The decoding properties of 22 structurally conservative base-pair and base-triple mutations in the anticodon hairpin and tertiary core of E. coli tRNAAlaGGC were determined under single turnover conditions using E. coli ribosomes. While all of the mutations were able to efficiently decode the cognate GCC codon, many showed substantial misreading of near-cognate GUC or ACC codons. Although all the misreading mutations were present in the sequences of other E. coli tRNAs, they were never found among bacterial tRNAAlaGGC sequences. This suggests that the sequences of bacterial tRNAAlaGGC have evolved to avoid reading incorrect codons.
Keywords: protein synthesis, ribosome
Abstract
The decoding properties of 22 structurally conservative base-pair and base-triple mutations in the anticodon hairpin and tertiary core of Escherichia coli tRNAAlaGGC were determined under single turnover conditions using E. coli ribosomes. While all of the mutations were able to efficiently decode the cognate GCC codon, many showed substantial misreading of near-cognate GUC or ACC codons. Although all the misreading mutations were present in the sequences of other E. coli tRNAs, they were never found among bacterial tRNAAlaGGC sequences. This suggests that the sequences of bacterial tRNAAlaGGC have evolved to avoid reading incorrect codons.
INTRODUCTION
Bacterial tRNAs possessing the same anticodon have a characteristic set of sequence restrictions distributed throughout the molecule that distinguish them from tRNAs with different anticodons (Saks and Conery 2007; Targanski and Cherkasova 2008). These multivalent tRNA consensus sequences reflect evolutionary adaptations that permit each tRNA to function with its particular combination of amino acid and anticodon without compromising translational rate and accuracy. Different parts of a tRNA consensus “tune” different steps in translation. tRNA identity residues that are required for specific aminoacylation of each tRNA by its associated aminoacyl synthetase tend to be conserved among bacteria and usually include the anticodon and several residues at other locations in the molecule (Saks et al. 1994; Giege et al. 1998; Ardell 2010). Three base pairs in the T-stem adjust the affinity of each aminoacyl-tRNA to elongation factor Tu to be tight enough to form a complex but weak enough to release from the protein during translation (Schrader et al. 2011). These T-stem base pairs are not conserved among bacteria, but each tRNA species uses different combinations of the three base pairs to achieve the requisite affinity (Schrader and Uhlenbeck 2011). In this communication, we show that many of the consensus residues present in the anticodon hairpin and the tertiary core of Escherichia coli tRNAAlaGGC have evolved to minimize misreading of incorrect codons during decoding on the ribosome.
RESULTS AND DISCUSSION
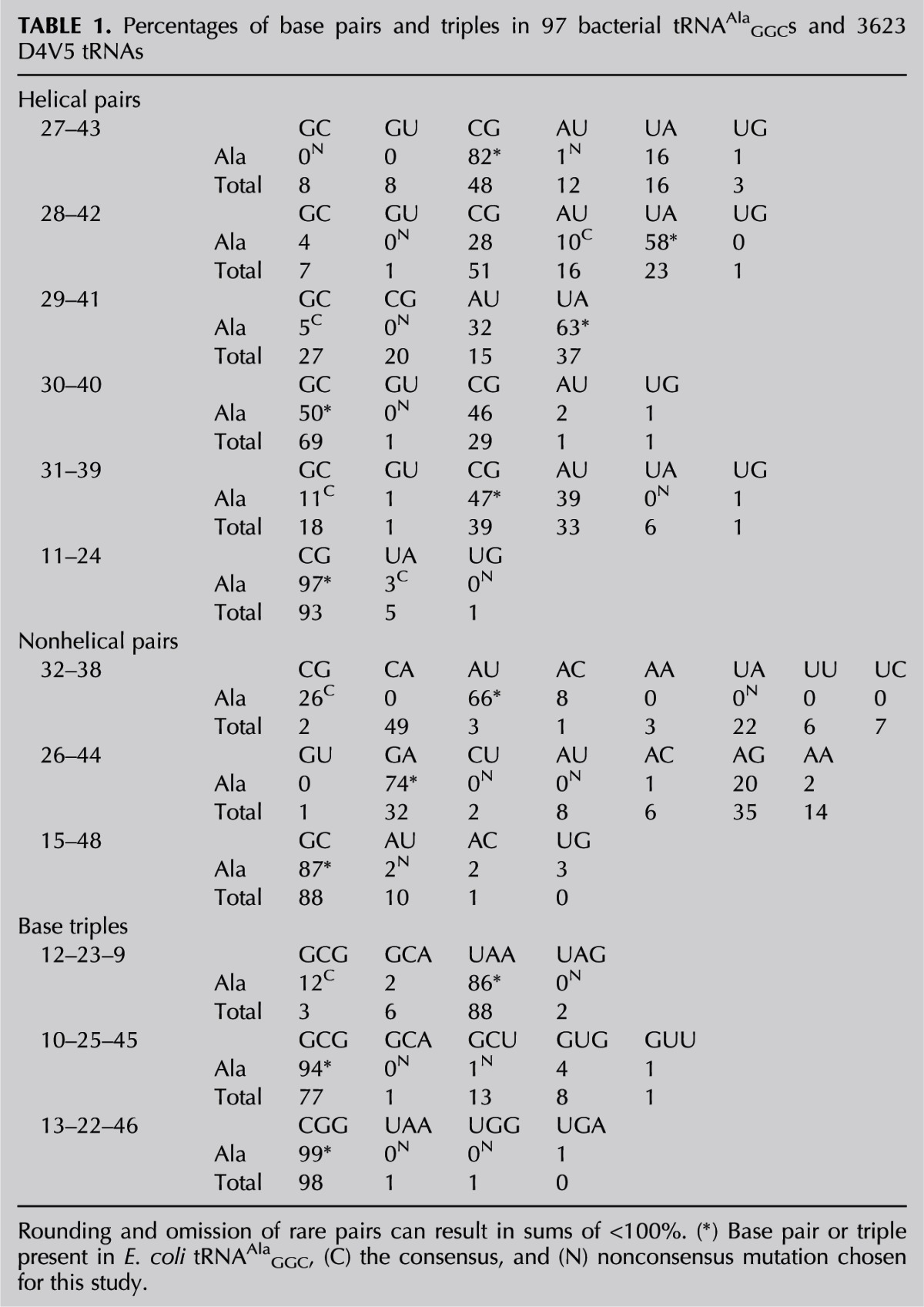
Mutations of E. coli tRNAAlaGGC were evaluated for their ability to decode their correct (cognate) GCC codon and incorrect (near-cognate) GUC and ACC codons. This tRNA was chosen because it is a member of the most common and best studied D4V5 structural class and because it is one of the few in E. coli that lacks post-transcriptional modifications in the anticodon hairpin, which are known to affect both the rate and accuracy of ribosomal decoding (Hagervall et al. 1990; Harrington et al. 1993; Li et al. 1997; Urbonavicius et al. 2001, 2003; Murphy et al. 2004; Agris 2008). Previous experiments have shown that unmodified tRNAAlaGGC is able to decode with a similar rate and accuracy as the fully modified form, permitting the functional analysis of unmodified mutant tRNAs prepared by in vitro transcription (Ledoux et al. 2009). The expected tertiary structure and the consensus of tRNAAlaGCC were used together to guide the selection of mutations of the five Watson-Crick and two noncanonical base pairs in the anticodon hairpin and the five base pairs and triples in the tertiary core of the molecule (Fig. 1; Table 1). Two general classes of mutations were chosen. Consensus mutations changed residues in the E. coli tRNAAlaGGC sequence to residues present at the corresponding positions in other bacterial tRNAAlaGGC. In contrast, nonconsensus mutations are changes in E. coli tRNAAlaGGC to residues that are either never or very rarely (<2%) found among other bacterial tRNAAlaGGC sequences but are present in other tRNAs of the D4V5 structural class. For example, E. coli tRNAAlaGCC has a C31-G39 base pair, while G31-C39 is present in this tRNA in many other bacteria, so the introduction of G31-C39 into E. coli tRNAAla is termed a consensus mutation. However, the U31-A39 base pair is never found in any bacterial tRNAAlaGGC but is present in many other D4V5 tRNAs, so introducing U31-A39 is termed a nonconsensus mutation. Since both consensus and nonconsensus mutations introduce residues present in other active tRNAs at the homologous position, the mutant tRNAs are likely to adopt a very similar structure. Combinations of residues 31-39 that are never found in any tRNA, such as most mismatched pairs, were not studied since they could potentially misfold. The same strategy was used to make mutations in the base pairs and base triples present in the core of tRNA, although this part of the molecule is so conserved among bacterial tRNAAlaGGC that consensus mutations could not always be identified. Furthermore, since the tertiary core is quite conserved for all tRNAs of the D4V5 class, even the choice of potential nonconsensus mutations was limited. This evolutionary approach for choosing mutations maximizes the chance of finding functional changes in tRNA while minimizing disruption of the folded structure of tRNA. Six consensus and 16 nonconsensus mutations throughout the anticodon helix and core of Ala-tRNAAlaGGC were chosen in this study (Table 1). When multiple choices for mutations in each category were available, no systematic secondary criterion was used.
FIGURE 1.
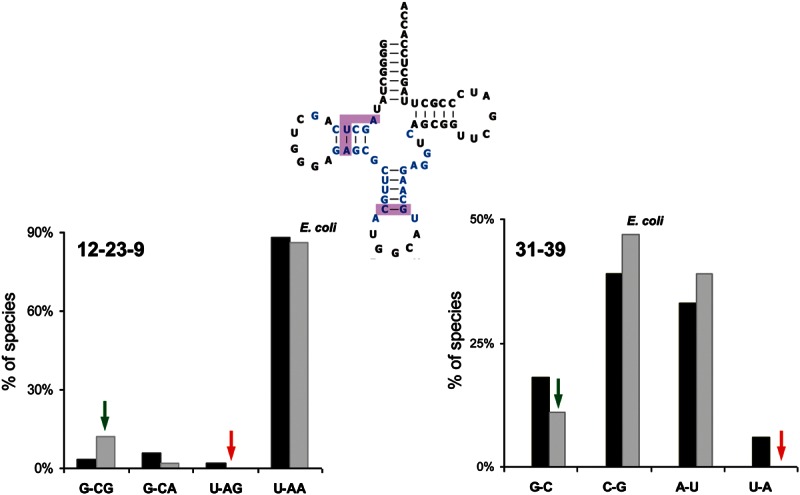
Using sequence conservation to direct the mutagenesis of nine base pairs and three base triples in the anticodon hairpin and tertiary core of E. coli tRNAAlaGGC (blue). The bar graphs compare the frequency of different 12-23-9 base triple sequences (left) and 31-39 base pair sequences (right) present among 97 bacterial tRNAAlaGGC (gray bars) with the corresponding sequences present among 3623 bacterial tRNAs of the D4V5 structural class (black bars). Nonconsensus mutations of E. coli tRNAAlaGGC (red arrows) are not present in any bacteria tRNAAlaGGC but are used in other tRNAs. Consensus mutations (green arrows) are present in tRNAAlaGGC from other species. Sequence conservation data and mutations chosen for all the base pairs and triples are in Table 1.
TABLE 1.
Percentages of base pairs and triples in 97 bacterial tRNAAlaGGCs and 3623 D4V5 tRNAs
Native tRNAAlaGGC, the unmodified wild type, and the 22 mutations were evaluated for their ability to decode cognate and near-cognate codons by measuring the rate and extent of dipeptide formation using E. coli ribosomes. Since peptide bond formation is the final step in decoding, any mutation that causes a defect at an earlier step can potentially also be detected. The assay uses a rapid mixing device to combine a 0.2 μM ternary complex containing [3′-P32]-labeled Ala-tRNA with 1 μM 70S ribosomes containing an mRNA fragment with the appropriate codon in the A site and fMet-tRNAfMet in the P site (Ledoux and Uhlenbeck 2008a). Reactions were performed in a “high fidelity” buffer containing polyamines and a low concentration of magnesium ions to ensure accurate decoding (Gromadski and Rodnina 2004). At differing times, the reactions were quenched, treated with P1 nuclease, subjected to thin-layer chromatography to separate the P32-labeled Ala-AMP starting material from the fMet-Ala-AMP product and quantitated using a phosphorimager. The initial concentrations of ternary complex and ribosomes were chosen to be subsaturating for both cognate and near-cognate codons in order to maximize detection of an altered rate or extent of decoding when screening the mutant tRNAs. Since an excess of EF-Tu•GTP over aa-tRNA was used to ensure efficient formation of ternary complex, and the ternary complex was not purified, a high concentration of a noncognate aa-tRNA was included to sequester any free EF-Tu•GTP after mixing and thereby prevented any Ala-tRNA released from ribosomes during proofreading from rebinding EF-Tu•GTP and undergoing a second round of decoding (Supplemental Fig. 1; Pan et al. 2008).
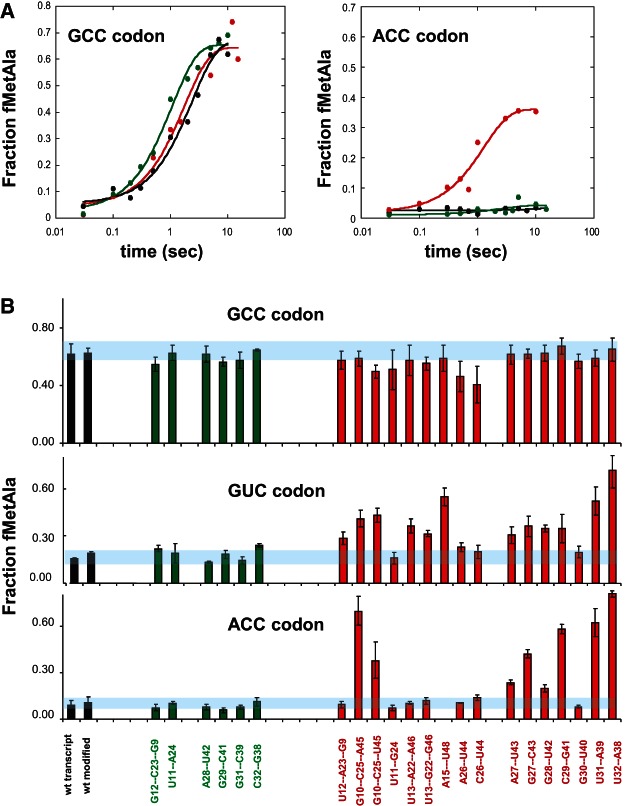
A sample data set for the two 31-39 mutations is shown in Figure 2A. As had been found previously (Ledoux et al. 2009), the unmodified wild-type tRNAAlaGGC shows rapid and efficient decoding on the cognate GCC codon, but little dipeptide is detected with the near-cognate GUC codon even after long incubation times. Such accurate in vitro decoding has been observed for several other aa-tRNAs (Cochella and Green 2005; Gromadski et al. 2006; Schrader et al. 2011; Johansson et al. 2012) and has been found to be a consequence of a combination of weak binding of the ternary complex to the near-cognate codon, a reduced rate of GTP hydrolysis due to poor induced fit, and efficient rejection of the near-cognate tRNA after GTP hydrolysis (Pape et al. 1999; Gromadski and Rodnina 2004; Rodnina 2012). Results using the consensus G31-C39 mutation are essentially identical to the wild-type tRNA. In contrast, the nonconsensus U31-A39 mutation functions normally on the GCC codon but shows substantial amounts of product accumulating at a similar rate on the near-cognate GUC codon. This behavior is consistent with a substantial fraction of the mutant tRNA molecules being able to escape proofreading by having a faster rate of accommodation and/or a slower rate of rejection compared to wild-type tRNA (Wohlgemuth et al. 2011). While additional experiments will be required to determine the individual rate constants for these mutations, more thorough kinetic analyses of other tRNA misreading mutations indicate that changes in the rate constants of several steps in the decoding mechanism are altered (Cochella and Green 2005; Ledoux et al. 2009). This enhanced misreading of the U31-A39 mutation presumably accounts for its absence among the sequences of bacterial tRNAAlas.
FIGURE 2.
Decoding of tRNAAlaGGC mutations on cognate and near-cognate codons. (A) Time course of conversion of [3′-P32] Ala-tRNAAla to fMet-Ala-tRNAAla for unmodified wild type (black), G31-C39 mutation (green), and U31-A39 mutation (red) using the cognate GCC codon (left) or near-cognate ACC codon (right). (B) Fraction of fMet-Ala-tRNAAla formed at completion (10 sec) for native and unmodified tRNAAla (black), six consensus mutations (green), and 16 nonconsensus mutations (red) using GCC, GUC, and ACC codons. Horizontal blue bars indicate range of values observed for wild-type tRNA.
The results of screening all the tRNAAla mutations on the cognate and two near-cognate codons are summarized in Figure 2B by plotting the fraction of labeled tRNAAlaGCC converted to dipeptide at completion. With the cognate GCC codon, native tRNAAla, the unmodified wild-type tRNA and all 22 mutations produce similar high levels of dipeptide. In addition, they all show rates of dipeptide bond formation in the order of 1 sec−1 at 0.5 μM 70S ribosomes (data not shown). These rates are similar to kpep values observed for a number of native E. coli tRNAs and unmodified tRNA transcripts (Cochella and Green 2005; Ledoux and Uhlenbeck 2008b; Schrader et al. 2011). Thus, since all of the mutant tRNAAlas are able to undergo ribosomal decoding in a manner similar to the wild-type control, it appears that their tertiary structures are not significantly disrupted by any of the structurally conservative mutations. tRNA mutations predicted to disrupt folding often show impaired rates and extents of decoding (Harrington et al. 1993; Pan et al. 2008).
The tRNAAla mutations show very different behaviors when decoding the near-cognate ACC and GUC codons in a manner that correlates closely with the tRNAAla consensus. All six consensus mutations formed very low levels of dipeptide on both noncognate codons, similar to what is observed for native and wild-type tRNAAla. Accurate decoding by the consensus mutations is consistent with their presence in the sequences of tRNAAla in other bacteria. In striking contrast, 12 of the 16 nonconsensus mutations produced significantly more dipeptide than wild-type tRNA with at least one of the two near-cognate codons. In every case where the amount of dipeptide was great enough to be accurately measured, the observed kpep was close to 1 sec−1, confirming them as misreading mutations. These nonconsensus misreading mutations were distributed throughout the tRNAAla structure, including four of the five base pairs in the anticodon stem and five of the six tertiary interactions tested. It is noteworthy that, for many of the tRNAAla misreading mutations, the amount of dipeptide formed differed significantly between the two near-cognate codons. For example, the G10-C25-A45 tRNA misreads the ACC codon more than the GUC codon, while the U13-A22-A46 tRNA acts oppositely. Finally, there were four nonconsensus mutations (U11-G24, A26-U44, C26-U44, and G30-U40) that showed little product with either near-cognate codon and thus do not appear to be misreading mutations. These mutations may represent exceptions to the otherwise strong correlation between the tRNAAla consensus and misreading. Alternatively, a different near-cognate codon may be required to detect misreading with these four tRNA mutations. The observed correlation between the tRNAAla consensus and translational accuracy strongly suggests that a substantial portion of the bacterial tRNAAla sequence has evolved to avoid reading incorrect codons.
How do the structurally conservative nonconsensus tRNAAla mutations cause ribosomal misreading? It is likely that different mutations are explained in different ways. Previous experiments have shown that the nonconsensus U32-A38 mutation enhances binding of the ternary complex to near-cognate codons, thereby promoting misreading (Ledoux et al. 2009). A plausible explanation for many of the remaining nonconsensus mutations is based on the observation that the structure of tRNA distorts during decoding (Valle et al. 2002; Frank et al. 2005; Li et al. 2008). Crystal structures of ternary complexes of E. coli tRNAThr or tRNATrp bound to their cognate codons on Thermus thermophilus 70S ribosomes that are blocked just before or after GTP hydrolysis reveal a distorted tRNA conformation in order to accommodate the simultaneous binding of codon with anticodon in the 30S subunit and the binding of EF-Tu to its site on the 50S subunit (Schmeing et al. 2009, 2011). In this kinetically trapped decoding intermediate, the anticodon helix is underwound by 14°, and the locations of numerous residues in the tertiary core are altered compared to those expected for the corresponding free-ternary complexes. Many of these residues coincide with the positions where the tRNAAla misreading mutations were identified (Fig. 3). Based upon the crystal structures of similar ternary complexes with two different misreading mutations of tRNATrp, it was proposed that tRNA misreading mutations can access the distorted state more easily, either by selectively stabilizing the structure of the distorted form or destabilizing the structure of the free form (Schmeing et al. 2011). The tRNA sequence changes, therefore, allow the weaker-binding near-cognate codons to have a longer residence time in the bent intermediate state, thereby enhancing their decoding efficiency (Ogle et al. 2002; Yarus et al. 2003; Ogle and Ramakrishnan 2005). Although the precise explanation for how each nonconsensus tRNAAla mutation can cause enhanced misreading will require additional kinetic and structural data, they could function in a similar manner as those in tRNATrp. The details of RNA helix geometry are expected to be affected by the sequence of base pairs, and the flexibility of nucleic acid helices also depends upon sequence (Faustino et al. 2010; Peckham and Olson 2011). Thus, the altered stacking interactions resulting from the base-pair changes in the anticodon helix could alter either the structure or the dynamics of the anticodon helix such that it favors the bent state more easily than the wild-type sequence. A similar explanation could be applied to the core mutations.
FIGURE 3.
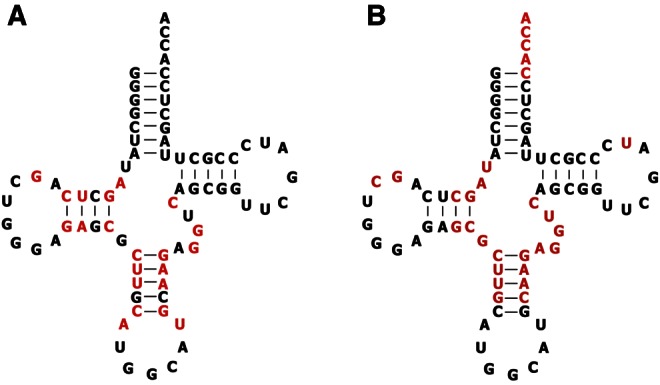
(A) Residues in tRNAAlaGGC where misreading mutations were found (red). (B) Residues in the tRNAThr ternary complex that are distorted upon ribosome binding (Schmeing et al. 2009), mapped onto the sequence of tRNAAlaGGC (red).
The idea that the distortability of a tRNA can explain a substantial portion of the consensus sequence of each individual tRNA has many attractive features. First, since the distortion involves such a large portion of the molecule, it explains why so many nonconsensus tRNAAla misreading mutations were identified, including several that do not directly contact the ribosome during decoding. Secondly, the extensive region of bending permits the contribution of many different combinations of residues to be integrated to achieve the appropriate overall distortability, thereby explaining why the consensus sequences are so redundant in this region of the molecule. While we have not analyzed the effects of making multiple nonconsensus tRNAAla mutations on decoding accuracy in much detail, preliminary experiments suggest that the effects of mutations are not additive, indicating that a simple set of sequence rules for misreading may be difficult to deduce. Finally, the bendability concept can also explain the observation that certain base-pair changes in the anticodon helix of tRNA alter its ability to decode its cognate codon (Raftery and Yarus 1987). It seems plausible that the sequence of a tRNA is “tuned” to optimize its bendability for decoding. A tRNA sequence that is too “stiff” will not be able to access the bent state sufficiently easily to decode its cognate codon, while a tRNA sequence that is too flexible will read near-cognate codons too well. The offsetting needs of speed and accuracy is a common theme in the design of the translational machinery (Thompson and Karim 1982; Wohlgemuth et al. 2011; Johansson et al. 2012), so it is not surprising that it is critical in dictating the evolution of tRNA sequences.
If all aa-tRNAs must bend into a similar structure during decoding, why have different tRNA species evolved to have a distinct set of consensus residues in the anticodon helix and core regions? A reasonable possibility is that each consensus evolved in response to the thermodynamic properties of the codon–anticodon interaction. tRNAAla and other tRNAs that form stable, GC-rich codon-anticodon complexes will have no difficulty binding ribosomes but have the potential to bind certain near-cognate codons too well. As a result, the consensus sequence of tRNAAla and other members of this class have primarily evolved to avoid misreading by possessing sequences that either weaken binding or stiffen the tRNA structure to avoid access to the bent state. This would explain the large number of misreading mutations discovered for tRNAAla. In contrast, tRNAs with weaker codon-anticodon interactions would be less prone to misreading near-cognate codons, so their consensus sequences evolved to promote binding and enhance bendability. In addition, the tRNAs in this group have all evolved post-transcriptional modifications in the anticodon loop that stabilize the codon-anticodon interaction (Grosjean et al. 2010). Nonconsensus mutations for this second class of tRNAs would be expected to reduce the reading of cognate codons, and relatively few misreading mutations would be found.
The experiments presented here suggest that many of the consensus residues that define the observed sequence diversity of bacterial tRNAs have evolved to maximize accuracy during decoding. However, since the evolution of each tRNA consensus reflects the superposition of the sequence requirements for all the steps in the synthesis, processing, function, and degradation of tRNA, it remains possible that certain consensus residues are also important for some other step in their function. It is interesting that, in the structure of tRNAPhe bound to E. coli 70S ribosomes in the hybrid P/E site, the anticodon helix is also in a distorted configuration, but the direction and angle of the bend is different than in the decoding complex (Dunkle et al. 2011). Since the entry of tRNAs into the E site also shows a distinct sequence specificity (Fei et al. 2011), it is possible that some of the tRNA consensus has evolved to optimize this step as well. Since tRNAs bound to near-cognate codons in the P site are also subject to proofreading (Zaher and Green 2009, 2010), it may be that that much of the tRNA consensus has evolved to maintain translational accuracy.
MATERIALS AND METHODS
Tightly coupled 70S ribosomes from exponential E. coli MRE600 were purified, stored in small aliquots in 50 mM HEPES (pH 7.5), 10 mM MgCl2, 70 mM NH4Cl, 1 mM DTT buffer at −80°C, and heat activated before reaction, as previously described (Fahlman et al. 2004). E. coli EF-Tu was purified from a His6-TEV expression clone (Chapman et al. 2012). mRNA fragments (IDT) are derivatives of the T4 gp32 mRNA with the following sequence: (5′-GGCAAGGAGGUAAAAAUGXXXGAACGU-3′), where XXX indicates the codon of interest in the A site. Unmodified E. coli tRNAAlaGGC and its mutants were prepared by in vitro transcription by T7 RNA polymerase from templates generated by primer extension of overlapping DNA oligonucleotides (IDT) and purified via 10% denaturing PAGE. Native tRNAAla was isolated from unfractionated E. coli tRNA using an oligonucleotide hybridization protocol (Fahlman et al. 2004). E. coli tRNAfMet was purchased from Sigma-Aldrich. tRNAs were [3′-32P]-labeled by tRNA nucleotidyltransferase (Ledoux and Uhlenbeck 2008a) and aminoacylated with purified E. coli AlaRS. Anthraniloyl-tRNAPhe was prepared from yeast tRNAPhe as described (Nawrot and Sprinzl 1998).
EF-Tu was converted into its GTP-bound active form by incubating 0.6 µM EF-Tu with 60 µM GTP, 3 µM phosphoenolpyruvate, 12 units/mL pyruvate kinase in HiFi buffer (50 mM HEPES, pH 7.0, 70 mM NH4Cl, 30 mM KCI, 3.5 mM MgCl2, 2 mM DTT, 0.5 mM spermidine, 8 mM putrescine) for 20 min at 37°C. Ternary complex was formed by mixing 0.3 mM activated EF-Tu with 25 nM [3′-32P]-labeled tRNA, incubating at 22°C for 10 min and 0° for 90 min. The rate of peptide bond formation at 22°C was performed as described previously (Ledoux and Uhlenbeck 2008a), except that HiFi buffer was used. Equal volumes of 25 nM ternary complex was mixed in a quench-flow device (Kintek) with 0.5 µM 70S ribosomes preloaded with 1.0 µM fMet-tRNAfMet and 1.5 µM mRNA. As described in Supplemental Figure 1, the 70S ribosomes in most reactions also contained 2–8 µM of a noncognate aa-tRNA (either Phe-tRNAPhe or anthraniloyl-tRNAPhe) to sequester any free EF-Tu•GTP into ternary complex and thereby prevent multiple turnovers. Each reaction was quenched with 5 mM NaOAc, 50 mM EDTA (pH 5.0). Samples were digested by P1 nuclease, separated and analyzed on PEI cellulose TCL plates in glacial acetic acid/1 M NH4Cl/H2O (5:10:85) buffer. The observed rate of reaction (kobs) and extent of dipeptide formation were determined by fitting each curve with a single exponential.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Margaret Saks and Jared Schrader for useful discussions. This work was supported by NIH grant GM037552-25 to O.C.U.
REFERENCES
- Agris PF 2008. Bringing order to translation: The contributions of transfer RNA anticodon-domain modifications. EMBO Rep 9: 629–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardell DH 2010. Computational analysis of tRNA identity. FEBS Lett 584: 325–333 [DOI] [PubMed] [Google Scholar]
- Chapman SJ, Schrader JM, Uhlenbeck OC 2012. Histidine 66 in Escherichia coli elongation factor Tu selectively stabilizes aminoacyl-tRNAs. J Biol Chem 287: 1229–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochella L, Green R 2005. An active role for tRNA in decoding beyond codon:anticodon pairing. Science 308: 1178–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkle JA, Wang L, Feldman MB, Pulk A, Chen VB, Kapral GJ, Noeske J, Richardson JS, Blanchard SC, Cate JH 2011. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science 332: 981–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahlman RP, Dale T, Uhlenbeck OC 2004. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol Cell 16: 799–805 [DOI] [PubMed] [Google Scholar]
- Faustino I, Perez A, Orozco M 2010. Toward a consensus view of duplex RNA flexibility. Biophys J 99: 1876–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei JY, Richard AC, Bronson JE, Gonzalez RL 2011. Transfer RNA-mediated regulation of ribosome dynamics during protein synthesis. Nat Struct Mol Biol 18: 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Sengupta J, Gao H, Li W, Valle M, Zavialov A, Ehrenberg M 2005. The role of tRNA as a molecular spring in decoding, accommodation, and peptidyl transfer. FEBS Lett 579: 959–962 [DOI] [PubMed] [Google Scholar]
- Giege R, Sissler M, Florentz C 1998. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res 26: 5017–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromadski KB, Rodnina MV 2004. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol Cell 13: 191–200 [DOI] [PubMed] [Google Scholar]
- Gromadski KB, Daviter T, Rodnina MV 2006. A uniform response to mismatches in codon-anticodon complexes ensures ribosomal fidelity. Mol Cell 21: 369–377 [DOI] [PubMed] [Google Scholar]
- Grosjean H, de Crecy-Lagard V, Marck C 2010. Deciphering synonymous codons in the three domains of life: Co-evolution with specific tRNA modification enzymes. FEBS Lett 584: 252–264 [DOI] [PubMed] [Google Scholar]
- Hagervall TG, Ericson JU, Esberg KB, Li JN, Bjork GR 1990. Role of tRNA modification in translational fidelity. Biochim Biophys Acta 1050: 263–266 [DOI] [PubMed] [Google Scholar]
- Harrington KM, Nazarenko IA, Dix DB, Thompson RC, Uhlenbeck OC 1993. In vitro analysis of translational rate and accuracy with an unmodified tRNA. Biochemistry 32: 7617–7622 [DOI] [PubMed] [Google Scholar]
- Johansson M, Zhang JJ, Ehrenberg M 2012. Genetic code translation displays a linear trade-off between efficiency and accuracy of tRNA selection. Proc Natl Acad Sci 109: 131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledoux S, Uhlenbeck OC 2008a. [3′-32P]-labeling tRNA with nucleotidyltransferase for assaying aminoacylation and peptide bond formation. Methods 44: 74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledoux S, Uhlenbeck OC 2008b. Different aa-tRNAs are selected uniformly on the ribosome. Mol Cell 31: 114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledoux S, Olejniczak M, Uhlenbeck OC 2009. A sequence element that tunes Escherichia coli tRNAAlaGGC to ensure accurate decoding. Nat Struct Mol Biol 16: 359–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Esberg B, Curran JF, Bjork GR 1997. Three modified nucleosides present in the anticodon stem and loop influence the in vivo aa-tRNA selection in a tRNA-dependent manner. J Mol Biol 271: 209–221 [DOI] [PubMed] [Google Scholar]
- Li W, Agirrezabala X, Lei J, Bouakaz L, Brunelle JL, Ortiz-Meoz RF, Green R, Sanyal S, Ehrenberg M, Frank J 2008. Recognition of aminoacyl-tRNA: A common molecular mechanism revealed by cryo-EM. EMBO J 27: 3322–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy FV IV, Ramakrishnan V, Malkiewicz A, Agris PF 2004. The role of modifications in codon discrimination by tRNALysUUU. Nat Struct Mol Biol 11: 1186–1191 [DOI] [PubMed] [Google Scholar]
- Nawrot B, Sprinzl M 1998. Aminoacyl-tRNA analogues; synthesis, purification and properties of 3′-anthraniloyl oligoribonucleotides. Nucleosides Nucleotides 17: 815–829 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Ramakrishnan V 2005. Structural insights into translational fidelity. Annu Rev Biochem 74: 129–177 [DOI] [PubMed] [Google Scholar]
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732 [DOI] [PubMed] [Google Scholar]
- Pan D, Zhang CM, Kirillov S, Hou YM, Cooperman BS 2008. Perturbation of the tRNA tertiary core differentially affects specific steps of the elongation cycle. J Biol Chem 283: 18431–18440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape T, Wintermeyer W, Rodnina M 1999. Induced fit in initial selection and proofreading of aminoacyl-tRNA on the ribosome. EMBO J 18: 3800–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckham HE, Olson WK 2011. Nucleic-acid structural deformability deduced from anisotropic displacement parameters. Biopolymers 95: 254–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftery LA, Yarus M 1987. Systematic alterations in the anticodon arm make tRNAGlu-Suoc a more efficient suppressor. EMBO J 6: 1499–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV 2012. Quality control of mRNA decoding on the bacterial ribosome. Adv Protein Chem Struct Biol 86: 95–128 [DOI] [PubMed] [Google Scholar]
- Saks ME, Conery JS 2007. Anticodon-dependent conservation of bacterial tRNA gene sequences. RNA 13: 651–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saks ME, Sampson JR, Abelson JN 1994. The transfer RNA identity problem: A search for rules. Science 263: 191–197 [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FV IV, Weir JR, Ramakrishnan V 2009. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326: 688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Ramakrishnan V 2011. How mutations in tRNA distant from the anticodon affect the fidelity of decoding. Nat Struct Mol Biol 18: 432–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader JM, Uhlenbeck OC 2011. Is the sequence-specific binding of aminoacyl-tRNAs by EF-Tu universal among bacteria? Nucleic Acids Res 39: 9746–9758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader JM, Chapman SJ, Uhlenbeck OC 2011. Tuning the affinity of aminoacyl-tRNA to elongation factor Tu for optimal decoding. Proc Natl Acad Sci 108: 5215–5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targanski I, Cherkasova V 2008. Analysis of genomic tRNA sets from Bacteria, Archaea, and Eukarya points to anticodon-codon hydrogen bonds as a major determinant of tRNA compositional variations. RNA 14: 1095–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RC, Karim AM 1982. The accuracy of protein biosynthesis is limited by its speed: High fidelity selection by ribosomes of aminoacyl-tRNA ternary complexes containing GTP[γS]. Proc Natl Acad Sci 79: 4922–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR 2001. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J 20: 4863–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbonavicius J, Stahl G, Durand JM, Ben Salem SN, Qian Q, Farabaugh PJ, Bjork GR 2003. Transfer RNA modifications that alter +1 frameshifting in general fail to affect −1 frameshifting. RNA 9: 760–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Sengupta J, Swami NK, Grassucci RA, Burkhardt N, Nierhaus KH, Agrawal RK, Frank J 2002. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J 21: 3557–3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlgemuth I, Pohl C, Mittelstaet J, Konevega AL, Rodnina MV 2011. Evolutionary optimization of speed and accuracy of decoding on the ribosome. Philos Trans R Soc Lond B Biol Sci 366: 2979–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarus M, Valle M, Frank J 2003. A twisted tRNA intermediate sets the threshold for decoding. RNA 9: 384–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R 2009. Quality control by the ribosome following peptide bond formation. Nature 457: 161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher HS, Green R 2010. Kinetic basis for global loss of fidelity arising from mismatches in the P-site codon: Anticodon helix. RNA 16: 1980–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.