold that coordinates targeting of PRC2 and LSD1 to chromatin, thereby coupling H3K27 methylation with loss of H3 lysine 4 methylation, a modification associated with gene activity.
Genome-scale analysis of PRC2 targeting by lncRNAs
The evidence implicating lncRNAs in targeting PRC2 to defined loci led to the idea that lncRNAs could play a wider role, directing PRC2 complexes to target loci genome-wide. PRC2 target loci have been mapped in several cell types using ChIP-seq methods. Peaks of occupancy number ∼1–2 × 103 and occur in relatively broad domains that correspond closely with CpG islands of the associated genes (Ku et al. 2008). A recent study identified a novel type of promoter-associated short RNAs transcribed from CpG island promoters (Kanhere et al. 2010). A large proportion of the short RNAs were predicted to form stem–loop structures resembling those seen with the A-repeat region in Xist RNA. Consistent with such a link, EMSA assays indicated binding of the short RNAs to PRC2, and RIP assays demonstrated that short promoter RNAs co-IP with PRC2 subunits. The EMSA assays indicated an interaction between A-repeat/short promoter RNAs and the SUZ12 PRC2 subunit but not the EZH2 subunit, as was reported in studies on Xist and HOTAIR ncRNAs (Kaneko et al. 2010; Tsai et al. 2010). Further studies are required to define the domain in Suz12 responsible for ncRNA interaction and to resolve the apparent discrepancy regarding which PRC2 subunit interacts with the ncRNAs (see further discussion below).
Genome-scale approaches have been adopted to identify RNAs associated with PRC2 in ES cells using RIP coupled to high-throughput sequencing (Zhao et al. 2010). In this study, some 104 individual transcripts, comprising from 10% to 25% of the ES cell transcriptome, were identified as binding to PRC2. There was no evidence for a bias toward ncRNAs and, although some defined PRC2 RNA targets were found (for example, Xist and the antisense RNA Tsix), others were not (Kcnq1ot1). One concern, in view of the large number of transcripts identified, is the extent to which the RIP assay discriminates specific vs. nonspecific interactions. Evidence for specificity was provided for a small subset of candidate ncRNAs using a variation on the cross-linking IP (CLiP) assay. However, CLiP steps employed by others to purify only RNA that is UV cross-linked to protein were omitted, and given the low efficiency of UV cross-linking (2%–5%), these assays essentially recapitulate the non-UV cross-link RIP experiments. Challenges associated with discriminating specific and nonspecific ncRNA-protein interactions are explored further in the discussion below.
High-throughput approaches have also been applied to determine the link between PRC targeting and long intergenic ncRNAs, i.e., those that were mapped based on transcription-associated chromatin signatures (Khalil et al. 2009). In this study, RNA isolated by PRC2 RIP was hybridized to custom tiling arrays comprising lincRNA sequences and sequences from a similar number of protein-coding mRNAs. The analysis demonstrated that lincRNAs, notably those that were most strongly expressed, were preferentially associated with PRC2 relative to mRNA. Functional analysis of novel lincRNAs associated with PRC2 suggested a role in trans, similar to that proposed for HOTAIR.
In addition to PRC2 binding by lincRNAs, the aforementioned study also reported a significant interaction of lincRNAs with CoREST, a distinct chromatin-modifying complex linked to gene repression. Building on this, a subsequent study demonstrated that, in ES cells, several chromatin-modifying complexes, including factors involved in reading, writing, and erasing specific histone modifications, are associated with lincRNAs (Guttman et al. 2011). These lincRNAs were found to synergize in regulating ES cell pluripotency and differentiation, and it was suggested that this was attributable to their role in guiding chromatin-modifying complexes to defined target loci. In a departure from other studies discussed above, RIP was carried out following formaldehyde cross-linking. This step helps to ensure that only interactions occurring in vivo are preserved. However, formaldehyde cross-links protein-to-protein as well as protein-to-nucleic acid and, as such, does not discriminate direct and indirect protein–RNA interactions that may occur, for example, at the loci from which lincRNAs are transcribed.
A role for ncRNA in recruitment of PRC1 complexes
Although there has been considerable focus on potential interactions between PRC2 proteins and ncRNA, there is also evidence linking ncRNAs to the recruitment of PRC1 complexes. As discussed above, PRC1 recruitment has generally been attributed to interaction of the chromodomain (CD) present in the core PRC1 protein PC (CBX2/4/6/7/8 in mammals) with PRC2-mediated H3K27me3 (Fig. 2C). Although the CD domain is generally associated with binding methylated lysine residues, there are examples in which the CD domain found in other proteins has been linked to RNA binding (for review, see Bernstein and Allis 2005). This prompted an analysis of RNA binding activity of the mammalian PC homologs (Bernstein et al. 2006). In vitro EMSA assays demonstrated CD binding to ssRNA in all cases, except CBX2, albeit without apparent sequence specificity. In a cellular context, CBX7 was found to localize to Xi domains in XX cells, and this association was disrupted following treatment of cells with RNase, lending support to the idea that ncRNA (Xist) has a direct role in localizing PRC1 complexes.
In a more recent study, the CBX7 CD has been shown to interact with the ncRNA ANRIL (Yap et al. 2010). Prior studies had demonstrated that PRC1 complexes, and specifically CBX7, are important for repression at the human cyclin-dependent kinase inhibitor locus INK4b/ARF/INK4a, a master regulator of cellular senescence in response to cellular stress (Gil et al. 2004). This was initially attributed solely to the interaction of CBX7 with PRC2-mediated H3K27me3. However, subsequent studies identified an antisense ncRNA at the INK4b/ARF/INK4a locus, termed ANRIL (Pasmant et al. 2007), and depletion of ANRIL RNA and/or H3K27me3 was found to impact on the ability of CBX7 to repress the INK4b/ARF/INK4a locus (Yap et al. 2010). EMSA assays were used to show that the CBX7-PRC1 complex binds ANRIL RNA in a specific manner, and candidate sequences/ structures within ANRIL RNA were then identified. Interaction of these sequences/structures, and also of the H3K-27me3 peptide with the CBX7 CD was analyzed by biophysical methods, including NMR. Thus, it was possible to identify key residues required for contacting RNA or H3K27me3 peptide and, from there, to determine specific mutations that disrupted either ANRIL or H3K27me3 binding. Analysis of these mutants in vivo demonstrated that ablation of either H3K-27me3 or RNA binding compromises the capacity of CBX7 to repress INK4b/ARF/INK4a repression. It should be noted that a costructure of the CBX7 CD with RNA was not reported, and further analysis is needed to substantiate the proposed interactions.
Although there is evidence supporting a link between specific ncRNAs and PRC1 recruitment, this is unlikely to be pivotal. Thus, ChIP-seq analysis in PRC2 mutant ES cells has demonstrated that CBX7 occupancy at target loci is entirely dependent on H3K27me3, indicating that CD-mediated interactions with ncRNA, where they exist, are not sufficient to target PRC1 complexes (Tavares et al. 2012).
As discussed above, recruitment of PRC1 also occurs through an H3K27me3-independent pathway (Schoeftner et al. 2006; Tavares et al. 2012). Thus, a distinct subfamily of PRC1-related complexes in which the CBX subunit is replaced by the protein RYBP (see Fig. 2B,C) is targeted to closely overlapping target genes, both in the presence and absence of PRC2-mediated H3K27me3. Moreover, RYBP-PRC1 complexes were shown to account for H3K27me3-independent deposition of H2AK119u1 on Xi. The fact that PRC2 and RYBP-PRC1 localize to essentially the same target loci, including Xi, suggests that both of these complexes could be recruited by the same signal. The most parsimonious model is that this signal is common to Xi and to PcG target genes genome-wide, and in this regard, ncRNAs are a good candidate. Consistent with this idea, RYBP has a RanBP2-zinc finger, a domain that, in some other proteins, has been shown to bind RNA (Loughlin et al. 2009; Nguyen et al. 2011). However, sequence comparisons indicate that key RNA contact residues in RanBP2-ZnF, as determined from cocrystal structures, are not conserved in RYBP, suggesting that the RYBP RanBP2-ZnF more likely belongs to a distinct subset of RanBP2-zinc fingers, likely those that interact with ubiquitin (Arrigoni et al. 2006). This does not rule out that RYBP-PRC1 complexes interact with ncRNA by some other unidentified mechanism.
DISCUSSION
The aforementioned examples, when viewed collectively, provide a compelling argument that ncRNAs recruit PcG repressors by a direct mechanism and, as such, using both in cis and in trans mechanisms, play a key role in directing PcG complexes to defined target loci. The development of these ideas owes much to observations made on the role of Xist RNA in recruiting PRC2 in cis to Xi. As such, it should be viewed, with perhaps more than a little concern, that there are confounding observations in the X inactivation field that are difficult to reconcile with the direct recruitment model. Specifically, there are circumstances in which recruitment of PcG proteins does not occur following expression and accumulation of wild-type Xist RNA. Thus, in early mouse preimplantation embryos, Xist expression is first seen at the two-cell stage, coincident with activation of the embryonic genome (Nesterova et al. 2001). However, recruitment of PRC2 and appearance of associated H3K27me3 is not evident until the 8- to 16-cell stage, some 36 h later (Okamoto et al. 2004). Additionally, although induction of Xist RNA transgene expression in undifferentiated ES cells and in cells in early stages of differentiation leads to PRC2/PRC1 recruitment, if transgenes are induced after this time, or in fibroblast cell lines, PRC2/PRC1 recruitment and associated chromatin modifications do not occur (Kohlmaier et al. 2004). In both of these examples, Xist RNA coats the Xi territory, forming large domains in interphase nuclei, although in the latter case, expression of transgenes in differentiated cells, Xist RNA expression does not elicit chromosome silencing (Wutz and Jaenisch 2000). Importantly, in both instances, PRC1/2 apparently localizes and functions normally at other PcG target loci. It could be argued that because PcG levels are significantly lower in differentiated cells, induced Xist RNA expression is not sufficient to recruit detectable levels. However, as mentioned above, even the relatively low levels of PcG recruitment to Xi occurring normally in differentiated cells are sufficient to establish H3K27me3 on Xi (Plath et al. 2003; Silva et al. 2003).
The aforementioned considerations do not disprove direct interaction between Xist RNA and core PRC2 proteins but rather highlight the need to remain open to alternative models. With this in mind, how might we view some of the data obtained for other models that invoke a direct interaction between ncRNA and PcG proteins? At the heart of many of these studies is the use of in vitro EMSA assays and RIP to demonstrate interactions in vitro and in vivo, respectively. It is notable that none of the core PcG proteins, with the possible exception of CBX7, has a previously identified RNA binding domain (RBD). As discussed above, a domain in EZH2 has been implicated in ncRNA interaction (Kaneko et al. 2010), although whether or not we should consider this a bona fide RBD remains a matter of debate. Indeed, preliminary evidence suggests that UV cross-linking assays do not detect the interaction between PRC2 subunits and A-repeat RNA (Maenner et al. 2010). The issue of unidentified RBDs extends to other chromatin-modifying factors suggested to interact directly with ncRNAs, specifically G9a (Nagano et al. 2008), LSD1 (Terranova et al. 2008), DNMT1 (Mohammad et al. 2010), and the numerous chromatin complexes implicated in lincRNA binding in ES cells (Guttman et al. 2011). While it is possible that these factors have all evolved novel RBDs, an alternative explanation is that the assays employed to date do not sufficiently discriminate specific and nonspecific interactions.
Could nonspecific interactions account for data obtained for PRC2-ncRNA interactions observed using EMSA and RIP assays? In the case of EMSA assays, RNAs antisense to the ncRNA, or in some cases, mutated RNAs in which predicted secondary structures were disrupted, were used as negative controls. Thus, the interaction of A-repeat RNA with EZH2 occurs using antisense RNA but not with an RNA in which predicted A-repeat stem–loop structures are disrupted (Zhao et al. 2008). Conversely, EMSA experiments with HOTAIR ncRNA demonstrate interaction with PRC2 and sense, but not antisense, RNAs (Rinn et al. 2007). Either of these findings could be interpreted to show specific interactions but could also be attributed to nonspecific interactions that favor more structured RNAs.
In the case of RIP assays, there are related, and also other, issues. Prior to RIP, cell/nuclei are disrupted using specific buffers and sonication, and soluble material is then subjected to RIP. Because there is no cross-linking involved, it cannot be easily determined whether association of a given RNA is present in intact cells or occurs during subsequent processing, for example, via nonspecific binding to basic patches on protein surfaces. It is not obvious how to control for this. Thus, use of IgG IPs or IPs in cells deficient for the protein of interest, as has been described, only define the nonspecific background of the assay, not the nonspecific binding to the protein of interest. Parallel RIP, using antibodies to an unrelated protein, may give some indication of nonspecific binding, but it is not obvious how to select a protein suitable for this purpose.
A further issue with RIP assays relates to recovery. There is an assumption in many cases that soluble extracts recover ncRNAs efficiently. However, Xist RNA has been shown to remain associated with the nuclear matrix following extraction of chromatin (Clemson et al. 1996) and, as such, may be strongly associated with the insoluble pellet fraction that is discarded during preparation of extracts. Thus, any conclusions as to which proteins associate with soluble Xist RNA have to be tempered with the caveat that this may not reflect what occurs with the bulk of Xist RNA complexes that remain insoluble. Further biochemical studies are needed to assess the extent to which this caveat may apply to other ncRNAs linked to recruitment of chromatin-modifying factors.
If we consider that current data supporting a direct interaction of ncRNA with PRC2 proteins is open to interpretation, what are the killer experiments that could ultimately prove or disprove this hypothesis? Clearly, the first step must be to define the putative RBD domains present in these factors. Progress toward this end has been achieved in the case of EZH2, with the definition of the short region of the protein, residues 342–370, required for interaction with A-repeat and HOTAIR ncRNA (Kaneko et al. 2010). The region includes a number of arginine and lysine residues, consistent with it having a role in charge-mediated interactions with RNA, and, as discussed above, interaction between this region and ncRNAs in vitro is enhanced by phosphorylation of threonine 345. With this information in hand, it should be possible to identify key residues required for the ncRNA interaction and then to investigate how their mutation affects recruitment by ncRNA. A simple experiment would be to determine if wild-type or mutant proteins localize to Xi domains in cells that are expressing Xist RNA.
A second important direction for future experiments will be to investigate PRC2 interactions with ncRNA using CLiP (Ule et al. 2003) or the related PAR-CLIP (Hafner et al. 2010) or iCLiP (Konig et al. 2010) methods in which use of UV cross-linking circumvents reassociation artifacts that occur in RIP assays. This is distinct from the use of formaldehyde for cross-linking which, as discussed above, has the disadvantage that it captures protein-protein as well as protein-nucleic acid interactions. A further advantage with using CLiP methods is that the methodology can be used to define the regions of target RNAs that interact with RBDs, potentially down to nucleotide resolution in the case of PAR-CLiP and iCLiP. Moreover, when coupled to high-throughput sequencing, CLiP assays can be used to survey target RNAs on a genome-wide basis. Such analyses would be further enhanced with knowledge of the putative RBDs present in PRC2 proteins, as discussed above. Thus, CLiP assays could be carried out for wild-type and RBD mutant proteins, reducing background from nonspecific binding. An obvious caveat to this suggestion is that studies to date have failed to detect direct interactions between PRC2 and Xist A-repeats following UV cross-linking (Maenner et al. 2010).
The above considerations indicate that the hypothesis invoking direct interaction between PRC2 proteins and specific ncRNAs should be treated with some caution at this time. But what are the alternatives? How else could we account for key observations such as the strict requirement for ongoing Xist RNA expression to bring about PRC2 recruitment to Xi? As discussed above, studies analyzing PcG recruitment using inducible Xist transgenes lacking the A-repeat provide a strong argument favoring direct over indirect recruitment mechanisms. Specifically, A-repeat-deficient Xist RNA localizes to the chromosome and recruits PcG proteins in the absence of gene silencing, (Kohlmaier et al. 2004), indicating that indirect recruitment via silencing-associated chromatin modifications is unlikely. However, recent data obtained using this experimental model suggests that this conclusion may have been premature (Pullirsch et al. 2010). In the initial studies, it was demonstrated that induction of A-repeat mutant Xist transgenes results in PcG recruitment to Xist RNA territories in the absence of overt gene silencing. PcG recruitment was relatively weak in undifferentiated ES cells but increased significantly as cells underwent differentiation. As is the case for wild-type Xist RNA, PcG recruitment did not occur if the transgene was induced in fully differentiated cells. Moreover, PcG recruitment was lost when Xist transgene expression was discontinued. Curiously, when transgene expression was induced during early differentiation stages, discontinued, and then reinduced in late differentiation stages, PcG recruitment did occur (see Fig. 3). This indicates that Xist transgene expression during early differentiation stages must confer a memory on the transgene-bearing chromosome such that reinduction of Xist RNA now recruits PcG proteins. On first inspection, these observations probably seem perplexing. Xist RNA lacking the A-repeats fails to silence, so how does it establish a chromosomal memory? In the recent study, it was found that, although Xist RNA lacking the A-repeats fails to silence X-linked genes, specific chromosomal histone modifications associated with X inactivation do occur (Pullirsch et al. 2010). Thus, immunofluorescence analysis of metaphase chromosomes demonstrated that loss of histone H4 acetylation and H3K4me2/3, modifications associated with gene activity, is clearly evident. When transgene expression was discontinued, H3K4me2/3 was restored, but H4 hypoacetylation remained for a number of days, suggesting that this could constitute the memory mark required for PcG recruitment in response to re-expression of the Xist transgene in late-stage differentiated cells.
FIGURE 3.
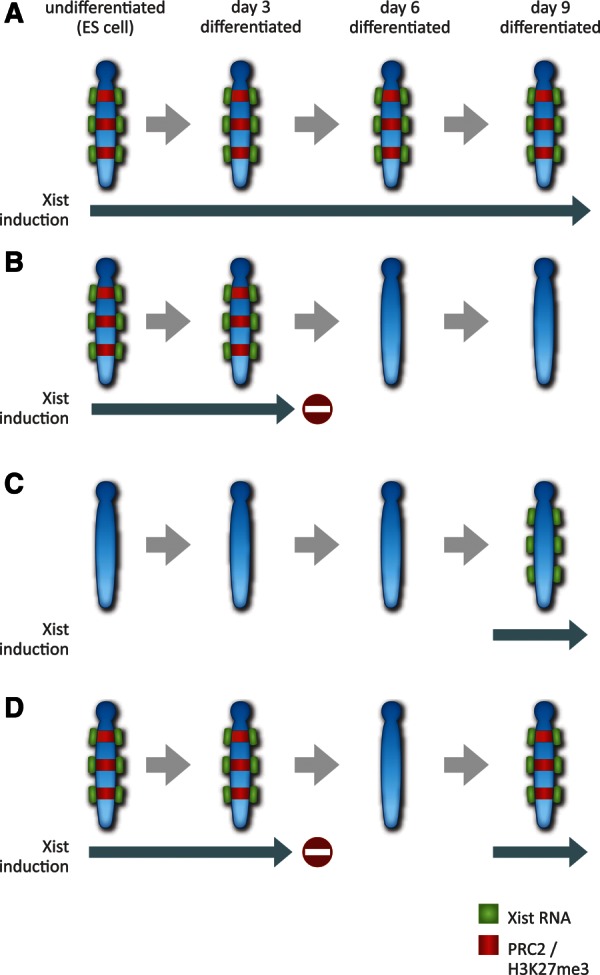
Polycomb recruitment to the inactive X chromosome in response to A-repeat-deficient Xist transgene expression. Schematic illustrates the outcome of experiments inducing A-repeat-deficient Xist RNA transgenes in differentiating ES cells (Kohlmaier et al. 2004; and see text). (A) Induction of Xist RNA and chromosome coating in undifferentiated ES cells, with maintenance throughout subsequent differentiation, results in continual chromosomal localization of PRC2 and associated H3K27me3. (B) Cessation of Xist induction after ∼3 d results in rapid loss of PRC2/H3K27me3. (C) Xist induction and chromosome coating in late-stage differentiated cells does not recruit PRC2 unless (D) there was a transient expression of Xist with associated PRC2 recruitment in early differentiating cells. Collectively, these results demonstrate that early expression of the Xist transgenes confers an epigenetic memory to the chromosome that is required in order for differentiated cells to recruit PRC2 proteins in response to Xist RNA coating.
The fact that A-repeat mutant Xist RNA induces chromosome-wide chromatin modifications other than those attributable to PcG protein function means that we cannot rule out the possibility that PcG recruitment by Xist RNA occurs by an indirect mechanism. Specifically, PcG recruitment could occur in response to Xist-dependent changes in underlying chromatin structure rather than via direct interaction with Xist RNA (Fig. 4). Reduced efficiency of PcG recruitment relative to wild-type Xist RNA could thus reflect that underlying chromatin modifications are deposited less robustly/widely. This model would better explain the memory effect, where PcG occupancy is established in differentiated cells only after an initial burst of Xist expression during early differentiation. Consistent with this idea, recent studies have begun to identify a role for specific histone modifications (Schmitges et al. 2011) and the compaction state of chromatin (Yuan et al. 2012) in defining sites of PcG protein occupancy.
FIGURE 4.
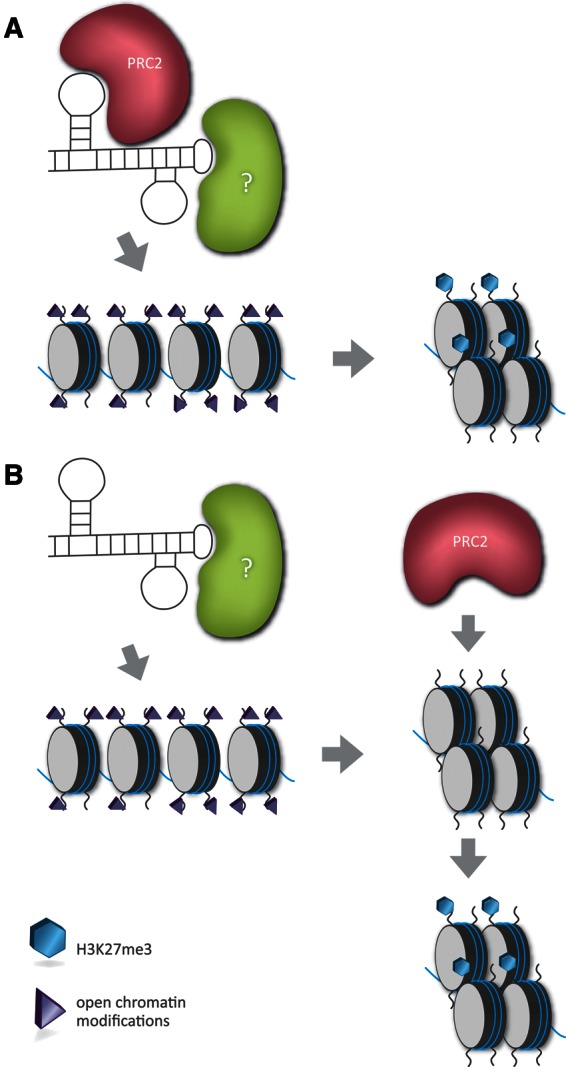
Direct vs. indirect models for PRC2 recruitment in response to ncRNA expression. In the direct model (A), PRC2 complex binds to the ncRNA via an RBD domain and then, together with other RNA-bound chromatin modifiers (?), remodels the chromatin at sites where the ncRNA localizes. PRC2-mediated H3K27me3 is a component part of the resultant compacted chromatin configuration and could potentially have a causative role in establishing the modified state. In the indirect model (B), the ncRNA first modifies the underlying chromatin structure, for example, by erasing active chromatin marks, and the resultant compacted/modified chromatin then functions as a signal to recruit PRC2. In this scenario, the primary function of PRC2/H3K27me3 may be in facilitating stable propagation of modified/compacted chromatin states that were established by primary silencing factors.
Could the indirect model account for data linking other ncRNAs, for example, Kcnq1ot1 or HOTAIR, to PcG recruitment? To put it another way, is it possible that PcG recruitment by ncRNAs is, in general, a secondary consequence of chromatin modifications that occur as a result of the ncRNA being expressed? In principle, probably yes, although, unlike the case of Xist RNA, there are as yet no specific experimental findings that cast doubt on the direct recruitment model. Extending the analysis of putative direct interactions as detailed above should further inform us as to whether or not this needs to be considered.
CONCLUDING REMARKS
In this review, I have discussed our current understanding of ncRNAs that are implicated in long-range gene silencing, either in cis or in trans, and on the emerging theme linking such ncRNAs with recruitment and targeting of chromatin-modifying complexes, notably PcG repressors. I have provided an overview of the evidence supporting this exciting new model for genome regulation by ncRNA. I have also highlighted contradictory observations and alternative interpretations of both genetic and biochemical studies that have been carried out to date. Somewhat like the little boy, I find myself wondering if the emperor is wearing any clothes? Perhaps more accurately, I am concerned that the emperor may be a bit underdressed for the occasion! This view may, of course, turn out to be entirely unfounded, and it may well be that further studies fully substantiate the direct recruitment model. If not, at least two important and interesting new questions will emerge: What are the indirect mechanisms that target PcG complexes and other chromatin-modifying factors? And, if PcG recruitment is an indirect consequence of ncRNA-mediated silencing, what are the primary mechanisms that first establish long-range gene silencing in cis or in trans?
ACKNOWLEDGMENTS
I thank Sarah Cooper and Andrew Bassett for critical reading of the manuscript and stimulating discussions. N.B. is a Wellcome Trust Principal Research Fellow (grant number 081385).
REFERENCES
- Arrigoni R, Alam SL, Wamstad JA, Bardwell VJ, Sundquist WI, Schreiber-Agus N 2006. The Polycomb-associated protein Rybp is a ubiquitin binding protein. FEBS Lett 580: 6233–6241 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Allis CD 2005. RNA meets chromatin. Genes Dev 19: 1635–1655 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD 2006. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol 26: 2560–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. 2004. Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246 [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S 1992. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71: 515–526 [DOI] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF 1991a. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349: 38–44 [DOI] [PubMed] [Google Scholar]
- Brown CJ, Lafreniere RG, Powers VE, Sebastio G, Ballabio A, Pettigrew AL, Ledbetter DH, Levy E, Craig IW, Willard HF 1991b. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature 349: 82–84 [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafreniere RG, Xing Y, Lawrence J, Willard HF 1992. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71: 527–542 [DOI] [PubMed] [Google Scholar]
- Brzeska K, Brzeski J, Smith J, Chandler VL 2010. Transgenic expression of CBBP, a CXC domain protein, establishes paramutation in maize. Proc Natl Acad Sci 107: 5516–5521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Cullen BR 2007. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 13: 313–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Casanova M, Preissner T, Cerase A, Poot R, Yamada D, Li X, Appanah R, Bezstarosti K, Demmers J, Koseki H, et al. 2011. Polycomblike 2 facilitates the recruitment of PRC2 Polycomb group complexes to the inactive X chromosome and to target loci in embryonic stem cells. Development 138: 1471–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J, Waters PD, Koina E, Gilbert C, Robinson TJ, Graves JA 2011. Evolution from XIST-independent to XIST-controlled X-chromosome inactivation: Epigenetic modifications in distantly related mammals. PLoS One 6: e19040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY 2011. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell 44: 667–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB 1996. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol 132: 259–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196 [DOI] [PubMed] [Google Scholar]
- De Lucia F, Crevillen P, Jones AM, Greb T, Dean C 2008. A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci 105: 16831–16836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. 2004. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7: 663–676 [DOI] [PubMed] [Google Scholar]
- Duret L, Chureau C, Samain S, Weissenbach J, Avner P 2006. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312: 1653–1655 [DOI] [PubMed] [Google Scholar]
- Duszczyk MM, Wutz A, Rybin V, Sattler M 2011. The Xist RNA A-repeat comprises a novel AUCG tetraloop fold and a platform for multimerization. RNA 17: 1973–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duthie SM, Nesterova TB, Formstone EJ, Keohane AM, Turner BM, Zakian SM, Brockdorff N 1999. Xist RNA exhibits a banded localization on the inactive X chromosome and is excluded from autosomal material in cis. Hum Mol Genet 8: 195–204 [DOI] [PubMed] [Google Scholar]
- Engemann S, Strodicke M, Paulsen M, Franck O, Reinhardt R, Lane N, Reik W, Walter J 2000. Sequence and functional comparison in the Beckwith–Wiedemann region: Implications for a novel imprinting centre and extended imprinting. Hum Mol Genet 9: 2691–2706 [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811 [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev 17: 1870–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick GV, Soloway PD, Higgins MJ 2002. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat Genet 32: 426–431 [DOI] [PubMed] [Google Scholar]
- Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forne T, Jammes H, Ainscough JF, Surani MA, Journot L, et al. 2009. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development 136: 3413–3421 [DOI] [PubMed] [Google Scholar]
- Gil J, Bernard D, Martinez D, Beach D 2004. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol 6: 67–72 [DOI] [PubMed] [Google Scholar]
- Grant J, Mahadevaiah SK, Khil P, Sangrithi MN, Royo H, Duckworth J, McCarrey JR, VandeBerg JL, Renfree MB, Taylor W, et al. 2012. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature 487: 254–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. 2011. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952 [DOI] [PubMed] [Google Scholar]
- Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM 2000. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature 405: 486–489 [DOI] [PubMed] [Google Scholar]
- Heo JB, Sung S 2011. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79 [DOI] [PubMed] [Google Scholar]
- Herzing LB, Romer JT, Horn JM, Ashworth A 1997. Xist has properties of the X-chromosome inactivation centre. Nature 386: 272–275 [DOI] [PubMed] [Google Scholar]
- Kalantry S, Magnuson T 2006. The Polycomb group protein EED is dispensable for the initiation of random X-chromosome inactivation. PLoS Genet 2: e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D 2010. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev 24: 2615–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araujo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. 2010. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell 38: 675–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP, Gingeras TR 2002. Large-scale transcriptional activity in chromosomes 21 and 22. Science 296: 916–919 [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. 2009. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A 2004. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol 2: E171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J 2010. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17: 909–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Jaenisch R 1997. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature 386: 275–279 [DOI] [PubMed] [Google Scholar]
- Lee JT, Strauss WM, Dausman JA, Jaenisch R 1996. A 450 kb transgene displays properties of the mammalian X-inactivation center. Cell 86: 83–94 [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D 1999. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet 21: 400–404 [DOI] [PubMed] [Google Scholar]
- Letswaart R, Wu Z, Dean C 2012. Flowering time control: Another window to the connection between antisense RNA and chromatin. Trends Genet 28: 445–453 [DOI] [PubMed] [Google Scholar]
- Lewis A, Mitsuya K, Umlauf D, Smith P, Dean W, Walter J, Higgins M, Feil R, Reik W 2004. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat Genet 36: 1291–1295 [DOI] [PubMed] [Google Scholar]
- Loughlin FE, Mansfield RE, Vaz PM, McGrath AP, Setiyaputra S, Gamsjaeger R, Chen ES, Morris BJ, Guss JM, Mackay JP 2009. The zinc fingers of the SR-like protein ZRANB2 are single-stranded RNA-binding domains that recognize 5′ splice site-like sequences. Proc Natl Acad Sci 106: 5581–5586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF 1961. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190: 372–373 [DOI] [PubMed] [Google Scholar]
- Maenner S, Blaud M, Fouillen L, Savoye A, Marchand V, Dubois A, Sanglier-Cianferani S, Van Dorsselaer A, Clerc P, Avner P, et al. 2010. 2-D structure of the A region of Xist RNA and its implication for PRC2 association. PLoS Biol 8: e1000276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager J, Montgomery ND, de Villena FP, Magnuson T 2003. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat Genet 33: 502–507 [DOI] [PubMed] [Google Scholar]
- Mahadevaiah SK, Royo H, VandeBerg JL, McCarrey JR, Mackay S, Turner JM 2009. Key features of the X inactivation process are conserved between marsupials and eutherians. Curr Biol 19: 1478–1484 [DOI] [PubMed] [Google Scholar]
- Mak W, Baxter J, Silva J, Newall AE, Otte AP, Brockdorff N 2002. Mitotically stable association of polycomb group proteins eed and enx1 with the inactive x chromosome in trophoblast stem cells. Curr Biol 12: 1016–1020 [DOI] [PubMed] [Google Scholar]
- Mak W, Nesterova TB, de Napoles M, Appanah R, Yamanaka S, Otte AP, Brockdorff N 2004. Reactivation of the paternal X chromosome in early mouse embryos. Science 303: 666–669 [DOI] [PubMed] [Google Scholar]
- Marques AC, Ponting CP 2009. Catalogues of mammalian long noncoding RNAs: Modest conservation and incompleteness. Genome Biol 10: R124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Mondal T, Guseva N, Pandey GK, Kanduri C 2010. Kcnq1ot1 noncoding RNA mediates transcriptional gene silencing by interacting with Dnmt1. Development 137: 2493–2499 [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P 2008. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322: 1717–1720 [DOI] [PubMed] [Google Scholar]
- Nesterova TB, Barton SC, Surani MA, Brockdorff N 2001. Loss of Xist imprinting in diploid parthenogenetic preimplantation embryos. Dev Biol 235: 343–350 [DOI] [PubMed] [Google Scholar]
- Nguyen CD, Mansfield RE, Leung W, Vaz PM, Loughlin FE, Grant RP, Mackay JP 2011. Characterization of a family of RanBP2-type zinc fingers that can recognize single-stranded RNA. J Mol Biol 407: 273–283 [DOI] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E 2004. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 303: 644–649 [DOI] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C 2008. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 32: 232–246 [DOI] [PubMed] [Google Scholar]
- Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I 2007. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: Identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res 67: 3963–3969 [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N 1996. Requirement for Xist in X chromosome inactivation. Nature 379: 131–137 [DOI] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A 2006. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell 127: 1209–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300: 131–135 [DOI] [PubMed] [Google Scholar]
- Plath K, Talbot D, Hamer KM, Otte AP, Yang TP, Jaenisch R, Panning B 2004. Developmentally regulated alterations in Polycomb repressive complex 1 proteins on the inactive X chromosome. J Cell Biol 167: 1025–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Ponting CP, Lunter G 2007. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res 17: 556–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullirsch D, Hartel R, Kishimoto H, Leeb M, Steiner G, Wutz A 2010. The Trithorax group protein Ash2l and Saf-A are recruited to the inactive X chromosome at the onset of stable X inactivation. Development 137: 935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redrup L, Branco MR, Perdeaux ER, Krueger C, Lewis A, Santos F, Nagano T, Cobb BS, Fraser P, Reik W 2009. The long noncoding RNA Kcnq1ot1 organises a lineage-specific nuclear domain for epigenetic gene silencing. Development 136: 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regha K, Sloane MA, Huang R, Pauler FM, Warczok KE, Melikant B, Radolf M, Martens JH, Schotta G, Jenuwein T, et al. 2007. Active and repressive chromatin are interspersed without spreading in an imprinted gene cluster in the mammalian genome. Mol Cell 27: 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY 2012. Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripoche MA, Kress C, Poirier F, Dandolo L 1997. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev 11: 1596–1604 [DOI] [PubMed] [Google Scholar]
- Sanchez-Elsner T, Gou D, Kremmer E, Sauer F 2006. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science 311: 1118–1123 [DOI] [PubMed] [Google Scholar]
- Schmitges FW, Prusty AB, Faty M, Stutzer A, Lingaraju GM, Aiwazian J, Sack R, Hess D, Li L, Zhou S, et al. 2011. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42: 330–341 [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A 2006. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J 25: 3110–3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorderet P, Duboule D 2011. Structural and functional differences in the long non-coding RNA Hotair in mouse and human. PLoS Genet 7: e1002071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N 2003. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell 4: 481–495 [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE 2009. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP 2002. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415: 810–813 [DOI] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C 2009. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802 [DOI] [PubMed] [Google Scholar]
- Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, et al. 2012. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148: 664–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, Orkin SH, Peters AH 2008. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell 15: 668–679 [DOI] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY 2010. Long noncoding RNA as modular scaffold of histone modification complexes. Science 329: 689–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB 2003. CLIP identifies Nova-regulated RNA networks in the brain. Science 302: 1212–1215 [DOI] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R 2004. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat Genet 36: 1296–1300 [DOI] [PubMed] [Google Scholar]
- Wagschal A, Sutherland HG, Woodfine K, Henckel A, Chebli K, Schulz R, Oakey RJ, Bickmore WA, Feil R 2008. G9a histone methyltransferase contributes to imprinting in the mouse placenta. Mol Cell Biol 28: 1104–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Chang HY 2011. Molecular mechanisms of long noncoding RNAs. Mol Cell 43: 904–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Mager J, Chen Y, Schneider E, Cross JC, Nagy A, Magnuson T 2001. Imprinted X inactivation maintained by a mouse Polycomb group gene. Nat Genet 28: 371–375 [DOI] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y 2004. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431: 873–878 [DOI] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R 2000. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell 5: 695–705 [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet 30: 167–174 [DOI] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM 2010. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38: 662–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RS, Marques AC, Tibbit C, Haerty W, Bassett AR, Liu JL, Ponting CP 2012. Identification and properties of 1,119 candidate lincRNA loci in the Drosophila melanogaster genome. Genome Biol Evol 4: 427–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Wu T, Fu H, Dai C, Wu H, Liu N, Li X, Xu M, Zhang Z, Niu T, et al. 2012. Dense chromatin activates Polycomb repressive complex 2 to regulate H3 lysine 27 methylation. Science 337: 971–975 [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT 2008. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322: 750–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT 2010. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 40: 939–953 [DOI] [PMC free article] [PubMed] [Google Scholar]