Proper regulation of gene expression during cell cycle entry assures the successful completion of proliferation, avoiding risks such as carcinogenesis. The microRNA (miRNA) network is an emerging molecular system regulating multiple genetic pathways. The authors demonstrate here that the global elevation of miRNAs is critical for proper control of gene expression program during cell cycle entry. Strikingly, Exportin 5 (XPO5) is promptly induced during cell cycle entry by a PI3K-dependent post-transcriptional mechanism. The data suggest that XPO5-mediated global miRNA elevation might be involved in a broad range of cellular events associated with cell cycle control.
Keywords: microRNA, Exportin 5, PI3-kinase, cell cycle
Abstract
Proper regulation of gene expression during cell cycle entry ensures the successful completion of proliferation, avoiding risks such as carcinogenesis. The microRNA (miRNA) network is an emerging molecular system regulating multiple genetic pathways. We demonstrate here that the global elevation of miRNAs is critical for proper control of gene expression program during cell cycle entry. Strikingly, Exportin 5 (XPO5) is promptly induced during cell cycle entry by a PI3K-dependent post-transcriptional mechanism. Inhibition of XPO5 induction interfered with global miRNA elevation and resulted in a proliferation defect associated with delayed G1/S transition. During cell cycle entry, XPO5 therefore plays a paramount role as a critical molecular hub controlling the gene expression program through global regulation of miRNAs. Our data suggest that XPO5-mediated global miRNA elevation might be involved in a broad range of cellular events associated with cell cycle control.
INTRODUCTION
The cell cycle consists of a series of distinct stages that allows cells to proliferate. The entry of cells into the cell cycle process triggers drastic and dynamic changes in gene expression programs. A variety of regulation checkpoints monitor progression through the cell cycle (Elledge 1996). At the G1/S checkpoint, a critical decision is taken whether to proceed with cycling or, alternatively, to enter into the quiescent phase. This step is of utmost importance since deregulation of the control mechanisms at the G1/S transition may lead to the accumulation of detrimental changes such as gene mutations and genetic instability, which are known to promote carcinogenesis (Kaelin 1997).
MicroRNAs (miRNAs) are a growing class of functional noncoding RNAs that play a critical role in the regulation of gene expression, mainly by repressing target genes at the post-transcriptional level (Bartel 2004). In a cell, dozens of coexpressed miRNAs constitute a molecular system called microRiboNucleome (miRNome) (Zhang 2008). Since each single miRNA may regulate numerous target mRNAs, the miRNome actually modulates a broad spectrum of the genes.
The biogenesis of functional mature miRNAs is based on the stepwise processing machinery (Fukao et al. 2007; Siomi and Siomi 2010). During the process, Exportin 5 (XPO5)–mediated nuclear export of pre-miRNAs has been shown as a rate-limiting step of the miRNA biogenesis (Yi et al. 2005) and, therefore, might be critical for the quantitative control of global miRNA levels.
Critical roles of miRNAs in the cell cycle have been highlighted by their potential involvement in cancer biology. The expression profiles of miRNAs in several tumors have been shown to significantly differ from those of the tissues from which they are derived and also among different tumor types (Lu et al. 2005). This indicates that the miRNome pattern is sensitive to tumorigenesis. Interestingly, recent studies have provided data showing the importance of global miRNA regulation in carcinogenesis. Profiling of the miRNA expression pattern demonstrated a defect in miRNA production and global down-regulation of miRNAs in human tumors (Lu et al. 2005). More recently, several reports have described defects in critical genes for general miRNA processing—such as Tarbp2, Dicer1, Drosha, and Xpo5—in tumor cells or as causative to enhanced carcinogenesis (Merritt et al. 2008; Kumar et al. 2009; Melo et al. 2009, 2010). This is consistent with the global reduction of miRNAs in tumor cells.
Despite growing knowledge in cancer genetics of miRNA and its processing machinery, the relevance of these genetic defects to the mechanisms of carcinogenesis is still elusive. For this issue, it is essential to elucidate how and when global miRNA control is critically involved in the regular cell cycle events of healthy primary cells. In this report, we explored the regulation of global miRNA expression and its effects on the gene expression program during cell cycle entry in healthy primary cells.
RESULTS
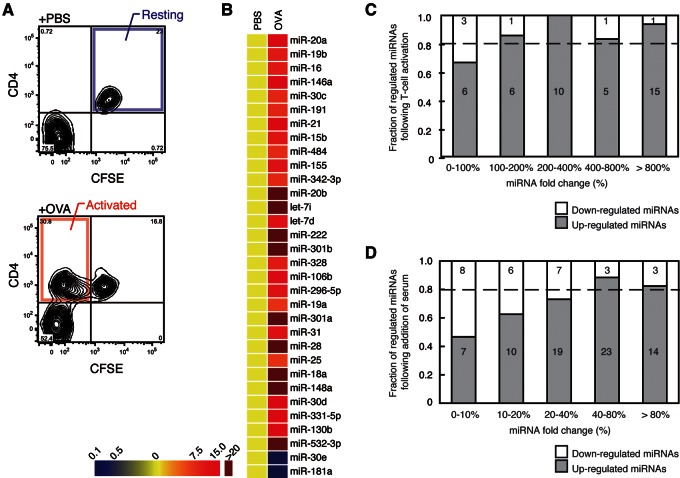
To explore the dynamics of the miRNome during the cell cycle entry, we compared the miRNA expression profiles between resting and antigen-activated lymphocytes (Fig. 1A; for details, see Supplementary Materials and Methods) (Bird et al. 1998). The most remarkable result from the profiling is the overall elevation of numerous miRNA species (for >80% [42 out of 48] of the detected miRNAs) in the activated T-cell population compared with resting T cells, although a few miRNAs were down-regulated (Fig. 1B,C; Supplemental Fig. S1A,B; Supplemental Table S2).
FIGURE 1.
Global miRNA elevation during cell cycle entry. (A) FACS pattern of the resting and activated helper T cells purified using CFSE and CD4 as detailed in the Supplementary Materials and Methods. (B) Heat map of miRNAs with a greater than threefold change in activated T cells (OVA) compared with resting T cells (PBS) is shown. The data are from the real-time PCR array. (C) The fractions of up-regulated (shaded) and down-regulated (open) genes were shown for each range of fold change. miRNAs were binned according to the fold change. (D) The fractions of miRNAs with different fold change for serum-starved and refed MEF cells. Figure shown as described in C.
In order to confirm the global miRNA elevation during cell cycle entry, we also employed primary mouse embryonic fibroblast (MEF) cultures as another experimental system (Lundberg and Weinberg 1999). We reproduced the cell cycle arrest and entry conditions by serum starvation and refeeding, respectively. Again, an overall increase of global miRNAs was observed during cell cycle entry (Fig. 1D; Supplemental Table S2).
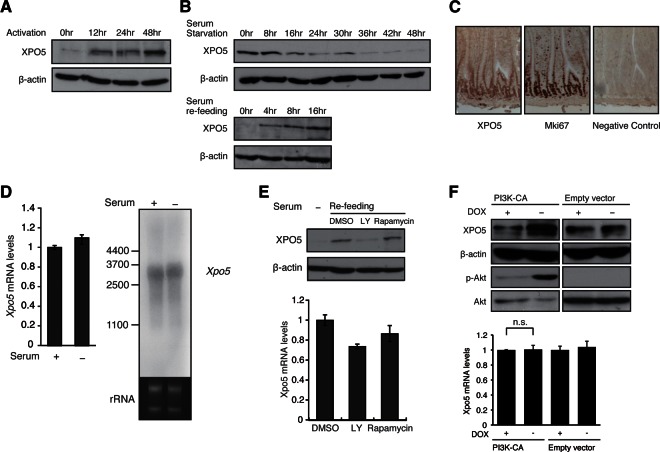
Given the global elevation of miRNAs, we presumed that the general pathway of miRNA biogenesis might be also regulated during cell cycle entry. Indeed, we observed a significant increase in XPO5 protein during T-cell activation in vitro (Fig. 2A). Similarly for MEF cells, the level of XPO5 protein decreased under conditions of cell cycle arrest and was elevated during cell cycle entry (Fig. 2B). Up-regulation of the XPO5 protein is remarkable in comparison to the other major molecules—such as Ago2, Dicer1, Drosha, and DGCR8—for the general pathway of miRNA biogenesis (Supplemental Fig. S2A). Strikingly, the level of XPO5 protein is also much higher in actively proliferating cells, such as cells in the intestinal crypts, in comparison to quiescent cells in the upper villi (Fig. 2C), suggesting that the XPO5 protein is induced in the proliferating cells in vivo.
FIGURE 2.
XPO5 is rapidly induced during cell cycle entry and is dependent on a PI3K-mediated post-transcriptional mechanism. (A) Western blot for XPO5 at the indicated time points after in vitro activation of T-cell samples as detailed in the Supplementary Materials and Methods. Reblotting for β-actin is a loading control. (B) Western blot for XPO5 at indicated time points after serum starvation or refeeding of MEF cells. (C) Immunohistochemical analysis in the sections of small intestine stained with antibodies against XPO5 and proliferation marker Mki67. Note the costaining of XPO5 and Mki67 in the same cell population, indicating that XPO5 protein is preferentially expressed in the proliferating cells. (D) RNA level of Xpo5 in serum-fed and -starved NIH3T3 cells was determined by real-time PCR and Northern blotting. β-Actin and rRNA were measured as the internal controls. (E) Western blot for XPO5 in MEF cells treated with dimethyl sulfoxide (DMSO), LY-294002, and Rapamycin during cell cycle entry. The inhibitors were added in the culture 2 h before serum refeeding. RNA levels of Xpo5 were determined by real-time PCR using the same samples. β-Actin served as the loading control. (F) XPO5 induction following PI3K activation. Doxycycline (DOX)- or DMSO-treated PI3K constitutively active (CA) and negative control Tet-off NIH3T3 lines were subjected to Western blotting for XPO5, β-actin, phosphorylated AKT, and AKT. RNA levels of Xpo5 were determined by real-time PCR using DOX- or DMSO-treated PI3K CA and negative control Tet-off NIH3T3 lines.
Although the XPO5 protein level significantly changes, the mRNA level of Xpo5 remains unchanged during cell cycle entry as the quantitative RT-PCR assay for Xpo5 mRNA showed no significant difference between the starved and refed conditions (Fig. 2D). Furthermore, a Northern blot also shows no significant change in the expression pattern of the full-length Xpo5 transcript and/or splicing variants (Fig. 2D; Supplemental Fig. S2B,C). Consistent with its rapid protein induction, this suggests that XPO5 expression is controlled at the post-transcriptional level.
To link the regulation of cell cycle entry and the XPO5 elevation, we then focused on phosphoinositide 3-kinase (PI3K) signaling (Bader et al. 2005). An inhibitor of PI3K, LY-294002 (Vlahos et al. 1994), strongly suppressed the up-regulation of XPO5 protein, whereas the mRNA level was unchanged (Fig. 2E). Rapamycin did not show any significant effect (Fig. 2E), indicating that XPO5 protein production is not mediated by the canonical PI3K-mTOR translational machinery. We also established a stable NIH3T3/MEF cell line with inducible PI3K-CA, a constitutively active form of PI3K (Zhao et al. 2005), by the Tet-Off system. This cell line, which expresses PI3K-CA upon doxycycline (DOX) deprivation, enabled us to observe significantly higher expression of XPO5 protein under PI3K active condition (Fig. 2F), further supporting PI3K-dependent XPO5 production. Again, the Xpo5 mRNA level unchanged under PI3K active condition in this model (Fig. 2F), supporting the notion that the amount of XPO5 is regulated at the post-transcriptional level.
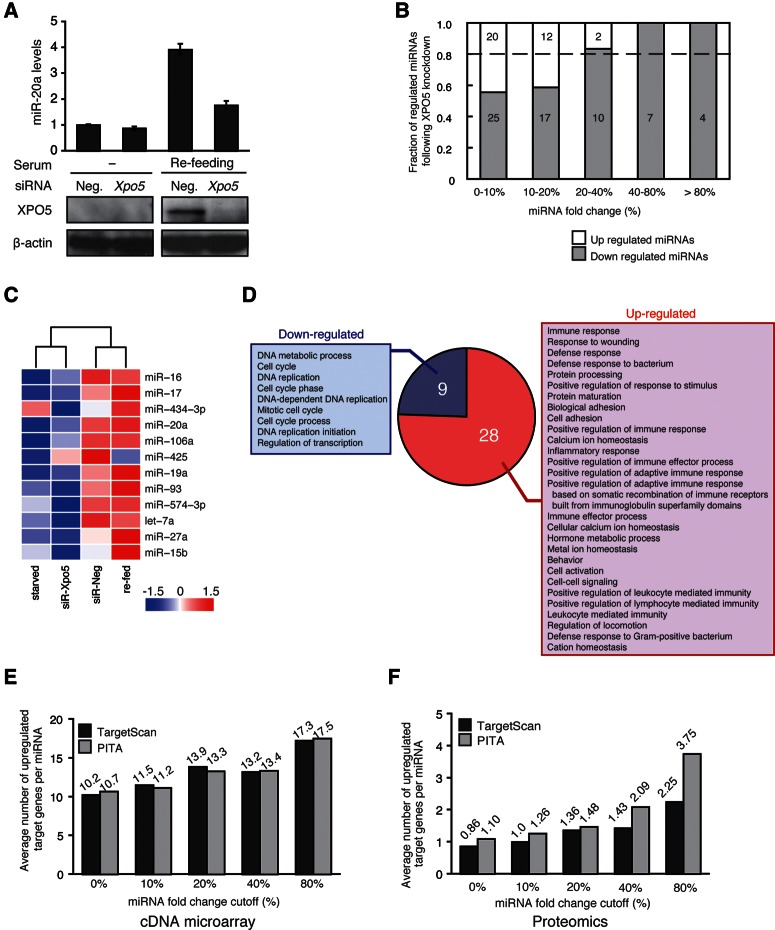
Given the role of XPO5 in miRNA biogenesis (Yi et al. 2005), we hypothesized that the regulation of XPO5 might be critical for the control of global miRNA levels during cell cycle entry. To test this, we introduced RNAi for Xpo5 mRNA into MEFs (Fig. 3A). The array-based profiling of miRNome patterns demonstrated suppression of global miRNA elevation in the XPO5 knockdown (KD) sample, especially for those miRNAs with larger changes (Fig. 3B). The effect of the specific RNAi against Xpo5 on mature miRNA levels was most probably due to changes at the post-transcriptional rather than the transcriptional level. Thus, increase in the ratio of mature to primary transcripts of up-regulated miRNAs, such as miR-20a, was significantly suppressed by the XPO5 KD (Supplemental Fig. S3B). The clustering analysis showed significant similarities in the expression patterns between the starved versus XPO5-siRNA transfected samples and between the refed versus negative control (NC) siRNA-treated samples (Fig. 3C; Supplemental Table S2). Taken together, these results indicate that induction of XPO5 is essential for the global miRNA elevation during cell cycle entry.
FIGURE 3.
Systems-wide effects of global miRNA elevation. (A) The RNA level of miR-20a in MEF cells (n = 3 mean ± SD) following transfection of Xpo5 or a negative control siRNA into serum-starved or refed cells. Western blotting shows the efficiency of the siRNA transfection; β-actin serves as a loading control. (B) The fractions of miRNAs with different fold changes were shown for XPO5 knockdown (KD) MEF cell samples as described in Figure 1A. (C) Clustering analysis of miRNA expression with fold change >20% in XPO5 KD, with serum refed and their control samples as detailed in the Supplementary Materials and Methods. (D) The fractions of GO terms with different P-value thresholds were shown for genes up-regulated (red) or down-regulated (blue) over 1.5-fold, according to microarray data. (E) Average number of up-regulated miRNA target transcripts predicted by TargetScan or PITA software is shown for each fraction of miRNAs for each different fold change cutoff following the XPO5 KD. The data are from the cDNA microarray. (F) Average number of up-regulated miRNA target proteins from proteomics analysis.
We then performed a cDNA microarray analysis with XPO5 KD and NC MEF cells to examine the possible transcriptome-wide effects of XPO5-mediated global miRNA elevation during cell cycle entry. The overall transcriptome pattern was reproducibly affected by XPO5 KD in MEF cells during cell cycle entry, about 2500 genes showing a significant change (greater than 1.5-fold) in expression levels. There was no significant difference between numbers of up- or down-regulated transcripts (Supplemental Fig. S3C). Nonetheless, by the Gene Ontology (GO) analysis (Ashburner et al. 2000), the number of extracted GO terms was over three times larger for up-regulated genes than for down-regulated genes (Fig. 3D). This indicates that up-regulated genes consist of the various genes with a broad range of cellular functions, whereas the functional characteristics of down-regulated genes have a narrower diversity and are enriched with GO terms, many of which are obviously key functions during cell cycle entry.
Since miRNAs repress target transcripts, we counted the number of potential miRNA target genes, predicted by the TargetScan (Lewis et al. 2005) or PITA (Kertesz et al. 2007) programs, that were up-regulated in the MEF cells following XPO5 KD (Fig. 3E; Supplemental Fig. S3D). The number of up-regulated genes/miRNA positively correlates with the degree of suppression for the corresponding miRNAs by XPO5 KD. Moreover, these effects of global miRNA elevation could be also confirmed by high-throughput mass spectrometry (MS)–based proteomics, although the coverage of genes is much less than that of the cDNA microarray (Supplemental Fig. S3E,F; Supplemental Table S3). Altogether, these results suggest a significant and dosage-dependent control by XPO5 and global miRNAs on the system-wide gene expression program during cell cycle entry.
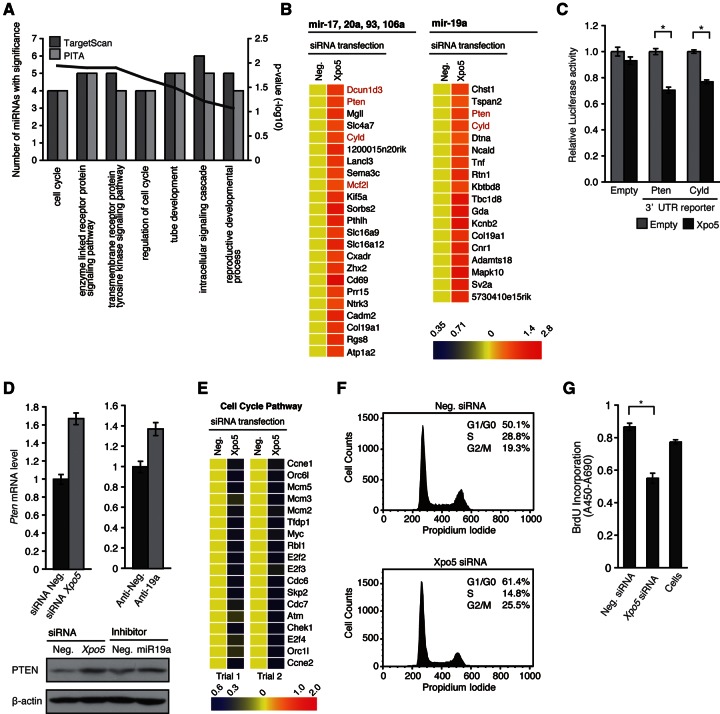
The computational characterization of the predicted targets and highly elevated miRNAs in MEF cells during cell cycle entry (Supplemental Fig. S4A; for detailed process, see Materials and Methods) indicated a significant overlap of the functional annotations assigned to the targets and the selected miRNAs, with highly significant probability (P < 0.005) (Fig. 4A). These common annotations shared by “predicted” and “validated” target functions may represent major roles of XPO5-mediated elevation of miRNA subsets. Strikingly, such common annotations include “cell cycle” and “regulation of cell cycle,” suggesting that those highly elevated miRNAs may be particularly involved in the mechanisms controlling cell cycle through regulation of the target genes.
FIGURE 4.
XPO5 KD during cell cycle entry causes defective proliferation associated with delayed G1/S transition. (A) Common functional annotation obtained from microarray data and miRNA target genes predicted using TargetScan (darker shade) or PITA (lighter shade) software. Bars, P-values of Gene Ontology (GO) terms in the cDNA array; the line shows the number of miRNAs with the indicated GO terms. For details, see the Supplementary Materials and Methods. (B) Heat map of miR-17, 20a, 93, 106a, and 19a target genes predicted using TargetScan, which were up-regulated by XPO5 KD. The genes known to be involved in cell cycle and cell proliferation are shown in red. (C) Reporter assay with luciferase reporter bearing 3′ UTRs of Pten and Cyld was quantified 48 h after cotransfection with an XPO5 expression vector (*P < 0.01). (D) Pten mRNA and protein levels following transfection of a negative control or Xpo5 siRNA, and a negative control or inhibitor for miR-19a (60 nM, 48 h). The RNA level was determined by real-time PCR, and the protein level was determined by Western blotting. β-Actin was used as internal control for both analyses. (E) Heat map of differentially expressed cell cycle genes in XPO5 KD and control MEF cells. GO category of “GO:0007049 cell cycle.” (F) Cell cycle analysis of XPO5 KD MEF cells. For details, see the Supplementary Materials and Methods. (G) Bromodeoxyuridine (BrdU) incorporation assay of XPO5 KD MEF cells (n = 3, mean ± SD, *P < 0.01).
To examine such potential mechanisms in the cDNA microarray data, we selected the predicted target genes of miR-17, 20a, 93, 106a, and 19a, which were highly down-regulated upon XPO5 KD (Fig. 4B). Several cell cycle- and/or proliferation-related genes such as Pten, Cyld, Dcun1d3, and Mcf2l (Yamauchi et al. 2002; Massoumi et al. 2006; Ma et al. 2008; Salmena et al. 2008) were found and confirmed using quantitative real-time PCR (Supplemental Fig. S4B). Among these validated genes, we are particularly interested in the tumor suppressors, Pten and Cyld (Massoumi et al. 2006; Salmena et al. 2008), which are likely to be suppressed during cell cycle progression. The regulation of Pten and Cyld by XPO5 is probably mediated through miRNAs targeting in their 3′ UTRs because XPO5 overexpression resulted in a significant down-regulation of luciferase reporters carrying the 3′ UTRs of each gene (Fig. 4C). To further extend this, we analyzed several miRNAs, such as miR-17, 20a, and 19a, predicted to regulate Pten (Supplemental Fig. S4C,D). The inhibition of miR-19a, but not other candidates, resulted in the significant up-regulation of the Pten 3′-UTR Luc reporter (Supplemental Fig. S4D), indicating that miR-19a is capable of repressing Pten gene. The quantitative real-time PCR and Western blotting confirmed this result. Of note, the effect of the miR-19a inhibitor on Pten regulation is significantly less than that of XPO5 KD (Fig. 4D). Many other highly inducible miRNAs were predicted to target Pten (Supplemental Fig. S4C), supporting the notion that Pten is one of the critical factors being regulated by multiple miRNAs during cell cycle entry (Supplemental Fig. S4E).
On the other hand, when down-regulated genes with the annotations were mapped to the cell cycle pathway based on the KEGG pathway map (Kanehisa and Goto 2000), most of the genes localized to the G1-to-S transition phase (Fig. 4E; Supplemental Fig. S4F). Such genes include E2F family members (e.g., E2f2, E2f3, E2f4, Tfdp1), CDC genes (e.g., Cdc6, Cdc7), cyclin genes (e.g., Ccne1, Ccne2), and variety of other cell cycle regulator genes (e.g., Myc, Rbl1, Skp2). Although the regulation of each gene function (and/or expression) might not be defined by the transcript levels, this significant enrichment of G1/S-related genes in the down-regulated group suggests a possible effect of XPO5 on the G1 phase. Indeed, XPO5 KD resulted in a significant increase in the cell proportion (up to 10%) at G0/G1 phase as judged by the cell cycle analysis (Fig. 4F). Proliferation is significantly and reproducibly suppressed by the XPO5 KD (Fig. 4G), suggesting that defective XPO5 expression during the cell cycle entry disturbs the progression of proliferation by causing a cycling delay at the G1/S transition.
DISCUSSION
Physiological significance of the global miRNA elevation during cell cycle entry
We demonstrated a global elevation of miRNA levels during cell cycle entry. The inhibition of this process by XPO5 KD resulted in up-regulation of numerous miRNA target genes with a broad spectrum of functions, suggesting a general control of the complex gene expression program by miRNAs during cell cycle entry. This implies that a significant number of genes might actually be overtranscribed during the cell cycle, even under physiological conditions. The miRNA network acts on such excessively transcribed genes to stabilize them to the proper expression levels. In this regard, the quantitative regulation of miRNAs in a global, rather than a specific, manner could be a more efficient way to deal with the system-wide dynamics of gene expression.
Through the identification of potential targets, we could show that Pten is one of the tumor suppressors repressed by miRNAs during cell cycle entry. This could suggest a unique regulatory circuit composed of PI3K, XPO5, miRNAs (e.g., miR-19a), and tumor suppressors (e.g., Pten and Cyld), which may positively regulate cell cycle progression by the miRNA-mediated repression of tumor suppressors during cell cycle entry (Supplemental Fig. S4E). Given a great many combinations of miRNAs and targets, similar molecular circuits or cascades may exist and constitute a robust regulatory network for the control of the cell cycle. In any of such regulatory models, XPO5 plays a paramount role as a critical molecular hub controlling the gene expression program through global regulation of miRNAs.
In this regard, the regulation of XPO5 by PI3K signaling is particularly intriguing since numerous growth stimuli trigger PI3K activation, implying that XPO5-mediated global miRNA elevation might be involved in a broad range of cellular events associated with cell cycle control. To further understand this critical event, the molecular mechanism underlying PI3K-dependent post-transcriptional regulation of XPO5 should be investigated in the future studies.
Defects of global miRNA regulation during cell cycle entry and cancer risks
Aberrant gene expression during the cell cycle significantly increases the risk of carcinogenesis (Hartwell and Kastan 1994). In this context and based on our observations, any abnormalities in the regulation and/or function of XPO5 might pose a cancer risk, as they may cause defects in the global elevation of miRNAs. This could cause deregulation of numerous target genes and consequently lead to aberrant gene expression. Intriguingly, a very recent study indeed reported mutations of Xpo5 gene in several tumor cell lines (Melo et al. 2010). Our current study may provide a mechanistic insight into the carcinogenesis by such XPO5 mutations, by showing the regulatory role of XPO5 during cell cycle entry in healthy primary cells. Since control of the global miRNA level may involve different molecular machineries, genes in such machineries might be also susceptible to cancer by a mechanistic analogous to the one described above.
Deregulation of global miRNA elevation and cell cycle checkpoints
The KD of XPO5 may mimic naturally occurring XPO5 defects that expose the cell to the danger of cancer risks. Strikingly, the XPO5 KD inhibited the proliferation of MEF cells by leading to a delayed G1/S transition. This implies that the up-regulation of XPO5 and the following global miRNA elevation might be the crucial steps to be assessed at the G1/S checkpoint.
For this G1 arrest, up-regulation of tumor suppressors due to their liberation from the miRNA control might be one of the causal events (Supplemental Fig. S4E). This mechanism may work as a safety net standing by to counteract the risk of cell cycling when expression of miRNAs is deregulated. Also in this context, any defects leading to a fatal miRNome abnormality may sensitize this safety net, and Xpo5 is one of the most susceptible candidate genes for such defects. According to the multistep theory (Vogelstein and Kinzler 1993), XPO5 dysfunction together with other genetic defects, such as mutations in tumor suppressors, may eventually cause carcinogenesis by overcoming this safety net.
Brief perspectives and related issues on cell cycle-associated XPO5 control
Rapid induction and functional up-regulation of XPO5 during cell cycle entry imply possible regulatory mechanisms for other nuclear transport processes during cell cycle entry. Indeed, a recent work reported accumulation of tRNAs in the nucleus due to down-regulation of their transportin, XPO-t, during nutrient deprivation (Huynh et al. 2010). Together with this report, our work highlights the nuclear transport machineries as critical control units for proper cell cycle regulation.
XPO5 is involved in the export of not only miRNA precursors but also other non-miRNA molecules (Wild et al. 2010). In this report, we argue that the major effect of XPO5 induction on cell cycle control is due to its function for the global miRNA elevation because our system biological approach clearly demonstrated that the major miRNAs up-regulated during cell cycle entry tend to target critical cell cycle regulators (Figs. 3D, 4A,E; Supplemental Fig. S4F). However, of course, we do not exclude the possibility for potential influences of non-miRNA XPO5-targets on cell cycle control. This possibility should be carefully examined in the future investigations.
CONCLUSION
Overall, our findings highlight the global miRNA regulation not only as a critical molecular event in the cell cycle but also as a risky step for carcinogenesis. Therefore, the molecules regulating the global miRNome, such as XPO5, could be potentially therapeutic and/or diagnostic targets in cancer medicine. Further investigations focusing on the mechanism of cell cycle arrest caused by global miRNA deregulation would be particularly crucial to understand the nature of carcinogenesis.
MATERIALS AND METHODS
Cell isolation
MEFs were isolated from 13.5 days post coitum (dpc) C57BL/6 mice. In vivo and in vitro T-cell isolation and activation are described in the Supplementary Materials and Methods.
High-throughput real-time PCR array
miRNA expression profiling was performed with the TaqMan Rodent microRNA Array A version 2.0 (Applied Biosystems). For comparison of the miRNA profile within fetal calf serum (FCS)starved and XPO5 KD condition, the R statistics package (http://www.r-project.org/) was used. Clustering was done by complete linkage clustering taking the Euclidean distance of miRNAs with a fold change >20%.
Microarray data analysis
The transcriptome analysis was conducted with Agilent 4 × 44K mouse genome expression assays (Agilent Technologies, 14868).
Miscellaneous information
Detailed methods for the all experiments are available in the Supplementary Materials and Methods.
DATA DEPOSITION
The microarray data have been submitted to Gene Expression Omnibus (GEO) database under accession no. GSE25498.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by the Max-Planck Gesellshaft and by funding from the DFG (SFB620 to T.F.) and by the Japan Society for the Promotion of Science (Y.W.I.).
REFERENCES
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG, Kang S, Zhao L, Vogt PK 2005. Oncogenic PI3K deregulates transcription and translation. Nat Rev Cancer 5: 921–929 [DOI] [PubMed] [Google Scholar]
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA, Sider JR, Gajewski TF, Wang CR, Reiner SL 1998. Helper T cell differentiation is controlled by the cell cycle. Immunity 9: 229–237 [DOI] [PubMed] [Google Scholar]
- Elledge SJ 1996. Cell cycle checkpoints: Preventing an identity crisis. Science 274: 1664–1672 [DOI] [PubMed] [Google Scholar]
- Fukao T, Fukuda Y, Kiga K, Sharif J, Hino K, Enomoto Y, Kawamura A, Nakamura K, Takeuchi T, Tanabe M 2007. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell 129: 617–631 [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Kastan MB 1994. Cell cycle control and cancer. Science 266: 1821–1828 [DOI] [PubMed] [Google Scholar]
- Huynh LN, Thangavel M, Chen T, Cottrell R, Mitchell JM, Praetorius-Ibba M 2010. Linking tRNA localization with activation of nutritional stress responses. Cell Cycle 9: 3112–3118 [DOI] [PubMed] [Google Scholar]
- Kaelin WG Jr 1997. Alterations in G1/S cell-cycle control contributing to carcinogenesis. Ann N Y Acad Sci 833: 29–33 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E 2007. The role of site accessibility in microRNA target recognition. Nat Genet 39: 1278–1284 [DOI] [PubMed] [Google Scholar]
- Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, Kirsch DG, Golub TR, Jacks T 2009. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev 23: 2700–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. 2005. MicroRNA expression profiles classify human cancers. Nature 435: 834–838 [DOI] [PubMed] [Google Scholar]
- Lundberg AS, Weinberg RA 1999. Control of the cell cycle and apoptosis. Eur J Cancer 35: 531–539 [PubMed] [Google Scholar]
- Ma T, Shi T, Huang J, Wu L, Hu F, He P, Deng W, Gao P, Zhang Y, Song Q, et al. 2008. DCUN1D3, a novel UVC-responsive gene that is involved in cell cycle progression and cell growth. Cancer Sci 99: 2128–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R 2006. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-κB signaling. Cell 125: 665–677 [DOI] [PubMed] [Google Scholar]
- Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C, et al. 2009. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet 41: 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, Fernandez AF, Davalos V, Villanueva A, Montoya G, et al. 2010. A genetic defect in Exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell 18: 303–315 [DOI] [PubMed] [Google Scholar]
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, et al. 2008. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med 359: 2641–2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmena L, Carracedo A, Pandolfi PP 2008. Tenets of PTEN tumor suppression. Cell 133: 403–414 [DOI] [PubMed] [Google Scholar]
- Siomi H, Siomi MC 2010. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell 38: 323–332 [DOI] [PubMed] [Google Scholar]
- Vlahos CJ, Matter WF, Hui KY, Brown RF 1994. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J Biol Chem 269: 5241–5248 [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW 1993. The multistep nature of cancer. Trends Genet 9: 138–141 [DOI] [PubMed] [Google Scholar]
- Wild T, Horvath P, Wyler E, Widmann B, Badertscher L, Zemp I, Kozak K, Csucs G, Lund E, Kutay U 2010. A protein inventory of human ribosome biogenesis reveals an essential function of Exportin 5 in 60S subunit export. PLoS Biol 8: e1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi J, Hirasawa A, Miyamoto Y, Kokubu H, Nishii H, Okamoto M, Sugawara Y, Tsujimoto G, Itoh H 2002. Role of Dbl’s big sister in the anti-mitogenic pathway from α1B-adrenergic receptor to c-Jun N-terminal kinase. Biochem Biophys Res Commun 296: 85–92 [DOI] [PubMed] [Google Scholar]
- Yi R, Doehle BP, Qin Y, Macara IG, Cullen BR 2005. Overexpression of Exportin 5 enhances RNA interference mediated by short hairpin RNAs and microRNAs. RNA 11: 220–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C 2008. MicroRNomics: A newly emerging approach for disease biology. Physiol Genomics 33: 139–147 [DOI] [PubMed] [Google Scholar]
- Zhao JJ, Liu Z, Wang L, Shin E, Loda MF, Roberts TM 2005. The oncogenic properties of mutant p110α and p110β phosphatidylinositol 3-kinases in human mammary epithelial cells. Proc Natl Acad Sci 102: 18443–18448 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.