An early step in HIV reverse transcription is the formation of the initiation complex between the RNA genome and a host-cell tRNA. The annealing process requires tRNA hybridization to the primer binding site sequence on the RNA genome, but additional interactions were proposed to occur, including the tRNA interaction with the primer activation signal, an 8-nt motif in the U5 region of the HIV RNA. The tRNA annealing process was investigated when induced by heat annealing or in the presence of the nucleocapsid protein using Fluorescence Resonance Energy Transfer, both at the ensemble and single-molecule level. Our results show that tRNA also interacts with the primer activation signal motif, which was proposed to play a regulatory role in reverse transcription initiation.
Keywords: HIV-1, tRNA annealing, ensemble FRET, single-molecule FRET, RNA conformation
Abstract
HIV-1 reverse transcription is primed by a cellular tRNAlys3 molecule that binds to the primer binding site (PBS) in the genomic RNA. An additional interaction between the tRNA molecule and the primer activation signal (PAS) is thought to regulate the initiation of reverse transcription. The mechanism of tRNA annealing onto the HIV-1 genome was examined using ensemble and single-molecule Förster Resonance Energy Transfer (FRET) assays, in which fluorescent donor and acceptor molecules were covalently attached to an RNA template mimicking the PBS region. The role of the viral nucleocapsid (NC) protein in tRNA annealing was studied. Both heat annealing and NC-mediated annealing of tRNAlys3 were found to change the FRET efficiency, and thus the conformation of the HIV-1 RNA template. The results are consistent with a model for tRNA annealing that involves an interaction between the tRNAlys3 molecule and the PAS sequence in the HIV-1 genome. The NC protein may stimulate the interaction of the tRNA molecule with the PAS, thereby regulating the initiation of reverse transcription.
INTRODUCTION
Reverse transcription in retroviruses is a complex process that results in the synthesis of a linear double-stranded DNA from the single-stranded genomic RNA template. An early step in the reverse transcription process is the formation of the initiation complex between the viral RNA (vRNA) genome and a host-cell tRNA used as a primer by reverse transcriptase (for review, see Isel et al. 2010). The annealing process requires the hybridization of the 3′ strand of the tRNA acceptor arm to the complementary primer binding site (PBS) sequence on the vRNA genome. In HIV-1, human tRNAlys3 is selectively packaged for use as the replication primer. After the primer/template complex is formed, reverse transcriptase extends the 3′ hydroxyl at the terminus of the tRNA to produce an initial cDNA product. Besides the tRNA–PBS interaction, additional interactions between the tRNA primer and the vRNA genome were proposed to play a role in reverse transcription (Murphy and Goff 1989; Aiyar et al. 1992; Isel et al. 1995; Liang et al. 1997; Miller et al. 2001). The different interactions (for review, see Sleiman et al. 2012) include an A-rich loop, a C-rich region, and the primer activation signal (PAS) motifs. A marked difference was observed between the HIV-1 MAL and NL4.3 stains. The A-rich loop interaction was found to play a role in the initiation of reverse transcription of the HIV-1 MAL strain, but not in the NL4.3 or HXB2 stains. The PAS is an 8-nt motif in the U5 region of the untranslated leader RNA that was found to stimulate tRNAlys3-mediated reverse transcription in the HIV-1 HXB2 and pNL4.3 strains (Beerens et al. 2001; Beerens and Berkhout 2002b). This U5 motif does not stimulate tRNA annealing, but rather activates the PBS-bound tRNA primer to initiate reverse transcription, and is therefore referred to as primer activation signal. The PAS motif was proposed to interact with the TΨC arm of tRNAlys3 (Fig. 1). A similar interaction was proposed for the Rous sarcoma virus (RSV) (Cobrinik et al. 1988; Aiyar et al. 1992, 1994; Miller et al. 1997; Morris and Leis 1999) and for HIV-2 (Berkhout and Schoneveld 1993; Freund et al. 2001). Thus, a common mechanism for the regulation of initiation of reverse transcription may be operating for different retroviruses.
FIGURE 1.
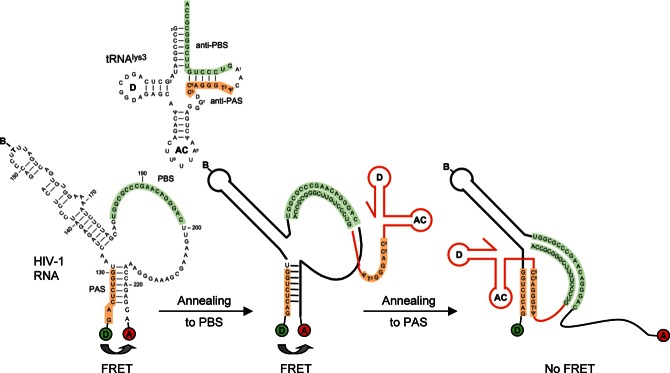
Schematic representation of the labeled vRNA template and the possible conformational changes upon tRNAlys3 annealing. The tRNAlys3 molecule is used as a primer for reverse transcription by HIV-1 and binds to the PBS (green). It was proposed that an additional interaction between the PAS element (orange) in the vRNA and the tRNA molecule activates the primer for reverse transcription. The vRNA is labeled with Cy3 (donor) at the 5′ end, and with Cy5 (acceptor) at the 3′ end, resulting in FRET. Annealing of the tRNA molecule onto the PBS is not expected to change the conformation of the stem region of the vRNA. Annealing of tRNAlys3 to the PAS motif is expected to disrupt base pairing in the stem region, resulting in loss of FRET.
Both the vRNA genome and the tRNA primer thus undergo extensive conformational changes upon initiation complex formation (Darlix et al. 2011). Previous studies have shown that the HIV-1 nucleocapsid protein (NC) acts as a nucleic acid “chaperone” that facilitates RNA rearrangements (for review, see Levin et al. 2010). NC stimulates conformational rearrangements into more stable base-paired structures, most likely by lowering the energy barrier for breakage and reformation of base pairs (Rein et al. 1998; Cruceanu et al. 2006). HIV-1 NC is a 55-amino-acid, highly basic, nucleic acid–binding protein. The mature form, also known as NCp7, is generated by the cleavage of the viral Gag precursor protein. The NC protein contains two zinc finger motifs that are critical to its nucleic acid chaperone activity (South et al. 1990; Williams et al. 2002), and appear to impart preferential binding to single-stranded nucleic acids. Due to these properties, NC plays key functions in both the early and late steps of the HIV-1 viral life cycle (Darlix et al. 2011). The NC protein assists the reverse transcriptase to convert the HIV-1 RNA genome into linear double-stranded DNA by promoting the annealing of the primer tRNA onto the PBS (Rong et al. 1998; Hargittai et al. 2004), and by directing the two DNA obligatory strand transfer reactions (Guo et al. 2000; Ramalanjaona et al. 2007). Little is known about the mechanism of tRNA annealing onto the viral genome, or about the tertiary structure of the vRNA–tRNA–NC complex. Previous studies have showed that NC does not unwind the acceptor stem of the tRNA primer in the absence of the template RNA (Chan et al. 1999). To directly investigate the tRNA annealing process and the role of the PAS interaction in the HIV-1 HXB2 strain, a fluorescence resonance energy transfer (FRET) assay was developed. In the assay, the fluorescent donor and acceptor molecules were covalently attached to an RNA template mimicking the region of the HIV-1 HXB2 genome in which the PBS is located (Fig. 1). Based on the strong dependency of FRET on the distance between the fluorescent molecules in the 2- to 10-nm range, FRET can be used as a “molecular ruler” to measure the conformational changes in the vRNA upon tRNA primer annealing. The vRNA conformation was studied both by ensemble FRET spectroscopy and by single-molecule FRET microscopy, which yield insight into the conformational heterogeneity of the RNA molecules. We also use these techniques to analyze the effects of NC on the conformation of the vRNA. The results are consistent with a model for tRNA annealing that involves a secondary interaction between the tRNAlys3 molecule and the PAS sequence in the HIV-1 genome. The interaction between the tRNA and the PAS motif was found to be dynamic, and the bound state is stimulated by the NC protein.
RESULTS
Generation of the HIV-1 RNA template and the viral RNA/tRNA complex
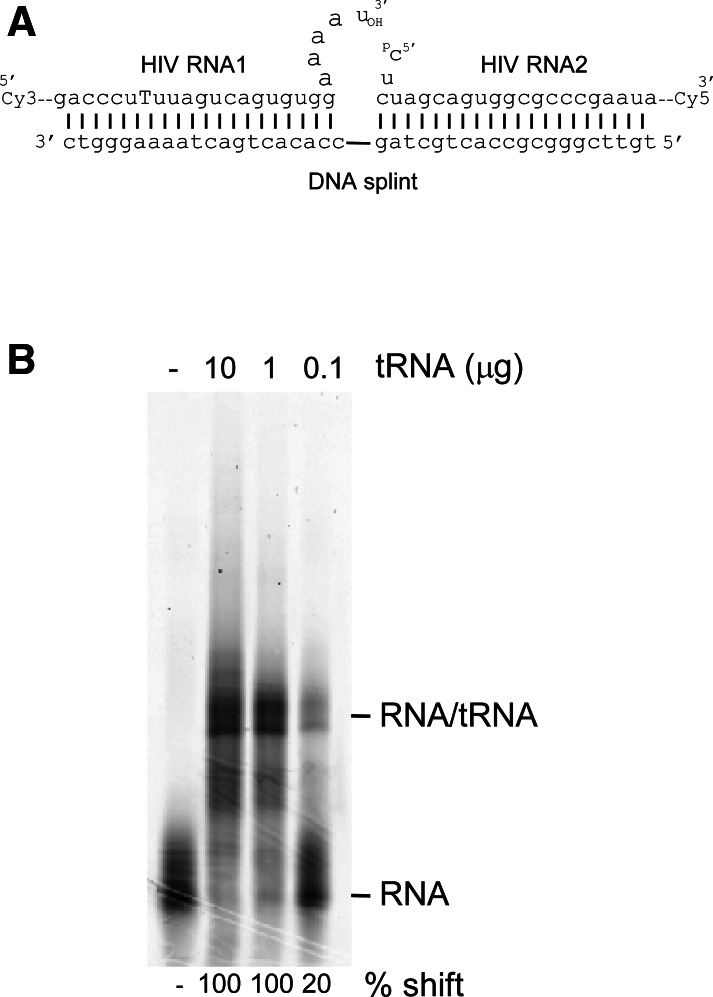
The vRNA template, encompassing nucleotides 123–225 of the genomic RNA transcript of the HXB2 molecular clone, was generated to mimic the PBS region of the HIV-1 genome. It was constructed by ligation from two synthetic RNA fragments (Fig. 2A). The RNA1 oligonucleotide contains a Cy3 dye at the 5′ end and a biotin-labeled T residue at position 154, and the RNA2 oligonucleotide has a Cy5 dye at the 3′ end and a phosphorylated 5′ end. A DNA splint was designed to hold the two RNA molecules in close proximity, while leaving single-stranded regions near the ligation junction for optimal T4 RNA ligase-mediated ligation. The reaction was analyzed by denaturing gel electrophoresis, and fluorescence scanning confirmed the production of a 103-nt product labeled with both Cy3 and Cy5, which was thereafter isolated from gel.
FIGURE 2.
(A) Schematic diagram showing generation of the Cy3- and Cy5-labeled vRNA template by ligation of two RNA oligonucleotides. (B) Gel shift assay showing the formation of the vRNA–tRNA complex upon heat annealing of increasing amounts of calf liver tRNA preparation onto the vRNA template.
The annealing of increasing amounts of the natural tRNAlys3 primer onto the PBS (nucleotides 182–199) of the HIV-1 vRNA template was studied by gel shift assays (Fig. 2B). Total calf liver tRNA was used as a source of tRNAlys3 primer. Previous studies demonstrated that this results in selective annealing of tRNAlys3, and even the related tRNAlys1,2 molecules do not act as primers (Oude Essink et al. 1996). A prominent shift of the labeled vRNA was observed upon incubation with the tRNA primer, demonstrating the formation of a vRNA/tRNA complex. Multiple bands are visible that may represent different conformations of the vRNA/tRNA complex. The percentage of complex formation was quantified by fluorescence scanning. With 3.5 ng of the HIV-1 vRNA template, complex formation was found to be 100% using 1 μg of calf liver tRNA.
Changes in HIV-1 RNA structure measured by ensemble FRET studies
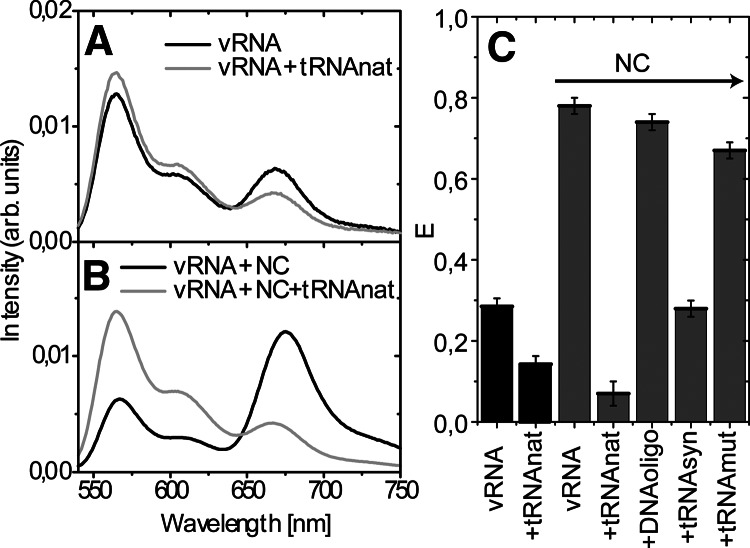
Conformational changes of the vRNA upon tRNA annealing in the absence or presence of NC were studied by ensemble FRET spectroscopy. Fluorescent probes were placed so that high FRET efficiency is expected for a correctly folded vRNA molecule (Fig. 1). Annealing of the tRNA molecule onto the PBS is not expected to change the FRET efficiency. However, annealing of tRNAlys3 to the PAS motif will disrupt base pairing in the stem region of the vRNA, which is expected to result in a strong decrease in FRET. Our assay is specifically designed to study the PAS interaction in HXB2 HIV-1. Other putative interactions with the C-rich region or A-rich loop will not disrupt base pairing in the stem region of the vRNA, and therefore cannot be studied using this assay. The emission spectrum of the vRNA upon excitation of Cy3 at 530 nm is shown in Figure 3A. The spectrum displays an emission band centered at 565 nm, originating from Cy3 fluorescence. The second emission band is centered around 670 nm and originates from Cy5 fluorescence. This band thus results from energy transfer from the Cy3 donor to the Cy5 acceptor dye. The calculated FRET value is shown in Figure 3C and can range from zero, when no energy is transferred from the donor to the acceptor, to one, when all energy is transferred. The FRET value measured for the vRNA was ∼0.25, indicating that not all vRNA molecules were found to display a high FRET efficiency. The experiments were repeated using various RNA renaturation protocols and buffers, but this did not significantly change the FRET value obtained for the vRNA sample (results not shown).
FIGURE 3.
Fluorescence spectra of the vRNA and vRNA/tRNAlys3 complex after donor excitation at 530 nm, upon heat annealing (A) and NC-mediated annealing (B) of the tRNAlys3 molecule. The vRNA was incubated with NC using a NC/nucleotide molar ratio of 1:7, and the tRNAlys3 annealing reaction was performed using a 1:35 ratio. All spectra were normalized by their integrated fluorescence intensity between 540 and 750 nm. (C) Summary of the relative FRET efficiencies measured for the vRNA upon tRNA or DNA primer annealing, in the absence (in black) or presence (in gray) of NC. Reactions with vRNA, DNA primer, and synthetic tRNA molecules were incubated with NC at an NC/nucleotide molar ratio of 1:4, and natural tRNAlys3 at a 1:20 ratio. (vRNA) Viral RNA; (tRNAnat) natural tRNAlys3; (NC) nucleocapsid protein; (DNAoligo) oligonucleotide complementary to PBS; (tRNAsyn) synthetic tRNAlys3 transcript; (tRNAmut) synthetic tRNA with PAS mutation.
Total calf liver tRNA was heat-annealed onto the vRNA template. Sample preparation for fluorescence measurements was similar to the gel-shift assays, resulting in 100% vRNA/tRNA complex formation. Annealing of the natural tRNAlys3 molecule resulted in lower FRET (i.e., lower fluorescence intensity at 670 nm) (Fig. 3A). This suggests that the average distance between the donor and acceptor dyes in the vRNA increased upon heat-annealing of the tRNA primer. This can either result from a small conformational change in all RNA molecules or from a few molecules undergoing a large conformational change.
During HIV-1 replication, the annealing of the tRNA molecule onto the vRNA genome is facilitated by the NC protein. To mimic this process, annealing reactions were performed by incubation with NCp7 at 37°C using a NC/nucleotide molar ratio of 1:7 to ensure complete coating of the RNA molecules. The vRNA template was incubated with NC; the emission spectrum of the vRNA/NC complex upon excitation at 530 nm is shown in Figure 3B. A significant increase in FRET efficiency of the HIV-1 vRNA in the presence of the NC protein was observed. This increase in signal in the presence of NC may result from more RNA molecules being folded correctly as a consequence of the chaperone activity of the protein. Alternatively, the formation of vRNA/NC aggregates may also affect FRET efficiency. Subsequently, the vRNA and natural tRNAlys3 molecules were incubated with NC (1:35 ratio), which resulted in a decrease in FRET efficiency (Fig. 3B). NC-mediated tRNA annealing resulted in a relatively larger decrease in FRET efficiency than heat-annealing of tRNAlys3 (Fig. 3A,B). A summary of the FRET values of the vRNA template alone and in complex with different primers and NC protein is shown in Figure 3C. Annealing of a DNA primer complementary to the PBS in the presence of NC did not significantly affect the FRET efficiency (Fig. 3C). To analyze the effect of NC concentration, experiments were performed using lower NC/nucleotide molar ratios. In these experiments, increasing amounts of the DNA primer were added to establish NC/nucleotide molar ratios of 1:4, 1:40, and 1:200. We measured FRET efficiencies of 0.72, 0.75, and 0.61, respectively (data not shown); thus, no significant effect of the NC/nucleotide ratio could be measured.
Annealing of a synthetic tRNAlys3 molecule in the presence of NC resulted in a small decrease in FRET compared with the natural tRNAlys3 molecule (Fig. 3C). This suggests that post-transcriptionally modified nucleotides in the natural tRNAlys3 molecule may play a role in the PAS interaction. Finally, NC-mediated annealing of a synthetic tRNA molecule with a point mutation in the tRNA anti-PAS sequence was studied. Point mutations in the PAS motif were previously reported to affect the initiation of reverse transcription (Beerens and Berkhout 2002a). The FRET efficiency was not affected by annealing of the mutant synthetic tRNA molecule (Fig. 3C), suggesting that this mutant is unable to establish the PAS interaction.
The results from the ensemble FRET assays are consistent with the model for tRNA annealing that involves secondary interaction between the tRNAlys3 molecule and the PAS sequence in the vRNA. However, insight into the dynamics and homogeneity of the different conformations of the RNA molecules cannot be obtained only by these ensemble measurements that only measure average FRET values of vRNA molecules.
Changes in HIV-1 RNA structure measured by single-molecule FRET microscopy
To resolve these issues, we performed single-molecule (SM) fluorescence microscopy experiments, which measure FRET from one molecule at a time and thus yield additional information beyond the ensemble average (for review, see Roy et al. 2008). A distinct advantage of the SM FRET technique is its capability to yield detailed insight into the conformational heterogeneity of biomolecules and detect transient states within single molecules. The technique can also distinguish single molecules from aggregates.
The conformational heterogeneity and dynamics of the vRNA molecules were investigated by SM FRET microscopy in the presence and absence of tRNA and NC. The obtained single-molecule fluorescence time traces can be classified in three different categories: zero FRET, constant FRET, and dynamic FRET, examples of which are shown in Figure 4A–C, respectively. Figure 4A shows donor and acceptor fluorescence after direct excitation of the Cy3 donor and single-step fluorescence bleaching, indicating that there are only one Cy3 donor and one Cy5 acceptor fluorophore present. No FRET signal is measured, which implies that the donor and acceptor dyes are colocalized but are too far apart for FRET (i.e., further apart than 10 nm). Figure 4B shows low donor fluorescence and high acceptor fluorescence following donor excitation, with a clear anti-correlation when the acceptor fluorescence bleaches in a single step. Here, the fluorophores are sufficiently close to each other for energy transfer to occur. The resulting FRET value is constant with time, and no dynamics are observed. Figure 4C shows an example of fluorescence time traces displaying several dynamic changes of conformations. The molecule is in a low FRET state at the start of the trace; it then passes through a high FRET state and a middle FRET state, followed by a high FRET state again returning every time to the initial low FRET state.
FIGURE 4.
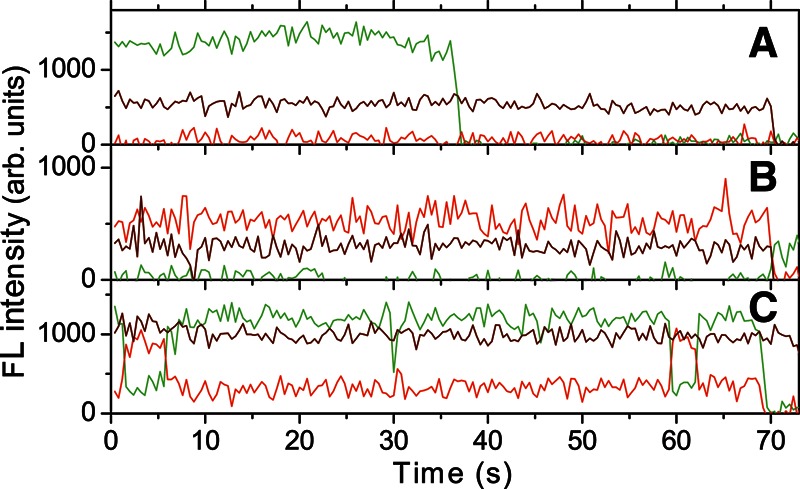
Examples of fluorescence time traces of three different single vRNA molecules with heat-annealed tRNAlys3. Fluorescence signals from the donor (green) and acceptor (red) fluorophores after donor excitation; fluorescence of the acceptor after acceptor excitation (brown). (A) Zero FRET: The two fluorophores are colocalized but are too far away to display a FRET effect. (B) Constant FRET: The molecule displays high FRET efficiency. (C) Dynamic FRET: Fluorescence time traces of a molecule showing dynamical changes in the FRET efficiency.
To visualize conformational heterogeneity, data from all selected fluorescence time traces are cut where one of the fluorophores bleaches and then plotted as FRET efficiency versus stoichiometry diagrams (Fig. 5A–D) or FRET efficiency histograms (Fig. 5E–H). FRET efficiency histograms give a direct view of the conformational diversity of the studied molecules. The same data are shown in the FRET efficiency versus stoichiometry (S) diagrams, which provide additional information about dye integrity and stoichiometry of the measurements (Kapanidis et al. 2004). Single molecules, containing only a donor, will have an S value of 1, and those containing only an acceptor, an S value of 0. Under our experimental conditions, single-molecule data with S ∼ 0.6 come from molecules that are doubly labeled and where neither the donor nor the acceptor is bleached or quenched. The single-molecule FRET histogram for the vRNA displayed a major peak centered around a high FRET efficiency of 1 (Fig. 5A,E). This FRET state is consistent with the value expected for correctly folded vRNA molecules. One-third of the vRNA molecules sampled at lower FRET values, which may be due to “breathing” or partial folding of the stem region of the vRNA.
FIGURE 5.
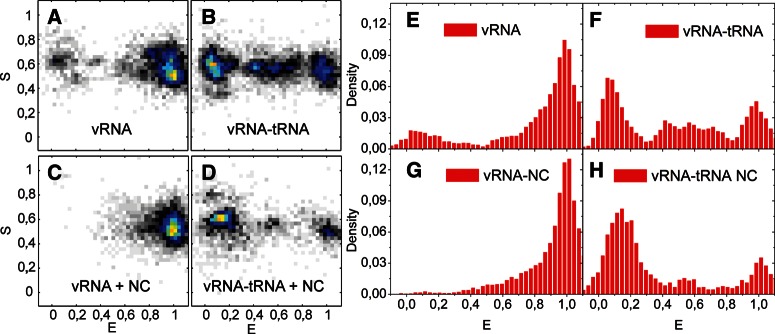
(A–D) Diagrams displaying stoichiometry versus FRET efficiency of all single-molecule events in SM buffer of doubly labeled vRNA molecules in different conditions. (E–H) FRET efficiency histograms showing single-molecule measurements in SM buffer of doubly labeled vRNA. (E) vRNA (97 molecules), (F) heat-annealed vRNA-tRNA (74 molecules), (G) vRNA in the presence of NC (88 molecules), and (H) NC-mediated vRNA-tRNA annealing (40 molecules). Data have been normalized by the number of events for comparison, and the y-axis displays which fractions of all measurements have a given FRET efficiency. Measurements in SM buffer and FRET buffer gave very similar results.
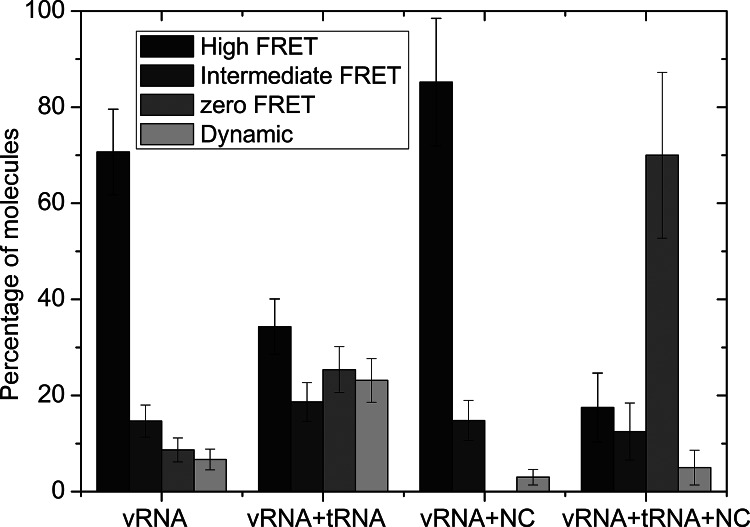
Analysis of the FRET time traces showed that ∼9% of the vRNA molecules displayed zero FRET (Fig. 6). Further analysis of the constant FRET time traces showed that the vRNA molecules can be either in a high (E = ∼1.0–0.8) or intermediate FRET state (E = ∼0.8–0.25), whereas only 6% of the vRNA molecules showed dynamic changes between these different FRET states (Fig. 6).
FIGURE 6.
Diagram showing the percentage of molecules from all single-molecule FRET data displaying a constant FRET time trace with either a high or an intermediate FRET, or a zero FRET state. Molecules that showed transition between different FRET states in their time traces are counted separately and appear in the Dynamic column. FRET values for the high FRET state were between ∼0.8 and 1, and for the intermediate state between ∼0.8 and 0.25.
This indicates that the observed broad distributions of FRET primarily arose from a relatively static heterogeneity of the vRNA, only infrequently undergoing conformational changes.
Data in the FRET efficiency versus stoichiometry diagram have a wide distribution in S ranging from 0.5 to 0.8 (Fig. 4A). A detailed analysis shows two different peaks in the S dimension at high FRET values that result from long-lived fluorescence fluctuations between discrete states. These can occur at short distances between the two dyes and originate from dye–dye fluorescence quenching (Cordes et al. 2010; Di Fiori and Meller 2010; Zhou et al. 2011). This effect has a distinct signature in fluorescence time traces, where the acceptor fluorescence intensity after direct acceptor excitation displays a decrease in fluorescence intensity when the donor and acceptor are in close proximity, i.e., for high FRET efficiencies (data not shown). The effect was also present under other experimental conditions (Fig. 5A–D).
Heat-annealing of the natural tRNAlys3 primer onto the vRNA resulted in a decrease in the number of molecules in the high FRET state compared with the vRNA alone. The major peak is now centered close to 0 and contained ∼35% of all molecules (Fig. 5B,F). In this peak, donor and acceptor dye molecules are colocalized but did not display any or very little FRET. This finding indicates that the conformational change in vRNA upon annealing of tRNAlys3 results in an increased donor–acceptor distance. Twenty-three percent of all molecules were found to display dynamics in their FRET time traces. Dynamic transitions were found both between the highest FRET peak and the zero FRET state, the highest and intermediate FRET states, and intermediate and lowest FRET states (Fig. 4C; data not shown).
To study the effect of the NC protein on the vRNA conformation, the HIV-1 RNA template was immobilized on the coverslip before incubation with NCp7 to reduce the formation of NC/RNA aggregates. The FRET histogram in the presence of NC displayed a major peak around 1, and only ∼15% of the molecules displayed lower FRET efficiencies (Fig. 5C,G). Annealing of the natural tRNAlys3 molecule in the presence of NC resulted in a decrease of molecules with high FRET and in the appearance of a major peak around 0, which contains 70% of all molecules (Fig. 5D,H). Thus, NC-mediated annealing of tRNAlys3 was found to increase the average distance significantly between the dyes in the vRNA. Fluorescence time traces of individual vRNA molecules in the presence of NC did not exhibit significant dynamics. Only 3% and 5% of the time traces showed any dynamics in the absence and presence of tRNAlys3, respectively. Using SM FRET microscopy experiments, both heat-annealing and NC-mediated annealing of tRNAlys3 were found to reduce the FRET efficiency of the vRNA. These results indicate that there is an interaction between the PAS motif in the vRNA and the tRNAlys3 molecule during primer annealing.
DISCUSSION
Reverse transcription of the HIV-1 RNA genome is primed by the cellular tRNAlys3 molecule. This highly specific process is regulated by interactions between the vRNA, tRNAlys3 molecule, and the viral RT enzyme and NC proteins. Besides binding to the PBS, additional interactions between the tRNA molecule and the viral genome were suggested to stimulate the process of reverse transcription. The interaction between the PAS motif in the viral HXB2 genome and the tRNAlys3 primer was reported to be important for the initiation of reverse transcription (Beerens et al. 2001; Beerens and Berkhout 2002a,b). An alternative interaction between the A-rich loop and the anticodon loop of the tRNAlys3 molecule was proposed specifically for the HIV-1 MAL isolate. In the HIV-1 HXB2 strain, the PAS element is located in a base-paired stem region near the PBS sequence. The binding of the tRNA molecule to the PAS motif thus requires structural rearrangements in both the vRNA and tRNA. This conformational change may be facilitated by the viral NC protein, which acts as an RNA chaperone (Rein et al. 1998). In this study, we further investigate the mechanism of tRNA annealing onto the HIV-1 HXB2 genome using a vRNA template that was specifically designed to change conformation as a response to the PAS interaction. This template was expected to display efficient energy transfer between a fluorescent donor and acceptor in a closed conformation, and a loss of energy transfer should occur only if the template is unwound by binding of the tRNAlys3 to the PAS motif.
The conformation of the HIV-1 RNA template was initially examined after heat renaturation and in the presence of NC, both by ensemble and single-molecule FRET. Both heat renaturation and the presence of NC led to the observation of a primary peak in the single-molecule experiments at a FRET value close to 1, consistent with correctly folded RNA molecules. However, the number of molecules in the major FRET peak was found to be higher in the presence of NC. For the vRNA alone, ∼70% of the molecules showed high FRET efficiency, versus 85% in the presence of NC. The addition of NC to the template thus caused a significantly higher average FRET efficiency. This finding suggests that NC stimulates folding of the fluorescently labeled vRNA into the predicted base-paired structure, resulting in efficient energy transfer. However, NC is also known to promote the aggregation of nucleic acids. This effect was clearly observed in the unsorted single-molecule events where the NC protein mediated aggregation of the vRNA into large-scale vRNA/NC complexes. Although aggregates were sorted out in the single-molecule FRET diagrams and histograms (Fig. 5), it should be noted that the single-molecule data are consistent with our ensemble FRET measurements that measure the average behavior over all molecules including aggregated and partially labeled molecules. This is also true for the experiments in the presence of tRNA and implies that our observations are not qualitatively influenced by aggregation. The observed SM FRET distributions of vRNA in the presence and absence of NC primarily arise from relatively static heterogeneity of the vRNA with only few RNA conformational changes observable in the explored time scale. This implies that complete dynamic unfolding of the vRNA stem containing the PAS motif was not observed to occur spontaneously in the absence of tRNA on the time scale of our measurements.
Both heat-annealing and NC-mediated annealing of the natural tRNAlys3 primer resulted in a loss of FRET, indicating an increase in the distance between the donor and acceptor dye, as seen by both ensemble and SM FRET. These results are consistent with the additional binding of tRNAlys3 to the PAS motif in the HIV-1 genomic RNA. The annealing of a DNA oligonucleotide onto the PBS did not change the ensemble FRET efficiency, which indicates that binding only to the PBS does not change the conformation of the vRNA. The annealing of synthetic tRNAlys3 resulted in a smaller effect on the ensemble FRET efficiency, compared with the natural tRNAlys3 molecule. We previously showed that modified nucleotides in the TΨC arm of the natural tRNAlys3 molecule are not critical for the stimulation of reverse transcription by the PAS interaction (Beerens and Berkhout 2002a). However, reduced reverse transcription activity was measured with a synthetic tRNA primer as compared with the natural primer. This suggests that modified nucleotides, although not essential for the PAS interaction, do contribute to optimal complex formation and reverse transcription. Finally, annealing of a synthetic tRNA molecule with a point mutation in the anti-PAS motif was analyzed. Point mutations in the PAS motif were previously reported to affect this interaction (Beerens and Berkhout 2002a). Annealing of the mutant tRNA molecule did not change the ensemble FRET efficiency. These combined results demonstrate that the decrease in FRET efficiency measured upon annealing of the natural or synthetic wild-type tRNA molecule results from conformational changes in the vRNA mediated by the PAS interaction.
Upon heat annealing of the tRNA primer onto the template, SM measurements showed that 35% of the molecules were in a completely open conformation, indicating that the PAS interaction had occurred, whereas 65% of the molecules displayed higher FRET efficiencies. While the high FRET efficiency state is consistent with our expectation for correctly folded RNA molecules, the intermediate FRET value states could arise from not optimally folded RNA molecules, as was shown for the vRNA template. Alternatively, these could arise from incomplete binding/dissociation of the tRNA molecule onto the PAS element. The presence of dynamics in the 23% of the FRET time trace indicates that tRNAlys3 can bind/dissociate from the PAS region during measurement of a single molecule, thereby mediating conformational changes in the vRNA.
A smaller amount of dynamic events is observed in the presence of NC in the FRET time traces, indicating that the tRNAlys3 possibly is more tightly bound to the PAS motif, probably as a consequence of NC annealing activity. In addition, ∼70% of the molecules showed a low FRET efficiency, and the conformational heterogeneity of the vRNA–tRNA–NC complex is smaller than for the heat-annealed vRNA–tRNA complex in the SM studies. This finding suggests that NC facilitates tRNA annealing to the PAS motif. NCp7 was previously suggested to stimulate the replication primer tRNALys3 annealing and minus-strand DNA synthesis and transfer (Lapadat-Tapolsky et al. 1997). In our study, we observe differences between the vRNA–tRNA complex formed by heat and by NC, while in a previous study these complexes were found to be identical (Brule et al. 2002).
The RNA structural rearrangements that occur during the reverse transcription process have previously been studied using various ensemble and single-molecule FRET assays. In one study, the structural rearrangements in the tRNA molecule during annealing onto the PBS were probed using ensemble FRET assays, demonstrating that the unwinding of the tRNA acceptor stem by NC is dependent on the presence of the vRNA template (Chan et al. 1999). Furthermore, the kinetics of the NC-chaperoned TAR–TAR interaction guiding minus-strand transfer were studied by SM microscopy (Liu et al. 2007). Recently, the initiation of HIV-1 reverse transcription was studied using SM fluorescence microscopy. This study demonstrated that the disruption of the stem–loop structure upstream of the PBS triggers the transition from initiation to the elongation phase (Liu et al. 2010). It will be very interesting to study the structural rearrangements in the PAS region of the vRNA occurring during complex formation with the RT protein, and upon subsequent initiation of reverse transcription, which is the subject of our ongoing studies.
Our results support the proposed additional interaction between the PAS sequence in the HIV-1 HXB2 genome and the tRNAlys3 primer. The PAS interaction was reported to activate the tRNA primer for the initiation of reverse transcription (Beerens et al. 2001). The virus may use the PAS interaction as a mechanism of reverse transcription regulation, where RT only recognizes or initiates reverse transcription if the vRNA/tRNA complex is in the conformation adopted upon tRNA annealing to PAS. Although binding of tRNAlys3 to the PBS may occur relatively early, e.g., in the virus-producing cells, the activation of the primer will require a structural rearrangement of the vRNA–tRNA complex to establish the PAS interaction. In this study, NC was found to stimulate the PAS interaction. Because NC is only released from the Gag polyprotein precursor during virus maturation, this will ensure proper timing for initiation of reverse transcription (Mougel et al. 2009).
MATERIALS AND METHODS
Generation of the HIV-1 RNA template
The studied HIV-1 RNA template was made by ligation of two oligonucleotides, RNA1 and RNA2. RNA1 represents nucleotides 123–172 in the HIV-1 genome and contains a Cy3 dye at the 5′ end and a biotin-labeled T residue at position 154. Nucleotide numbers refer to positions on the HIV-1 genomic RNA transcript of the HXB2 molecular clone, with +1 being the capped G residue. RNA2, encompassing nucleotides 173–225 in the HIV-1 genome, that contains a Cy5 dye at the 3′ end. A DNA splint oligonucleotide (TGTTCGGGCGCCACTGCTAGCCACACTGACTAAAAGGGTC) partially complementary to oligonucleotides RNA1 and RNA2 was used to guide the ligation reaction. The RNA oligonucleotides (Ribotask) were HPLC-purified for single-molecule fluorescence measurements.
The RNA2 oligonucleotide was phosphorylated by incubation with ATP and T4 polynucleotide kinase (NEB). The phosphorylated RNA2 oligonucleotide was mixed with the RNA1 oligonucleotide and the DNA splint oligonucleotide at a 1:1:1 molar ratio, at a final oligonucleotide concentration of 4 μM. Oligonucleotides were annealed by incubation for 10 min at 65°C, followed by slow cooling to room temperature in the presence of annealing buffer (83 mM Tris-HCl at pH 7.5, 125 mM KCl). Then T4 RNA ligase (NEB) was added to a final concentration of 1 unit/μL, and the reactions were incubated for 1 h at 37°C in the presence of ligase buffer (NEB). The ligation products were analyzed on a 12% denaturing polyacrylamide gel and visualized by Typhoon fluorescence scanning (Amersham Biosciences). The ligated RNA product was isolated from the gel by overnight elution in TE buffer (10 mM Tris-Cl at pH 7.5, 1 mM EDTA). The RNA was ethanol-precipitated and dissolved in renaturation buffer (10 mM Tris-HCl at pH 7.5, 100 mM NaCl). The RNA was renatured by incubation for 2 min at 85°C and for 10 min at 65°C, followed by slow cooling to room temperature. Aliquots were stored at −80°C.
Generation of synthetic tRNA primer
To generate a template for the production of synthetic tRNAlys3, the tRNA gene was PCR-amplified. We used the pUC-tRNAlys3 construct described previously (Oude Essink et al. 1995) with the sense primer 5′-T7-tRNA (tRNA positions 1–20) and the antisense primer 3′-tRNA (tRNA positions 54–76). This will yield the complete tRNA from nucleotide 1 to 76. A point mutation was introduced in the anti-PAS motif of the tRNA by PCR mutagenesis, changing the anti-PAS sequence from wild-type 5′-CCAGGGUU-3′ to mutant 5′-CCAGuGUU-3′. The PCR products were used directly for in vitro transcription using the MegaShortscript T7 transcription kit (Ambion), according to the manufacturer’s instructions. Transcripts were purified on 4% denaturing polyacrylamide gels and visualized by UV shadowing. Upon overnight elution in TE buffer, the RNA was ethanol-precipitated, dissolved in renaturation buffer, and quantified by UV spectrometry. The RNA was renatured by incubation for 2 min at 85°C and for 10 min at 65°C, followed by slow cooling to room temperature. Aliquots were stored at −20°C.
HIV-1 nucleocapsid protein
HIV-1 NCp7 was synthesized by the opfp chemistry and purified to homogeneity by HPLC as described previously (Lapadat-Tapolsky et al. 1997; Bampi et al. 2006). It was dissolved at 1 mg/mL in oxygen-free buffer: 25 mM HEPES (pH 6.9), 50 mM NaCl with 2.5 equivalents of ZnCl2 per mole of NCp7 so that the two zinc fingers were in the Zn2+-bound form.
Gel shift assays
The HIV-1 RNA template (3.5 ng) was incubated with 0.1, 1, or 10 μg of calf liver tRNA (total tRNA preparation; Novagen) in 10 µL of FRET buffer (10 mM Tris at pH 7.5, 100 mM NaCl, 2 mM Mg) for 2 min at 85°C and for 10 min at 65°C, followed by slow cooling to room temperature. Previous studies demonstrated that this results in selective annealing of tRNAlys3, and even the related tRNAlys1,2 molecules do not act as a primer (Oude Essink et al. 1996). After addition of 5 µL of nondenaturing gel loading buffer (30% glycerol with BFB dye), the samples were analyzed on a 4% native polyacrylamide gel. Gels were run at 100 V at room temperature, and analyzed by Typhoon fluorescence scanning (Amersham Biosciences). The percentage of complex formation was calculated by dividing the amount of shift by the total amount of labeled RNA in each lane.
Ensemble FRET measurements
Sample preparation for fluorescence measurements was similar to the gel-shift assays. Reactions were performed in 65 µL of annealing buffer. Seventy nanograms of the HIV-1 RNA template was incubated alone or with either 20 μg of calf liver tRNA, 50 ng of synthetic tRNA, or 15 ng of a 21-nt DNA primer complementary to the PBS (5′-CAAGTCCCTGTTCGGGCGCCA-3′) for 2 min at 85°C and for 10 min at 65°C, followed by slow cooling to room temperature. Reactions with the NC protein were incubated for 30 min at 37°C, where 70 ng of HIV-1 RNA template was incubated with 200 ng of NCp7, resulting in an NC/nucleotide molar concentration of 1:7.
The NC/nucleotide molar ratio for the different primer annealing reactions is described in the legend of Figure 3. Fluorescence spectra were recorded in FRET buffer at room temperature using a spectrofluorometer (FluoroMax3, Horiba Jobin Yvon) with an excitation wavelength of 530 nm. Relative FRET efficiencies were calculated as IA/(IA + ID), where IA is the peak fluorescence intensity of Cy5 excited through the FRET process, which has been corrected both for contribution from donor fluorescence and direct acceptor excitation, and ID is the peak fluorescence intensity of Cy3. Fluorescence measurements of the vRNA template in the absence and presence of tRNA were performed in several buffers including FRET buffer, FRET2 buffer (10 mM Tris at pH 7.5, 100 mM NaCl, 5 mM Mg) and SM buffer (20 mM Tris-HCl at pH 7.5, 100 mM KCl).
Single-molecule FRET microscopy
Experimental setup
Single-molecule FRET measurements were conducted using an inverted wide-field optical microscope and alternate laser excitation of the fluorescent donor (Cy3) and acceptor (Cy5); the setup has been described in more detail in Kruger et al. (2010). Fluorescence was collected through the objective in epi geometry and sent to a CCD (MicroMax; Roper scientific) for detection. Images were recorded with a 100- and a 200-msec integration time. Typical excitation intensities were 0.85 and 0.3 kW/cm2 for the green (514.5 nm) and red (630 nm) laser, respectively.
Sample preparation
For single-molecule studies, samples at a concentration of ∼50–100 pM were immobilized on a coverslip by biotin–streptavidin interaction. Silicon gaskets (Grace Bio-labs, USA), covered by an additional glass coverslip, were used to seal the imaging chamber from oxygen. Coverslips were first etched using 1% hydrofluoric acid for 1 min. The slides were then coated with BSA-biotin (Sigma-Aldrich) by 5 min of incubation at room temperature, followed by 5 min of incubation with streptavidin (Molecular Probes), both at a concentration of 0.1 mg/mL in SM buffer or in FRET buffer.
Samples containing the NC protein were immobilized on PEG-coated coverslips with a low density of biotin (MicroSurfaces Inc.) incubated with streptavidin as above.
Before immobilization, the HIV-1 RNA template was renatured as described previously. Heat-annealing of the tRNA primer was mediated by incubation of the HIV-1 RNA template (3.5 ng) with 1 μg of calf liver tRNA for 2 min at 85°C and for 10 min at 65°C, followed by slow cooling to room temperature. The RNA or RNA/tRNA samples were diluted about 50-fold in SM-buffer or in FRET buffer containing an oxygen scavenger system (1 mM Trolox [Sigma-Aldrich], 0.92 mg/mL glucose oxidase [Sigma-Aldrich], 0.04 mg/mL catalase [Sigma-Aldrich], and 4.5 mg/mL β-D-(+)glucose [Sigma Aldrich]). The sample was immobilized on streptavidin-coated coverslips by 5 min of incubation at room temperature and then brought to the microscope for detection.
To study the effects of NCp7, the HIV-1 RNA template was immobilized on the PEG-coated coverslips, before the addition of NCp7 to prevent the formation of NC/RNA aggregates. The renatured RNA template was incubated on the coverslip for 5 min, followed by rinsing with SM buffer. NCp7 (0.5 μg, to ensure complete coating) was then incubated for 10 min at room temperature in SM buffer containing the oxygen scavenger system, but not β-D-(+) glucose, which initiates the catalytic reactions of catalase and glucose oxidase. For samples containing only the RNA template and NCp7, β-D-(+)glucose was hereafter added. Otherwise, 100 ng of calf liver tRNA was incubated for 10 min at room temperature, and, finally, β-D-(+)glucose was added before recording the fluorescence intensity time trajectories.
Data analysis
Data analysis was performed by a software package written in MATLAB (MathWorks), and colocalized donor/acceptor fluorescence spots were found. The resulting fluorescence time traces were analyzed, and only those ending with single-step donor and/or acceptor photobleaching were selected for further analysis because this indicated that the source of the signal was a single doubly labeled molecule. Relative FRET efficiencies were calculated from the donor and acceptor fluorescence intensities as
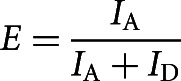 |
and the relative stoichiometry factor as
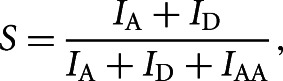 |
where IA and ID are the measured fluorescence intensities of acceptor and donor upon donor excitation, respectively, after correction for background, donor leakage, and direct acceptor excitation (Roy et al. 2008). IAA is the measured fluorescence intensity of the acceptor after acceptor excitation.
ACKNOWLEDGMENTS
We thank the mechanical workshop and the building group at the Department of Physics and Astronomy of Aarhus University for assistance, Rita Rosendahl Hansen for help with sample preparations, and Ebbe Sloth Andersen at the Department of Molecular Biology of Aarhus University for practical assistance in the early stages of the project. This work was supported by the Danish Council for Independent Research; the VKR foundation; the Lundbeck foundation; Aarhus University; and the Alfred Benzon foundation.
REFERENCES
- Aiyar A, Cobrinik D, Ge Z, Kung HJ, Leis J 1992. Interaction between retroviral U5 RNA and the TΨC loop of the tRNATrp primer is required for efficient initiation of reverse transcription. J Virol 66: 2464–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiyar A, Ge Z, Leis J 1994. A specific orientation of RNA secondary structures is required for initiation of reverse transcription. J Virol 68: 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bampi C, Bibillo A, Wendeler M, Divita G, Gorelick RJ, Le Grice SF, Darlix JL 2006. Nucleotide excision repair and template-independent addition by HIV-1 reverse transcriptase in the presence of nucleocapsid protein. J Biol Chem 281: 11736–11743 [DOI] [PubMed] [Google Scholar]
- Beerens N, Berkhout B 2002a. Switching the in vitro tRNA usage of HIV-1 by simultaneous adaptation of the PBS and PAS. RNA 8: 357–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerens N, Berkhout B 2002b. The tRNA primer activation signal in the human immunodeficiency virus type 1 genome is important for initiation and processive elongation of reverse transcription. J Virol 76: 2329–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerens N, Groot F, Berkhout B 2001. Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J Biol Chem 276: 31247–31256 [DOI] [PubMed] [Google Scholar]
- Berkhout B, Schoneveld I 1993. Secondary structure of the HIV-2 leader RNA comprising the tRNA-primer binding site. Nucleic Acids Res 21: 1171–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brule F, Marquet R, Rong L, Wainberg MA, Roques BP, Le Grice SF, Ehresmann B, Ehresmann C 2002. Structural and functional properties of the HIV-1 RNA–tRNA3Lys primer complex annealed by the nucleocapsid protein: Comparison with the heat-annealed complex. RNA 8: 8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan B, Weidemaier K, Yip WT, Barbara PF, Musier-Forsyth K 1999. Intra-tRNA distance measurements for nucleocapsid protein-dependent tRNA unwinding during priming of HIV reverse transcription. Proc Natl Acad Sci 96: 459–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobrinik D, Soskey L, Leis J 1988. A retroviral RNA secondary structure required for efficient initiation of reverse transcription. J Virol 62: 3622–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes T, Santoso Y, Tomescu AI, Gryte K, Hwang LC, Camara B, Wigneshweraraj S, Kapanidis AN 2010. Sensing DNA opening in transcription using quenchable Forster resonance energy transfer. Biochemistry 49: 9171–9180 [DOI] [PubMed] [Google Scholar]
- Cruceanu M, Urbaneja MA, Hixson CV, Johnson DG, Datta SA, Fivash MJ, Stephen AG, Fisher RJ, Gorelick RJ, Casas-Finet JR, et al. 2006. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res 34: 593–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darlix JL, Godet J, Ivanyi-Nagy R, Fosse P, Mauffret O, Mely Y 2011. Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J Mol Biol 410: 565–581 [DOI] [PubMed] [Google Scholar]
- Di Fiori N, Meller A 2010. The effect of dye–dye interactions on the spatial resolution of single-molecule FRET measurements in nucleic acids. Biophys J 98: 2265–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund F, Boulme F, Litvak S, Tarrago-Litvak L 2001. Initiation of HIV-2 reverse transcription: A secondary structure model of the RNA–tRNALys3 duplex. Nucleic Acids Res 29: 2757–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wu T, Anderson J, Kane BF, Johnson DG, Gorelick RJ, Henderson LE, Levin JG 2000. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J Virol 74: 8980–8988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargittai MR, Gorelick RJ, Rouzina I, Musier-Forsyth K 2004. Mechanistic insights into the kinetics of HIV-1 nucleocapsid protein-facilitated tRNA annealing to the primer binding site. J Mol Biol 337: 951–968 [DOI] [PubMed] [Google Scholar]
- Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R 1995. Initiation of reverse transcription of HIV-1: Secondary structure of the HIV-1 RNA/tRNA3Lys (template/primer) complex. J Mol Biol 247: 236–250 [DOI] [PubMed] [Google Scholar]
- Isel C, Ehresmann C, Marquet R 2010. Initiation of HIV reverse transcription. Viruses 2: 213–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapanidis AN, Lee NK, Laurence TA, Doose S, Margeat E, Weiss S 2004. Fluorescence-aided molecule sorting: Analysis of structure and interactions by alternating-laser excitation of single molecules. Proc Natl Acad Sci 101: 8936–8941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger AC, Raarup MK, Nielsen MM, Kristensen M, Besenbacher F, Kjems J, Birkedal V 2010. Interaction of hnRNP A1 with telomere DNA G-quadruplex structures studied at the single molecule level. Eur Biophys J 39: 1343–1350 [DOI] [PubMed] [Google Scholar]
- Lapadat-Tapolsky M, Gabus C, Rau M, Darlix JL 1997. Possible roles of HIV-1 nucleocapsid protein in the specificity of proviral DNA synthesis and in its variability. J Mol Biol 268: 250–260 [DOI] [PubMed] [Google Scholar]
- Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K 2010. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol 7: 754–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Li X, Rong L, Inouye P, Quan Y, Kleiman L, Wainberg MA 1997. The importance of the A-rich loop in human immunodeficiency virus type 1 reverse transcription and infectivity. J Virol 71: 5750–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HW, Zeng Y, Landes CF, Kim YJ, Zhu Y, Ma X, Vo MN, Musier-Forsyth K, Barbara PF 2007. Insights on the role of nucleic acid/protein interactions in chaperoned nucleic acid rearrangements of HIV-1 reverse transcription. Proc Natl Acad Sci 104: 5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Harada BT, Miller JT, Le Grice SF, Zhuang X 2010. Initiation complex dynamics direct the transitions between distinct phases of early HIV reverse transcription. Nat Struct Mol Biol 17: 1453–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JT, Ge Z, Morris S, Das K, Leis J 1997. Multiple biological roles associated with the Rous sarcoma virus 5′ untranslated RNA U5-IR stem and loop. J Virol 71: 7648–7656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JT, Ehresmann B, Hubscher U, Le Grice SF 2001. A novel interaction of tRNALys,3 with the feline immunodeficiency virus RNA genome governs initiation of minus strand DNA synthesis. J Biol Chem 276: 27721–27730 [DOI] [PubMed] [Google Scholar]
- Morris S, Leis J 1999. Changes in Rous sarcoma virus RNA secondary structure near the primer binding site upon tRNATrp primer annealing. J Virol 73: 6307–6318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mougel M, Houzet L, Darlix JL 2009. When is it time for reverse transcription to start and go? Retrovirology 6: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JE, Goff SP 1989. Construction and analysis of deletion mutations in the U5 region of Moloney murine leukemia virus: Effects on RNA packaging and reverse transcription. J Virol 63: 319–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oude Essink BB, Das AT, Berkhout B 1995. Structural requirements for the binding of tRNA3Lys to reverse transcriptase of the human immunodeficiency virus type 1. J Biol Chem 270: 23867–23874 [DOI] [PubMed] [Google Scholar]
- Oude Essink BB, Das AT, Berkhout B 1996. HIV-1 reverse transcriptase discriminates against non-self tRNA primers. J Mol Biol 264: 243–254 [DOI] [PubMed] [Google Scholar]
- Ramalanjaona N, de Rocquigny H, Millet A, Ficheux D, Darlix JL, Mely Y 2007. Investigating the mechanism of the nucleocapsid protein chaperoning of the second strand transfer during HIV-1 DNA synthesis. J Mol Biol 374: 1041–1053 [DOI] [PubMed] [Google Scholar]
- Rein A, Henderson LE, Levin JG 1998. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: Significance for viral replication. Trends Biochem Sci 23: 297–301 [DOI] [PubMed] [Google Scholar]
- Rong L, Liang C, Hsu M, Kleiman L, Petitjean P, de Rocquigny H, Roques BP, Wainberg MA 1998. Roles of the human immunodeficiency virus type 1 nucleocapsid protein in annealing and initiation versus elongation in reverse transcription of viral negative-strand strong-stop DNA. J Virol 72: 9353–9358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Hohng S, Ha T 2008. A practical guide to single-molecule FRET. Nat Methods 5: 507–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleiman D, Goldschmidt V, Barraud P, Marquet R, Paillart JC, Tisne C 2012. Initiation of HIV-1 reverse transcription and functional role of nucleocapsid-mediated tRNA/viral genome interactions. Virus Res 169: 324–339 [DOI] [PubMed] [Google Scholar]
- South TL, Blake PR, Sowder RC III, Arthur LO, Henderson LE, Summers MF 1990. The nucleocapsid protein isolated from HIV-1 particles binds zinc and forms retroviral-type zinc fingers. Biochemistry 29: 7786–7789 [DOI] [PubMed] [Google Scholar]
- Williams MC, Gorelick RJ, Musier-Forsyth K 2002. Specific zinc-finger architecture required for HIV-1 nucleocapsid protein’s nucleic acid chaperone function. Proc Natl Acad Sci 99: 8614–8619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Kunzelmann S, Webb MR, Ha T 2011. Detecting intramolecular conformational dynamics of single molecules in short distance range with subnanometer sensitivity. Nano Lett 11: 5482–5488 [DOI] [PMC free article] [PubMed] [Google Scholar]