Transfer RNAs (tRNAs) are typically considered housekeeping products with little regulatory function. However, misregulation of tRNA expression has been linked to cancer. Furthermore, overexpression of the initiator tRNA (tRNAiMet) in mouse fibroblasts has been shown to increase cell proliferation and promote tumor formation. To further investigate the cellular and physiological effects of tRNA overexpression, the authors overexpressed tRNAiMet in two human breast epithelial cell lines. Overexpression of tRNAiMet significantly altered the global tRNA expression profile and resulted in increased cell metabolic activity and cell proliferation.
Keywords: initiator methionine tRNA, tRNA, tRNA microarrays
Abstract
Transfer RNAs (tRNAs) are typically considered housekeeping products with little regulatory function. However, several studies over the past 10 years have linked tRNA misregulation to cancer. We have previously reported that tRNA levels are significantly elevated in breast cancer and multiple myeloma cells. To further investigate the cellular and physiological effects of tRNA overexpression, we overexpressed tRNAiMet in two human breast epithelial cell lines. We then determined tRNA abundance changes and performed phenotypic characterization. Overexpression of tRNAiMet significantly altered the global tRNA expression profile and resulted in increased cell metabolic activity and cell proliferation. Our results extend the relevance of tRNA overexpression in human cells and underscore the complexity of cellular regulation of tRNA expression.
INTRODUCTION
Misregulation of components of the translation machinery is characteristic of many types of tumor cells and can lead to malignant transformation (Bjornsti and Houghton 2004; Pandolfi 2004). Abnormally high levels of RNA polymerase III transcripts, including tRNA and 5S rRNA which are directly involved in translation, are found in a wide variety of transformed cell types (Marshall and White 2008). These cell types include cell lines transformed by DNA tumor viruses (such as hepatitis B virus), RNA tumor viruses (such as human T-cell leukemia virus 1), and chemical carcinogens. These observations have also been confirmed for tumors in situ by RT-PCR, Northern blot, and more recently by microarray analysis (Chen et al. 1997a,b; Winter et al. 2000; Pavon-Eternod et al. 2009). We have previously reported that tRNA levels are elevated in breast cancer and multiple myeloma cell lines (Pavon-Eternod et al. 2009; Zhou et al. 2009). Though abnormal RNA polymerase III activity has long been associated with cancer, it remains unclear whether it contributes to malignant transformation or is simply a byproduct of the cell’s cancer state.
Due to its unique function in translation initiation, we are particularly interested in the role of the initiator methionine tRNA (tRNAiMet) in cancer. An appealing possibility is that overexpression of tRNAiMet could alter the translational regulation of key genes involved in tumorigenesis. The effect may be both quantitative and qualitative: Overall protein synthesis may be increased, and mRNAs encoding cell-cycle or anti-apoptic proteins (such as Myc or cyclin D1) may be preferentially translated. To explore this question in the context of breast cancer, we set out to overexpress tRNAiMet in human breast epithelial cell lines. Overexpressing tRNAs in human cell lines, however, proved more challenging than we had expected. Here we present our experimental approach to tRNA overexpression and a phenotypic characterization of the resulting cell lines (Fig. 1). We find that tRNA overexpression in human cells requires the generation of stable cell lines, and that only modest increases (1.4- to 2.2-fold) can be achieved. Remarkably, overexpression of tRNAiMet in both epithelial cell lines changed the levels of other tRNAs, reprogramming the global tRNA expression profile. tRNAiMet overexpression also resulted in increased metabolic activity and cell proliferation. Our results underscore the need for caution in interpreting the effects of individual tRNA overexpression, as little is known about the regulation of individual tRNA expression in the cell.
FIGURE 1.
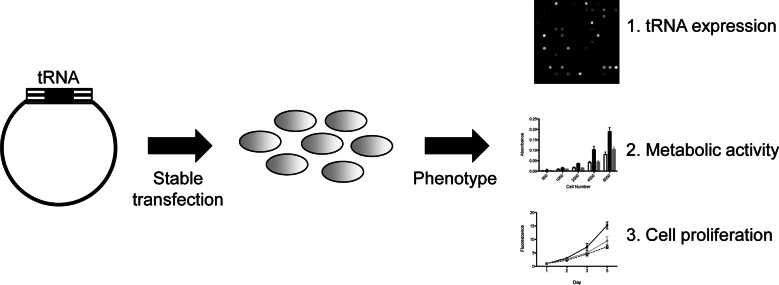
Experimental strategy for tRNA overexpression. The tRNA gene of interest with 200-bp flanking regions was cloned into a mammalian expression vector, then stably transfected into the human cell line. The stable cell line was then characterized in terms of tRNA expression profile, metabolic activity, and cell proliferation.
RESULTS AND DISCUSSION
Experimental strategies for tRNA overexpression
Because tRNAs are generally considered non-regulatory housekeeping genes, there are no well-established methods for manipulating the levels of specific tRNAs in mammalian cells. Furthermore, because tRNAs are highly abundant, overexpression even by twofold would require an additional transcription of ∼1,000,000 molecules of tRNAiMet per cell (Pavon-Eternod et al. 2009). In the past, exogenous tRNA has been introduced into cells either as DNA or directly as RNA (Carbon et al. 1983; Buvoli et al. 2000). We tried three different approaches to increase tRNAiMet levels in two human cell lines: transient transfection with tRNA transcripts, transient transfection with a DNA vector containing the tRNA gene, and stable transfection with a DNA vector containing the tRNA gene. We selected the human breast epithelial cell lines 184A1 and MCF10A for these experiments for the following reasons: (i) They are non-tumorigenic cell lines, (ii) our previous work has shown 184A1 and MCF10A have relatively low levels of tRNA (Pavon-Eternod et al. 2009), and (iii) they are readily transfectable.
Our first approach relied on transient transfection of tRNAiMet and tRNAeMet transcripts. In vitro transcribed tRNAs have been reported to be active in translation when transfected into eukaryotic cells. Indeed, they have been used to insert unnatural amino acids into proteins and to induce amino acid substitutions resulting in widespread proteome damage in mammalian cells (Kohrer et al. 2001; Geslain et al. 2010). In vitro transcribed tRNAs are simple to synthesize and allow direct control over the amount of tRNA transfected. However, the question remains whether they are truly fully functional in the cell. In vitro transcribed tRNAs lack the post-transcriptional modifications characteristic of endogenous tRNAs that serve as identity determinants, contribute to tRNA stability, and impact translational accuracy (Alexandrov et al. 2006; Agris et al. 2007; Waas et al. 2007; Phizicky and Hopper 2010). This is particularly relevant for our tRNA of interest: tRNAiMet transcripts lacking the m1A58 modification are subject to nuclear polyadenylation and rapid degradation (Kadaba et al. 2004, 2006; Vanacova et al. 2005). In any case, we found that in vitro transcribed tRNAs were toxic to MCF10A cells when transfected at high enough concentrations to detectably increase cellular tRNA levels (data not shown).
Our second approach relied on transient transfection of a DNA vector containing a tRNAiMet or tRNAeMet gene into MCF10A cells. Fragments containing the tRNA gene were PCR-amplified from human genomic DNA and cloned into the pTarget Mammalian Expression Vector. The fragments contained 200 base pairs each upstream of and downstream from the endogenous tRNA gene, which should include all the regulatory elements necessary for tRNA transcription (Geiduschek and Kassavetis 2001; Dieci et al. 2007). This approach is experimentally straightforward and allows the transfection of large amounts of DNA without inducing toxicity. High-copy plasmids have been successfully used to induce tRNA overexpression in yeast and bacteria (Borel et al. 1993; Anderson et al. 1998; Sorensen et al. 2005). Again, transfection with our tRNA vectors did not result in any detectable increase in tRNA levels (data not shown).
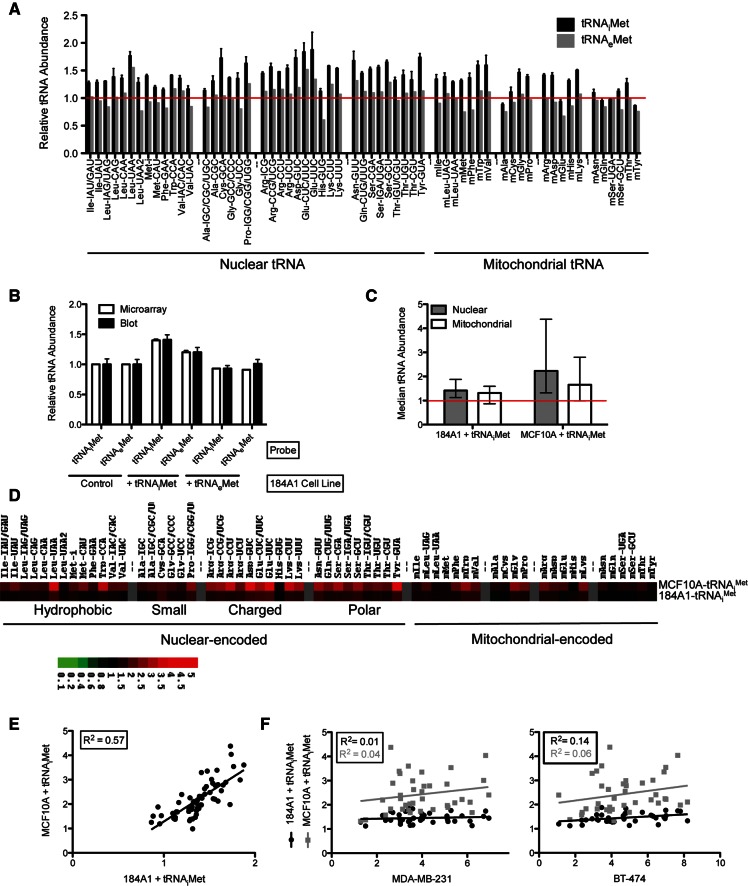
Our third approach involved the generation of stable cell lines after transfection with our tRNAiMet, tRNAeMet, and empty control vectors. Using this approach, we successfully generated an 184A1 cell line and an MCF10A cell line overexpressing tRNAiMet relative to the control cell line (Fig. 2). From this point forward, we designate these cell lines 184A1–tRNAiMet and MCF10A–tRNAiMet. The levels of tRNAiMet increased by 1.4- and twofold, respectively, as measured by tRNA microarrays. This level is comparable to that observed in breast cancer cell lines relative to non-tumorigenic breast epithelial cell lines (Pavon-Eternod et al. 2009). We were, however, unable to generate either a 184A1 or MCF10A cell line overexpressing tRNAeMet relative to the control cell line (Fig. 2A).
FIGURE 2.
tRNAiMet overexpression generates unique tRNA expression profiles. (A) Individual tRNA abundances in 184A1–tRNAiMet and 184A1–tRNAeMet cell lines. Individual tRNA abundance values are shown for 184A1–tRNAiMet (black) and 184A1-tRNAeMet (gray) relative to an empty vector control cell line (set to 1, red line). A value of 1 indicates no change, a value <1 indicates a decrease, and a value >1 indicates an increase in tRNA levels relative to the control cell line. Data are grouped according to amino acid type. The tRNAiMet and tRNAeMet probes are labeled Met-i and Met-CAU, respectively. Where error bars are present, values are averages from dye-swapped experiments and error bars indicate standard deviation. One sample t-test was performed to determine the statistical significance of the changes: *P-value <0.05. (B) Validation of microarray data by dot blot. As in A, relative tRNA abundance is defined as the ratio between the indicated cell line and the control cell line. Relative tRNA abundance values obtained by microarray (white) and dot blot (black, average of three replicates, error bars indicate standard deviation) are plotted for tRNAiMet and tRNAeMet in the three 184A1 cell lines generated for this study (control, 184A1–tRNAiMet and 184A1–tRNAeMet). (C) Median tRNA abundance upon tRNAiMet overexpression. Median values for 184A1–tRNAiMet and MCF10A–tRNAiMet relative to control cell lines (set to 1, red line). Median values for nuclear-encoded tRNAs (gray) and mitochondrial-encoded (white) tRNAs are shown. The upper and lower bars indicate the range of individual tRNA abundances. (D) Heat map of tRNA abundances upon tRNAiMet overexpression. Relative tRNA abundance levels of nuclear and mitochondrial-encoded tRNAs in 184A1–tRNAiMet and MCF10A–tRNAiMet are shown as TreeView images. Data are grouped according to amino acid type. Green indicates a decreased level of expression, red indicates an increased level of expression, and black indicates no change in expression level relative to the reference sample. (E) Individual tRNA abundances in 184A1–tRNAiMet compared with MCF10A–tRNAiMet. Individual tRNA abundance values for 184A1–tRNAiMet and MCF10A–tRNAiMet are relative to the corresponding control cell lines. (F) Individual nuclear-encoded tRNA abundances in two breast cancer lines, MDA-MB-231 (left) and BT-474 (right) compared with 184A1–tRNAiMet and MCF10A–tRNAiMet. Individual tRNA levels for the two breast cancer cell lines, MDA-MB-231 and BT-474, are relative to the breast epithelial MCF10A cell line. Individual tRNA levels for the 184A1–tRNAiMet and MCF10A–tRNAiMet cell lines are relative to the corresponding control cell lines.
To confirm our microarray data, we analyzed tRNAiMet and tRNAeMet content in our 184A1 cell lines (control, 184A1–tRNAiMet, and MCF10A–tRNAiMet) by dot blot (Fig. 2B). In all cases, the microarray and dot blot data were in very good agreement. Our selective fluorophore labeling method requires that all tRNAs measured by microarrays contain 3′CCA (Pavon-Eternod et al. 2009) which is characteristic of all mature tRNAs. The agreement between microarray, which measures mature tRNA, and dot blot data, which measure mature and precursor tRNA, indicates that the observed tRNAiMet overexpression is primarily derived from mature tRNA.
tRNAiMet overexpression generates unique tRNA expression profiles
Unexpectedly, tRNAiMet overexpression induced a significant change in the levels of other tRNAs. Compared with the corresponding control line, median nuclear-encoded tRNA abundance increased by 1.4-fold in 184A1–tRNAiMet and 2.2-fold in MCF10A–tRNAiMet, whereas median mitochondrial-encoded tRNA abundance increased by 1.3-fold in 184A1–tRNAiMet and 1.7-fold in MCF10A–tRNAiMet (Fig. 2C). These changes are more striking for individual tRNAs (Fig. 2A,D). Due to the nature of our microarray measurements, we express individual tRNA abundances in 184A1–tRNAiMet and MCF10A–tRNAiMet relative to the corresponding control cell line. While some tRNAs are increased up to fourfold upon tRNAiMet overexpression, others are not affected. Remarkably, tRNAiMet overexpression generates very similarly altered expression profiles for nuclear-encoded tRNAs in both 184A1 and MCF10A cell lines (R2 = 0.57, Fig. 2E).
We also compared whether the tRNA expression profiles induced by tRNAiMet overexpression in breast epithelial lines are similar to those measured in breast cancer cell lines. Our previous study of tRNA expression in breast cancer revealed that tRNA overexpression is characteristic of breast cancer cells, and that this overexpression is highly selective based on tRNA identity (Pavon-Eternod et al. 2009). We therefore plotted tRNA levels (relative to the breast epithelial cell line MCF10A) in two breast cancer cell lines, MDA-MB-231 and BT-474, vs. the tRNA levels (relative to the corresponding control cell line) in our 184A1–tRNAiMet and MCF10A–tRNAiMet cell lines. We find a very poor correlation between the tRNA levels in bona fide breast cancer cell lines and the tRNA levels induced by tRNAiMet overexpression (R2 < 0.15 in all cases) (Fig. 2F). While the tRNAs carrying charged and polar amino acids were consistently among the most overexpressed tRNAs in the breast cancer cell lines examined, we observe no such trends in our 184A1–tRNAiMet and MCF10A–tRNAiMet lines.
Initiator methionine tRNA overexpression leads to increased cell metabolism and proliferation
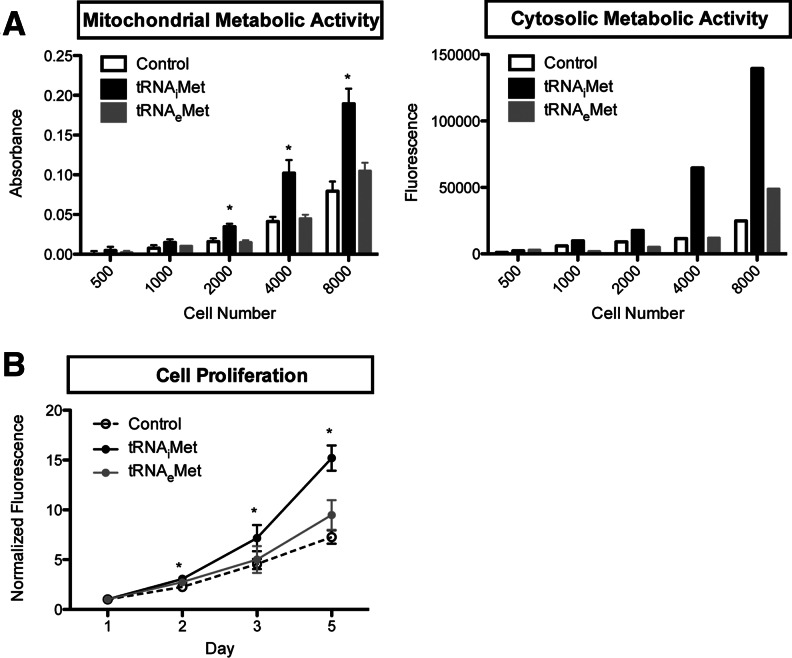
Elevated tRNA levels are characteristic of breast cancer cells (Pavon-Eternod et al. 2009) which often exhibit altered metabolic activity and unregulated growth compared with non-cancer cells. We therefore measured the metabolic activity and cell proliferation of 184A1–tRNAiMet relative to the 184A1–control cell line (Fig. 3). We also included the 184A1–tRNAeMet cell line as an additional control: Even though tRNAeMet expression was not increased in this line as we had expected, we detected punctual changes in the levels of several other tRNAs (such as tRNALeu(UAA) and tRNAGlu(CUC/UUC)) (Fig. 2A), which may have an effect. We first measured the metabolic activity using two assays: Calcein AM, which relies on the activity of cytoplasmic esterases, and WST1, which relies on the activity of mitochondrial dehydrogenases. Both assays showed increased metabolic activity for 184A1–tRNAiMet relative the control cell line, but no change in metabolic activity for 184A1–tRNAeMet. We also measured cell proliferation by Hoechst staining. Again, 184A1–tRNAiMet showed increased cell proliferation relative to the control cell line, but no significant change was seen for 184A1–tRNAeMet.
FIGURE 3.
tRNAiMet overexpression leads to increased cell metabolism and proliferation. Data are shown for the 184A1 cell lines 184A1–control, 184A1–tRNAiMet, and 184A1–tRNAeMet. T-tests were performed to determine the statistical significance of the differences observed relative to 184A1–control: *P-value <0.05. (A) Metabolic activity. Mitochondrial metabolic activity was measured by WST1, which relies on the activity of mitochondrial dehydrogenases. Cytosolic metabolic activity was measured by Calcein AM, which relies on the activity of cytoplasmic esterases. Where error bars are indicated, assays were performed in triplicate and the error bars indicate standard deviation. (B) Cell proliferation. Cell proliferation was measured over 5 d by Hoechst DNA staining. The assays were performed in triplicate; error bars indicate standard deviation.
Concluding remarks
In our experience, overexpressing a specific tRNA in human cell lines is not trivial. Of three possible approaches attempted by us, only one was successful: the generation of stable cell lines after transfection with a DNA vector containing the tRNA gene. Even so, we were able to generate cell lines stably overexpressing the desired tRNA for only one of two tRNAs. While we successfully generated stable cell lines overexpressing tRNAiMet, we were unable to generate stable cell lines overexpressing tRNAeMet to any detectable level. This may be due to some intrinsic properties of tRNAeMet, or other random factors such as the site of integration and copy number. Our results also indicate that manipulating the levels of one specific tRNA—in this case tRNAiMet—significantly affects the levels of other tRNAs in the cell, suggesting some kind of feedback regulatory mechanism in the cell. Remarkably, tRNAiMet overexpression in two different cell lines resulted in similar patterns of tRNA expression. We expect that the tRNA expression profile induced by overexpressing a specific tRNA is dependent on many factors, including but not limited to the identity of the tRNA being introduced, the genetic background of the cell, and the integration sites. Regardless, care must be taken in attributing phenotypic changes to overexpression of an individual tRNA. The increase in metabolic activity and cell proliferation we measure in our 184A1–tRNAiMet cell line may indeed be due to tRNAiMet overexpression, but also to overexpression of a number of other tRNAs or even to globally increased tRNA levels.
Our findings highlight the fact that little is known about the regulation of individual tRNA expression in the cell, and how cells respond to perturbations in tRNA levels. It is generally believed that tRNA transcription via RNA polymerase III is globally regulated in response to nutrient availability and other environmental signals, in coordination with rRNA transcription via RNA polymerase I. Current models hold that transcription at tRNA genes is coordinately regulated by shared transcription factors, acting at highly related promoter sequences (Phizicky and Hopper 2010). This view does not account for the tissue-specific differences in individual tRNA expression or the differential overexpression of individual tRNA species in breast cancer cells (Dittmar et al. 2006; Pavon-Eternod et al. 2009). A systematic study of individual tRNA expression is required to elucidate the functional significance and underlying regulatory mechanisms.
MATERIALS AND METHODS
DNA vectors
Fragments containing tRNA genes were PCR-amplified from human genomic DNA, using the following primer pairs: 5′-TGAGTTGGCAACCTGTGGTA and 5′-TTGGGTGTCCATGAAAATCA for tRNAiMet, 5′-AGCGACCTTCCCACA and 5′-GTCTCCCATTCCTACACG for tRNAeMet. These fragments were cloned into the pTarget Mammalian Expression vector (Promega) following the manufacturer’s instructions.
Cell lines
All cell lines were purchased from American Type Culture Collection (ATCC). MCF10A and 184A1 cells were cultured in 1:1 DMEM/F12 with 2.5 mM L-Gln and 15 mM HEPES (Thermo Scientific HyClone) supplemented with 10% FBS, 1% Penicillin/Streptomycin, 5 μg/mL insulin, 10 ng/mL EGF, and 0.5 μg/mL hydrocortisone. MDA-MB-231 and BT-474 were cultured in RPMI 1640 1× medium (Thermo Scientific HyClone) supplemented with 10% FBS and 1% Penicillin/Streptomycin.
To generate stable cell lines, cells were transfected using Amaxa Nucleofector technology (LonzaBio). After 48 h, medium was supplemented with 500 μg/mL G418 (Sigma) for selection. After 2–4 wk, G418 resistant colonies appeared and the G418 concentration was scaled down to 200 μg/mL. Medium was supplemented with 200 μg/mL G418 for routine culture.
Transfer RNA microarrays
Total RNA for each cell line was obtained at 80%–90% confluency using the miRVana miRNA Isolation Kit (Ambion). This procedure isolates RNA species as short as 15 nt and is therefore not biased against tRNA. Total RNA quality was verified by agarose gel electrophoresis.
The tRNA microarray experiment consists of four steps starting from total RNA: (1) deacylation to remove any amino acids still attached to the tRNA, (2) selective fluorophore labeling of tRNA, (3) hybridization, and (4) data analysis. The tRNA microarray method, including reproducibility and result validation by Northern blot, has been extensively described in previously published papers (Dittmar et al. 2004, 2006; Pavon-Eternod et al. 2009, 2010; Zhou et al. 2009).
Dot blots
The following DNA probes, identical to those spotted on the tRNA microarrays, were used to quantify tRNAiMet and tRNAeMet in total RNA: 5′-AGCAGAGTGGCGCAGCGGAAGCGTGCTGGGCCCATAACCCAGAGGTCGATGGATCGAAACCATCCTCTGCTA-3′ for tRNAiMet, and 5′-GCCYYCTTAGCGCAGYDGGCAGCGCGTCAGTCTCATAATCTGAAGGTCCTGAGTTCGAGCCTCAGAGRGGGCA-3′ for tRNAeMet. Probes were 5′-radiolabeled using T4 polynucleotide kinase and γ-32P-ATP (Perkin-Elmer), followed by purification on a denaturing urea polyacrylamide gel. To detect tRNAiMet and tRNAeMet in total RNA, 100 ng total RNA was spotted and UV crosslinked on a Hybond XL membrane (GE Healthcare). The membrane was pre-hybridized in hybridization buffer (300 mM NaCl, 1% SDS, 20 mM phosphate buffer pH 7) for 30 min at room temperature, then hybridized overnight at 60°C in hybridization buffer containing 100,000–300,000 cpm of the radiolabeled probe. The membrane was then washed three times for 20 min at room temperature in wash buffer (300 mM NaCl, 0.1% SDS, 20 mM phosphate buffer pH 7, 2 mM EDTA). Phosphorimaging was used to quantify the amount of tRNAiMet and tRNAeMet present in each sample.
Metabolic activity assays
Cells were plated in 100 μL medium in 96-well plate at the following cell densities: 500, 1000, 2000, 4000, and 8000 cells/well. For WST1 assays: 10 μL WST1 reagent (Roche 05 015 944 001) was added to each well. Absorbance at 440 nm was read after 1 h incubation at 37°C. For Calcein AM assays: Cells were incubated in 200 μL Calcein AM (BD 354216) working solution (1 μM in HBSS) for 1 h at 37°C. Fluorescence was measured at 490ex/520em.
Cell proliferation assays
Cell proliferation was measured over 5 d using Hoechst DNA staining (Invitrogen H1398). Cells were plated in 100 μL medium at 500 cells/well in 96-well plates. To stain DNA, cells were incubated in 100 μL of 0.1 μg/mL Hoechst solution in HBSS for 1 h at 37°C. After washing with HBSS to remove any unbound dye, fluorescence was measured 355ex/460em. Fluorescence is directly proportional to the number of cells present, regardless of cell type.
Statistical significance
T-tests and one-sample t-tests were performed using GraphPad QuickCalcs (http://www.graphpad.com/quickcalcs/contMenu/).
ACKNOWLEDGMENTS
This work was supported by a grant from CDMRP (W81XWH-10-1-0452, 10-1-0453 to T.P. and M.R.R.). M.P.-E. was supported in part by a Ruth Kirshstein Pre-doctoral Fellowship from the NIH (1F31CA139968).
REFERENCES
- Agris PF, Vendeix FA, Graham WD 2007. tRNA’s wobble decoding of the genome: 40 years of modification. J Mol Biol 366: 1–13 [DOI] [PubMed] [Google Scholar]
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM 2006. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 21: 87–96 [DOI] [PubMed] [Google Scholar]
- Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Bjork GR, Tamame M, Hinnebusch AG 1998. The essential Gcd10p-Gcd14p nuclear complex is required for 1-methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev 12: 3650–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsti MA, Houghton PJ 2004. Lost in translation: Dysregulation of cap-dependent translation and cancer. Cancer Cell 5: 519–523 [DOI] [PubMed] [Google Scholar]
- Borel F, Hartlein M, Leberman R 1993. In vivo overexpression and purification of Escherichia coli tRNAser. FEBS Lett 324: 162–166 [DOI] [PubMed] [Google Scholar]
- Buvoli M, Buvoli A, Leinwand LA 2000. Suppression of nonsense mutations in cell culture and mice by multimerized suppressor tRNA genes. Mol Cell Biol 20: 3116–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon P, Haumont E, Fournier M, de Henau S, Grosjean H 1983. Site-directed in vitro replacement of nucleosides in the anticodon loop of tRNA: Application to the study of structural requirements for queuine insertase activity. EMBO J 2: 1093–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Bocker W, Brosius J, Tiedge H 1997a. Expression of neural BC200 RNA in human tumours. J Pathol 183: 345–351 [DOI] [PubMed] [Google Scholar]
- Chen W, Heierhorst J, Brosius J, Tiedge H 1997b. Expression of neural BC1 RNA: Induction in murine tumours. Eur J Cancer 33: 288–292 [DOI] [PubMed] [Google Scholar]
- Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A 2007. The expanding RNA polymerase III transcriptome. Trends Genet 23: 614–622 [DOI] [PubMed] [Google Scholar]
- Dittmar KA, Mobley EM, Radek AJ, Pan T 2004. Exploring the regulation of tRNA distribution on the genomic scale. J Mol Biol 337: 31–47 [DOI] [PubMed] [Google Scholar]
- Dittmar KA, Goodenbour JM, Pan T 2006. Tissue-specific differences in human transfer RNA expression. PLoS Genet 2: e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiduschek EP, Kassavetis GA 2001. The RNA polymerase III transcription apparatus. J Mol Biol 310: 1–26 [DOI] [PubMed] [Google Scholar]
- Geslain R, Cubells L, Bori-Sanz T, Alvarez-Medina R, Rossell D, Marti E, Ribas de Pouplana L 2010. Chimeric tRNAs as tools to induce proteome damage and identify components of stress responses. Nucleic Acids Res 38: e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J 2004. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 18: 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Wang X, Anderson JT 2006. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 12: 508–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohrer C, Xie L, Kellerer S, Varshney U, RajBhandary UL 2001. Import of amber and ochre suppressor tRNAs into mammalian cells: A general approach to site-specific insertion of amino acid analogues into proteins. Proc Natl Acad Sci 98: 14310–14315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall L, White RJ 2008. Non-coding RNA production by RNA polymerase III is implicated in cancer. Nat Rev Cancer 8: 911–914 [DOI] [PubMed] [Google Scholar]
- Pandolfi PP 2004. Aberrant mRNA translation in cancer pathogenesis: An old concept revisited comes finally of age. Oncogene 23: 3134–3137 [DOI] [PubMed] [Google Scholar]
- Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T 2009. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res 37: 7268–7280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavon-Eternod M, Wei M, Pan T, Kleiman L 2010. Profiling non-lysyl tRNAs in HIV-1. RNA 16: 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK 2010. tRNA biology charges to the front. Genes Dev 24: 1832–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen MA, Elf J, Bouakaz E, Tenson T, Sanyal S, Bjork GR, Ehrenberg M 2005. Over expression of a tRNALeu isoacceptor changes charging pattern of leucine tRNAs and reveals new codon reading. J Mol Biol 354: 16–24 [DOI] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waas WF, Druzina Z, Hanan M, Schimmel P 2007. Role of a tRNA base modification and its precursors in frameshifting in eukaryotes. J Biol Chem 282: 26026–26034 [DOI] [PubMed] [Google Scholar]
- Winter AG, Sourvinos G, Allison SJ, Tosh K, Scott PH, Spandidos DA, White RJ 2000. RNA polymerase III transcription factor TFIIIC2 is overexpressed in ovarian tumors. Proc Natl Acad Sci 97: 12619–12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Goodenbour JM, Godley LA, Wickrema A, Pan T 2009. High levels of tRNA abundance and alteration of tRNA charging by bortezomib in multiple myeloma. Biochem Biophys Res Commun 385: 160–164 [DOI] [PMC free article] [PubMed] [Google Scholar]