The U2-U6 snRNA complex plays a critical catalytic role in pre-mRNA splicing. The authors use enzymatic structure probing, 19F NMR techniques, and analytical ultracentrifugation to characterize the secondary structural fold of the protein-free human spliceosomal U2-U6 snRNA complex. Results show that the complex predominantly adopts a four-helix conformation, consistent with results from previous genetic studies. However, there is a lesser fraction of a three-helix conformer, which had previously been identified by genetic studies in yeast, in equilibrium with the four-helix fold. The low energy barrier between the two may facilitate conformational exchange as part of the splicing process.
Keywords: spliceosome, U2-U6 snRNA, secondary structure, enzymatic structure probing, 19F-NMR, conformational heterogeneity
Abstract
The complex formed between the U2 and U6 small nuclear (sn)RNA molecules of the eukaryotic spliceosome plays a critical role in the catalysis of precursor mRNA splicing. Here, we have used enzymatic structure probing, 19F NMR, and analytical ultracentrifugation techniques to characterize the fold of a protein-free biophysically tractable paired construct representing the human U2-U6 snRNA complex. Results from enzymatic probing and 19F NMR for the complex in the absence of Mg2+ are consistent with formation of a four-helix junction structure as a predominant conformation. However, 19F NMR data also identify a lesser fraction (up to 14% at 25°C) of a three-helix conformation. Based upon this distribution, the calculated ΔG for inter-conversion to the four-helix structure from the three-helix structure is approximately −4.6 kJ/mol. In the presence of 5 mM Mg2+, the fraction of the three-helix conformation increased to ∼17% and the Stokes radius, measured by analytical ultracentrifugation, decreased by 2%, suggesting a slight shift to an alternative conformation. NMR measurements demonstrated that addition of an intron fragment to the U2-U6 snRNA complex results in displacement of U6 snRNA from the region of Helix III immediately 5′ of the ACAGAGA sequence of U6 snRNA, which may facilitate binding of the segment of the intron adjacent to the 5′ splice site to the ACAGAGA sequence. Taken together, these observations indicate conformational heterogeneity in the protein-free human U2-U6 snRNA complex consistent with a model in which the RNA has sufficient conformational flexibility to facilitate inter-conversion between steps of splicing in situ.
INTRODUCTION
The excision of noncoding intervening sequences (introns) from eukaryotic precursor messenger (pre-m)RNA molecules and ligation of flanking coding regions (exons), a process called pre-mRNA splicing, is a critical step in the maturation of nascent mRNA transcripts and in the generation of alternative products from polycistronic genes. This process involves two sequential transesterification reactions catalyzed by the spliceosome, a dynamic nuclear ribonucleoprotein complex that comprises five recyclable small nuclear (sn)RNA molecules and at least 70 small nuclear ribonucleoprotein particle (snRNP)-associated proteins and more than 100 non-snRNP proteins (Stark and Luhrmann 2006). In the first transesterification reaction, the 2′OH of a conserved adenosine residue in the intron, called the branch site because of the branched lariat intermediate it forms, executes a nucleophilic attack at the 5′ splice site and results in formation of a free 3′OH. The second reaction is characterized by attack of this free 3′OH at the 3′ splice site, thus joining the two exons and releasing the lariat intron.
The snRNA components of the spliceosome are implicated in key splicing roles, including recognition, pairing, and catalysis. Of the five snRNAs, only U2 and U6 snRNA are directly involved in both splicing steps and participate in generation of the catalytic core (Fabrizio and Abelson 1990; Madhani and Guthrie 1992; McPheeters and Abelson 1992). The complex formed between U2 and U6 snRNA molecules, comprising extensive intra- and intermolecular base-pairing sequences, assists in positioning both the 5′ splice site and the branch site region of the intron for the first cleavage reaction (Black et al. 1985; Parker et al. 1987; Lesser and Guthrie 1993).
In addition, the complex is implicated in catalytic activity, which depends upon several specifically bound Mg2+ ions (Sontheimer et al. 1997; Yean et al. 2000; Huppler et al. 2002; Valadkhan and Manley 2002; Yuan et al. 2007), one of which is located within the highly conserved AGC triad of U6 snRNA. Catalytic activity, albeit at a low rate and yield, by the protein-free human U2-U6 snRNA complex underscores the importance of the snRNA components in the splicing reaction (Valadkhan and Manley 2001; Valadkhan et al. 2007, 2009). However, the Prp8 protein, an integral component of the U5 snRNP, contains an RNase H domain near the C terminus, suggesting that a protein component contributes to the reaction chemistry (Pena et al. 2008; Ritchie et al. 2008; Yang et al. 2008).
Mechanistic parallels between reactions catalyzed by the spliceosome and the self-splicing group II intron (Weiner 1993; Sontheimer et al. 1999; Valadkhan and Manley 2002), as well as sequence and structural similarities between functionally analogous RNA sequences in the two systems (Gordon et al. 2000; Keating et al. 2010), suggest that the metal ion-binding sites in the U2-U6 snRNA complex are brought into close proximity to each other (Steitz and Steitz 1993), as noted in the Group II intron of Oceanobacillus iheyensis (Toor et al. 2008). The important role of these metal ions implies the need for a specific structural context for each site and for any interaction between them, which may differ for the two steps of splicing. Thus, the ability of the U2-U6 snRNA complex to undergo conformational change is likely to be important.
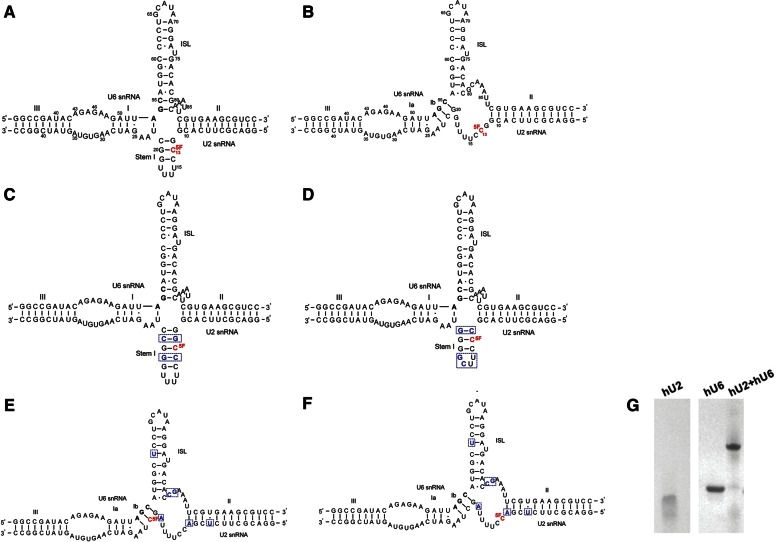
Functional and structural studies suggest different conformational models of the U2-U6 snRNA complex. Each of the proposed folds comprises U2-U6 snRNA intermolecular Helices II and III, as well as a U6 intramolecular stem–loop (U6 ISL); the features that vary include subhelices of Helix I, as well as lengths of the other helices and appearance or absence of an intramolecular U2 snRNA Stem I. Importantly, the models depict the genetically conserved AGC triad in different pairing environments, which may have an impact on folding and function. Genetic studies in yeast Saccharomyces cerevisiae support the importance of formation of Helix Ib, in which the AGC triad forms three intermolecular base pairs with U2 snRNA and U2 Stem I is opened, resulting in formation of a three-helix structure (Madhani and Guthrie 1992). This model was reinforced by mutational studies that implicated the requirements of the 3-bp-Helix Ib in at least one (Hilliker et al. 2007) and possibly both (Mefford and Staley 2009) cleavage steps. Additionally, results of cross-linking assays in yeast identified tertiary interactions only possible in the three-helix model (Ryan et al. 2004). On the other hand, the finding by McPheeters and Abelson that formation of certain base pairs corresponding to Stem I was important for splicing in yeast suggests the possibility that the alternative four-helix model is active at some point (McPheeters and Abelson 1992). In human cells, however, interactions identified by genetic suppression assays were consistent only with formation of an extended U6 snRNA ISL and formation of U2 Stem I (Fig. 1A; Sun and Manley 1995), characteristics of the four-helix model.
FIGURE 1.
Models for the conformation of the protein-free human (h) U2-U6 snRNA complex. (A) Four-helix junction model proposed by Sun and Manley (1995). (B) Three-helix model with data adapted from the model proposed by Madhani and Guthrie (1992) for the yeast U2-U6 snRNA complex. The sequences shown are fragments of hU2 and hU6 snRNAs with several changes made to the native sequences to increase the transcription yield and pairing efficiency as specified in Materials and Methods. For 19F NMR studies, C13 of U2 snRNA was substituted with 5-fluoro-cytidine (5-19F-C; red nucleotide). (C) Sequence and proposed secondary structure of a mutation that favors the four-helix model in which hU6 snRNA is paired with a mutated hU2 snRNA with two extra G-C/C-G base pairs to increase the stability of the Stem I. The extra G-C/C-G base pairs are in blue, and the 5-19F-C substitution is in red. (D) Sequence and proposed secondary structure of a mutation favoring the four-helix model. In this mutant, the UUUU tetra-loop of U2 snRNA was mutated to a hyperstable UUCG loop, and the top base pair in Stem I was mutated from G-C to C-G in order to disfavor the formation of Helix Ib. The mutated nucleotides from the original sequences are in blue and the 5-19F-C substitution is in red. (E) Sequence and proposed secondary structure of a mutation that favors the three-helix model. The mutated nucleotides from the original sequences are in blue and the 5-19F-C substitution is in red. (F) Same sequence as E but with different 5-19F-C substitution (red nucleotide). (G) Pairing of RNA oligomers representing hU2 and U6 snRNA analyzed by nondenaturing gel electrophoresis. Lanes 1 and 2 show relative migration of fragments representing U2 and U6 snRNA fragments, respectively; lane 3 demonstrates retarded migration upon annealing of the two snRNA fragments. All samples were electrophoresed in a single gel; lanes were cut to delete lanes of unrelated samples.
Biochemical studies by Moore and Sharp, using splicing substrates that contained a chiral phosphorothioate, demonstrated that both steps of splicing are strongly inhibited by one diastereomer but not by the other, suggesting that the active site undergoes conformational change between steps (Moore and Sharp 1993). Interestingly, most genetic studies in yeast support the formation of Helix Ib, associated with the three-helix model; in contrast, genetic studies in human cells support the formation of the extended ISL and Stem I, associated with the four-helix model. Since all evidence indicates that the chemical mechanism of splicing is identical in yeast and human systems (Moore and Sharp 1993), inter-conversion between these two structures may be a possibility. However, no direct evidence of inter-conversion between conformers has been shown in any one system in situ. It is, thus, tempting to speculate that the differences in conformations observed in the yeast and human systems are related to differences in experimental design, modulating effects of spliceosomal proteins, differences in energetics of stem formation by different RNA sequences, and/or observation of different events.
Conformational features of the U2-U6 snRNA complex have also been examined in vitro. NMR investigation of a truncated yeast complex demonstrated that U6 ISL is extended to include the AGC triad, and U2 snRNA forms the intramolecular Stem I, therefore creating a four-helix junction; no spectral evidence was reported for an alternative conformation (Sashital et al. 2004). However, a more recent model of the protein-free yeast U2-U6 snRNA complex derived from a combination of solution NMR, small angle X-ray scattering, and computer modeling of a sequence in which an additional 5 bp of the native sequence in Helix II were included, indicated formation of a three-helix junction structure consistent with the conformation identified in cellular studies of yeast (Burke et al. 2012). Thus, it appears that the complex is capable of forming multiple conformations in the region of the junction under different conditions.
Supporting these findings, Cao and Chen (2006), using computational studies, have demonstrated that both human and yeast U2-U6 snRNA complex can form multiple conformations. The distribution between them can be affected by minor changes in the sequence and/or presence of Mg2+ and/or spliceosomal proteins. Consistent with this notion, single-molecule fluorescence data of a model yeast U2-U6 snRNA complex have given a strong evidence of inter-conversion between two folds attributed to four- and three-helix conformers that was highly dependent upon Mg2+ concentration (Guo et al. 2009).
In order to analyze conformational changes associated with catalytic activity, it is essential to know the lowest energy conformation in the absence of metal ions, RNA, or protein components that may induce structural changes. To address this question, we used biochemical structure probing and solution NMR to investigate conformational features of a set of biophysically tractable constructs representing the human U2-U6 snRNA (and mutations thereof) in vitro (sequence changes described in Materials and Methods). Our results are consistent with the formation of a four-helix junction characterized by the presence of U2 Stem I, rather than the three-helix structure, as the predominant fold. However, our NMR studies also identified a small but significant fraction of the U2-U6 snRNA complex forming an alternative conformation, which may be the three-helix structure in equilibrium with the major conformation. These findings suggest that the energetic barrier for conformational change is low, consistent with the findings of Guo et al. (2009), thus facilitating inter-conversion between the steps of splicing.
RESULTS
To investigate the lowest energy (ground state) conformation of the human (h) U2-U6 snRNA complex in solution, we used enzymatic probing and NMR on the complexes formed by U2 and U6 snRNA fragments representing the functionally important sequences. Strand pairing to form a single complex with a stoichiometry of 1:1 was confirmed by nondenaturing gel electrophoresis under the conditions similar to those used for the different experiments (Fig. 1G; details in Materials and Methods).
Enzymatic structure probing
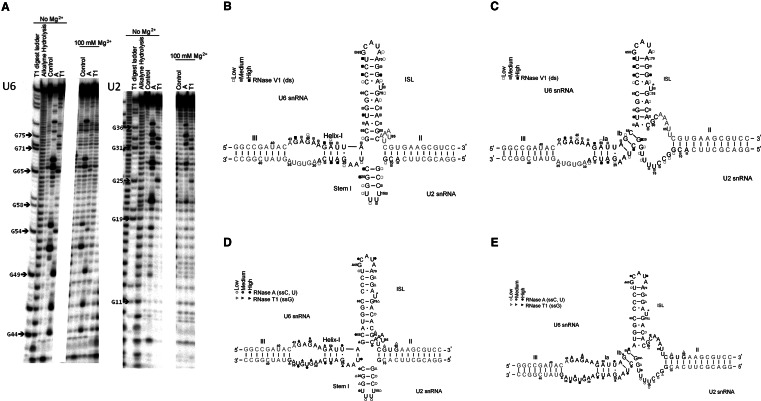
The products of the RNase V1 digestion, which preferentially cleaves at regions that are double-stranded or highly stacked, in the presence of 1 mM MgCl2 were resolved and quantified to generate cleavage profiles (representative gel in Fig. 2A). We observed the greatest cleavage of the labeled hU2 (*hU2) strand at nucleotides 9–11, 14–16, 20–21, and 26–28; *hU6 exhibited the most intense cleavage at nucleotides 44–51, 56–58, 62–65, 71–72, and 77–79. Very short fragments were not visible due to gel artifacts; however, these sequences are predicted to be double-stranded in either model. Formation of an appropriate number of Watson-Crick base pairs, likely to be associated with Helix III and Helix II, were verified in separate 1H NMR experiments by characteristic imino-imino and imino-amino patterns (data not shown).
FIGURE 2.
(A) Representative sequencing gel exhibiting cleavage patterns of 32P-labeled (*)hU2 and *hU6 snRNA paired with respective counter-strands subjected to enzymatic probing by ribonucleases RNase A and T1 (see Materials and Methods for experimental details). Lanes are labeled on top with the ribonuclease used and the concentration of Mg2+. (B,C) Normalized cleavage intensities of complex between *hU6 and hU2 snRNAs and the complex between *hU2 and hU6 snRNAs following reaction with RNase V1 were mapped onto the two possible folds (B: four-helix model; C: three-helix model) of the U2-U6 snRNA complex. Cleavage by RNase V1 at the corresponding nucleotides is represented by squares, with open, gray, and black squares corresponding to low, medium, and high intensity, respectively. (D,E) Normalized cleavage intensities of complex between *hU6 and hU2 snRNAs and the complex between *hU2 and hU6 snRNAs following reaction with RNases A and T1 were mapped onto the two folds as in Figure 2, B and C. Cleavage by RNase A and RNase T1 at corresponding nucleotides is represented by open, gray, and black circles or triangles, respectively, corresponding to low, medium, and high cleavage by the ribonuclease, respectively. Nucleotides in gray represent those for which information was not collected because of gel artifacts.
The cleavage profile was mapped onto schematic secondary structural models corresponding to the four-helix and three-helix folds (Fig. 2B,C). The cleavage pattern for the U6 strand is consistent with expected formation of the U6 intra-molecular stem–loop (ISL), a common feature in both models. In addition, the intense cleavage patterns of several residues in the segment G12-C21 of U2 snRNA suggest formation of a short stem corresponding to the position of the proposed U2 snRNA Stem I, a prominent feature of the four-helix junction model.
We probed for single-stranded regions by reaction with RNase A (unpaired C and U) and RNase T1 (unpaired G); assignment of “high,” “medium,” or “low” cleavage is based upon quantification of band intensity by SAFA software (Laederach et al. 2008). Cleavage profiles indicate the greatest intensities for *hU2 at nucleotides 18, 21–25, and 28–36, with low cleavage observed at nucleotides 12–14 and 19–20; the most intense cleavage for *hU6 was noted at nucleotides 44–49, 52–55, 64–69, and 80–81. These locations, mapped onto secondary structural models (Fig. 2D,E), coincide with the expected unpaired residues in the hU6 ISL and the ACAGAGA loop, features that are common to both models. However, we also observed some cleavage at residues proposed to reside in Helix I or Ib, which are not expected to be single-stranded. Other cleavage sites correspond to regions whose secondary structure differs in the two models: for example, when mapped onto the schematic figures of the two models, nucleotides 12–14 and 19–20 of U2 snRNA map to a segment anticipated to be single-stranded in the three-helix structure but double-stranded in the four-helix model. The lack of extensive cleavage in these regions, along with evidence for formation of Stem–Loop I from the RNase V1 cleavage patterns, favors formation of a four-helix junction by the U2-U6 snRNA complex under these conditions. However, presence of a low but reproducible amount of cleavage by RNases A and T1 in the putative Stem I, as well as other observed inconsistencies in the data as mapped on the two secondary structural models, suggests that there may be a fraction of the alternative three-stemmed model present.
19F-NMR studies
In order to analyze the possibility of the presence of a minor conformational fraction and, if so, the distribution between the two, we pursued 19F NMR studies of the human U2-U6 snRNA complex. Since Stem I only appears in the four-helix junction model, monitoring its presence or absence provides a straightforward “handle” to distinguish between the two proposed conformation models for the human U2-U6 snRNA complex. We measured the formation of Stem I in U2 snRNA from the fluorine chemical shift of a single 5-19F-cytidine (5-19F-C) at position C13 of U2 snRNA as a reporter for single- or double-stranded status. This substituted nucleotide residue is base-paired if Stem I forms or single-stranded if Stem I does not form.
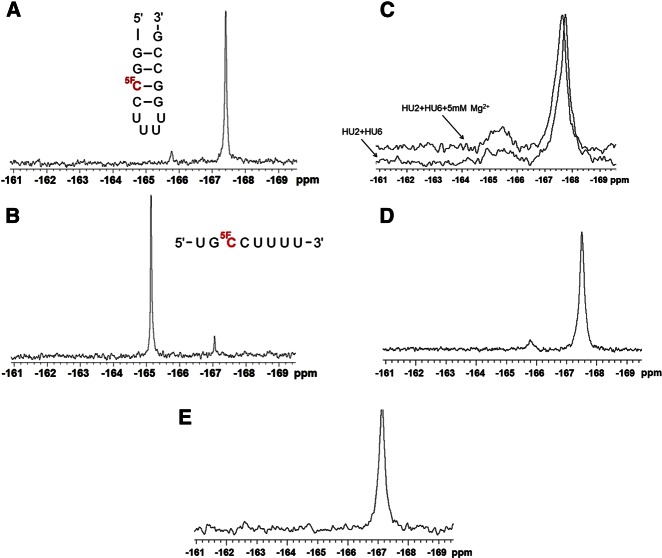
We acquired 19F spectra for two controls containing 5-19F-C, one within a stem in a short stem–loop, and the other as a single-stranded fragment, each with a sequence context matching that of the equivalent region of U2 snRNA, to provide reference spectra for the two alternative conformations (Fig. 3A,B, respectively). The predominant 19F resonance peaks for the single 5-19F-C in each oligomer were at −167.4 and −165.1 ppm for the double- and single-stranded controls, respectively, values that correspond with previous measurements (Puffer et al. 2009). We noted small peaks (∼5%–7% of the total) at −165.8 and −167.1 ppm in the controls, respectively, suggesting some distribution between paired and unpaired states in each case. A spectrum of exchangeable 1H of the double-stranded control at 10°C revealed four imino protons in the 12–15 ppm range, consistent with the anticipated four base pairs in the stem (data not shown). A spectrum of exchangeable 1H for the single-stranded control, however, did not reveal any hydrogen-bonded imino protons, suggesting that any protons involved in base-pairing in the single-stranded control were in rapid exchange with the solvent at 10°C and did not form a stable base-paired structure (data not shown).
FIGURE 3.
One-dimensional 19F NMR spectra of control RNA oligomers and the U2-U6 snRNA complex or its mutations with single 5-19F-C substitutions. (A) Double-stranded control oligomer, and (B) single-stranded control oligomer. The sequences were indicated by each spectrum, and the position of 5-19F-C substitution is shown in red. (C) Overlaid one-dimensional 19F NMR spectra of the human U2-U6 snRNA complex with (upper trace) and without (lower trace) 5 mM Mg2+ at 25°C in 95% H2O/5% 2H2O. Each spectrum was labeled in the figure. (D) One-dimensional 19F NMR spectrum of the extended U2 snRNA pairing with U6 snRNA (mutation 1) at 25°C in 95% H2O/5% 2H2O. (E) One-dimensional 19F NMR spectrum of the mutated UUCG tetra-loop U2 snRNA pairing with U6 snRNA (mutation 2) at 25°C in 95% H2O/5% 2H2O. All spectra were acquired on a Varian INOVA 500 MHz spectrometer using the acquisition parameters outlined in Materials and Methods.
The proton-coupled one-dimensional 19F spectrum of the U2-U6 snRNA complex acquired at 25°C displayed a large peak at approximately −167.8 ppm, suggesting that the 5-19F-C at position 13 is predominantly within a duplex (Fig. 3C). This peak appears to include a small “shoulder” of a second peak with a very similar chemical shift (also in the range consistent with a double-stranded environment), suggesting conformational heterogeneity, perhaps associated with different orientations of the stem–loop. However, we also observed a very broad (and most likely, multicomponent) peak with a center at approximately −165.4 ppm, with an area ∼14% of the total, implying that this fraction of the U2-U6 snRNA complex is single-stranded in this region. Whether this structure corresponds to the three-helix conformation proposed by Madhani and Guthrie (1992) or dissociation of the intramolecular base-pairing cannot be distinguished from these data alone.
In order to distinguish between the two alternatives and help identify the conformation associated with the single-stranded broad peak, we designed several mutations to favor either the four-helix or the three-helix structure, respectively. In the first mutation, formation of putative Stem I in U2 snRNA is strengthened by addition of two base pairs to either side of the 19FC-G base pair (Fig. 1C). Because the chemical shift of the 19F-substituted nucleotide is sensitive to the sequence context, the identity of the neighboring nucleotides was maintained. The one-dimensional 19F spectrum of this mutated complex displayed a major peak at approximate −167.5 ppm and a minor peak (∼5%) centered at approximately −165.8 ppm (Fig. 3D). The area of the peak with single-stranded chemical shift decreases from 14% to 5%, accompanied by significant narrowing compared with the very broad (and perhaps combined) resonance of the wild-type complex; there is also precise overlap with the single-stranded peak observed as the minor conformation of the stem–loop control. These data suggest that the downfield-shifted single-stranded peak of the mutant sequence with the extended Stem I is a different species from that seen in the one-dimensional 19F spectrum of the wild-type U2-U6 snRNA complex. Based upon similarities with the downfield peak in the double-stranded control, we anticipate that this peak also represents a minor species associated with transient opening of Stem I and that this species is also likely to contribute (as the upfield region) to the combined single-stranded broad peak observed in the spectrum of the wild-type complex.
To investigate this possibility further, we designed and tested another mutant in which we changed the hairpin loop sequence of the putative Stem I of U2 snRNA from UUUU to the hyperstable UUCG sequence, which is likely to enhance the thermal stability of Stem I, and the top base pair of Stem I from G-C to C-G, which we predicted would disfavor formation of Helix Ib (a key feature of the three-helix model), both of which would be expected to favor the four-helix structure (Fig. 1D). For this mutated complex, we observed no peak at the single-strand chemical shift (Fig. 3E). These findings support our hypothesis that the major component of the broad peak we observed in the wild-type complex represents an alternative conformation, such as the three-helix conformation, and not only “breathing” of Stem I or dissociation of the two strands in the complex.
Observations from the 19F-NMR studies are limited to the local conformation around the labeled nucleotide. In order to verify that the global structure was not altered by the Stem I mutations, we repeated the enzymatic structure probing assays on the two mutants designed to enhance formation of Stem I. The experiments were performed in the same buffer with 1 mM MgCl2 for all three enzymes RNase A, RNase T1, and RNase V1.
Cleavage patterns following incubation of both the mutant with the extended Stem I and with the UUCG-substituted Stem I with RNase V1 were very similar to those measured for the wild-type U2-U6 snRNA sequence. However, based upon visual comparison of the intensity of cleavage patterns, those regions associated with marked cleavage of the wild-type complex demonstrated a small increase in intensity in each of the mutant sequences for *hU2 (nucleotides 9–14, 19–23, and 26–28) compared to regions expected to be common to both models. Cleavage patterns of the mutant sequences by RNase A and RNase T1 were also very similar to those of the wild type, although with a relatively small decrease in the cleavage intensity of the nucleotides 13–19 in the mutant with the UUCG Stem I with respect to invariant regions. These data suggest that the mutations designed to favor formation of Stem I and the four-stemmed structure do not result in global conformational change, but that nucleotides in the region of U2 Stem I are less likely to be single-stranded in the mutant sequences, i.e., that formation of a four-stemmed conformation was, indeed, favored.
Formation of Helix Ib has not been previously demonstrated in the human U2-U6 snRNA complex. In order to verify the ability of the human complex to form a three-stemmed conformation analogous to that proposed in yeast, we also designed a mutated complex to favor Helix Ib formation. This was accomplished by disrupting base-pair formation in the lower region of the U6 ISL (G80C, C81G of U6 snRNA) and Stem I (C12A, G19A) (Fig. 1E,F). Presence of an upfield-shifted 19F peak at −168.1 ppm is consistent with a double-stranded environment for the substituted nucleotide in the mutant complex shown in Figure 1E. Moreover, presence of several downfield-shifted peaks in the range of −164.6 ppm implies a somewhat heterogeneous single-stranded environment for the substituted nucleotide in a complex with the same sequence mutations shown in Figure 1F. These findings suggest that a three-helix structure containing Helix Ib is stable and can be favored by disrupting U2 Stem I and lower U6 ISL (data not shown).
Effect of Mg2+
To investigate whether high Mg2+ concentrations affect the secondary structure of the human U2-U6 snRNA complex, we repeated the enzymatic structure probing experiments on the wild-type sequence in the presence of 100 mM MgCl2 under otherwise identical conditions. The overall cleavage patterns were very similar to those conducted in 0–1 mM Mg2+; however, we noted a small increase (5%–15%) in the cleavage intensity of nucleotides 78–82 of *hU6 relative to cleavages in the ISL and ACAGAGA regions, as well as for nucleotides 12–18 in *hU2. These data may be consistent with a small shift toward opening of U2 snRNA Stem–Loop I at high Mg2+concentrations.
We also repeated the 19F NMR experiment in the presence of 5 mM MgCl2. Our result showed that both conformations were present and that the fraction of the single-stranded peak increased slightly to 17% at 25°C, suggesting a minor shift in distribution induced by Mg2+ (Fig. 3C, upper trace).
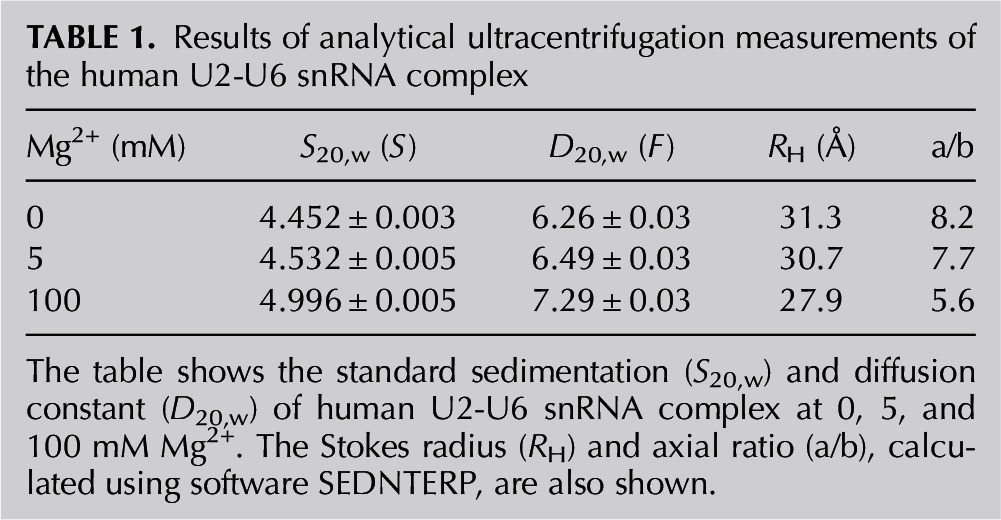
To detect changes in the global conformation of the human U2-U6 snRNA complex as a function of Mg2+ concentration, sedimentation velocity measurements were obtained. U2 and U6 snRNA fragments were annealed and folded in buffer containing 0, 5, and 100 mM Mg2+, and the sedimentation and diffusion coefficients (S20,w and D20,w) were determined. Each sample is characterized by a single symmetric peak in the g (s*) vs. s* distributions, consistent with a single species. The Stokes radius (RH) and the axial ratio (a/b) for human U2-U6 snRNA complex in the absence of Mg2+ were calculated to be 31.3 Å and 8.2 (Table 1). These values decreased to 30.7 Å/7.7 and 27.9 Å/5.5 in the presence of 5 and 100 mM Mg2+, respectively, suggesting that Mg2+ induces a slightly more compact and less asymmetric tertiary structure.
TABLE 1.
Results of analytical ultracentrifugation measurements of the human U2-U6 snRNA complex
Interaction of the intron strand with U2 snRNA
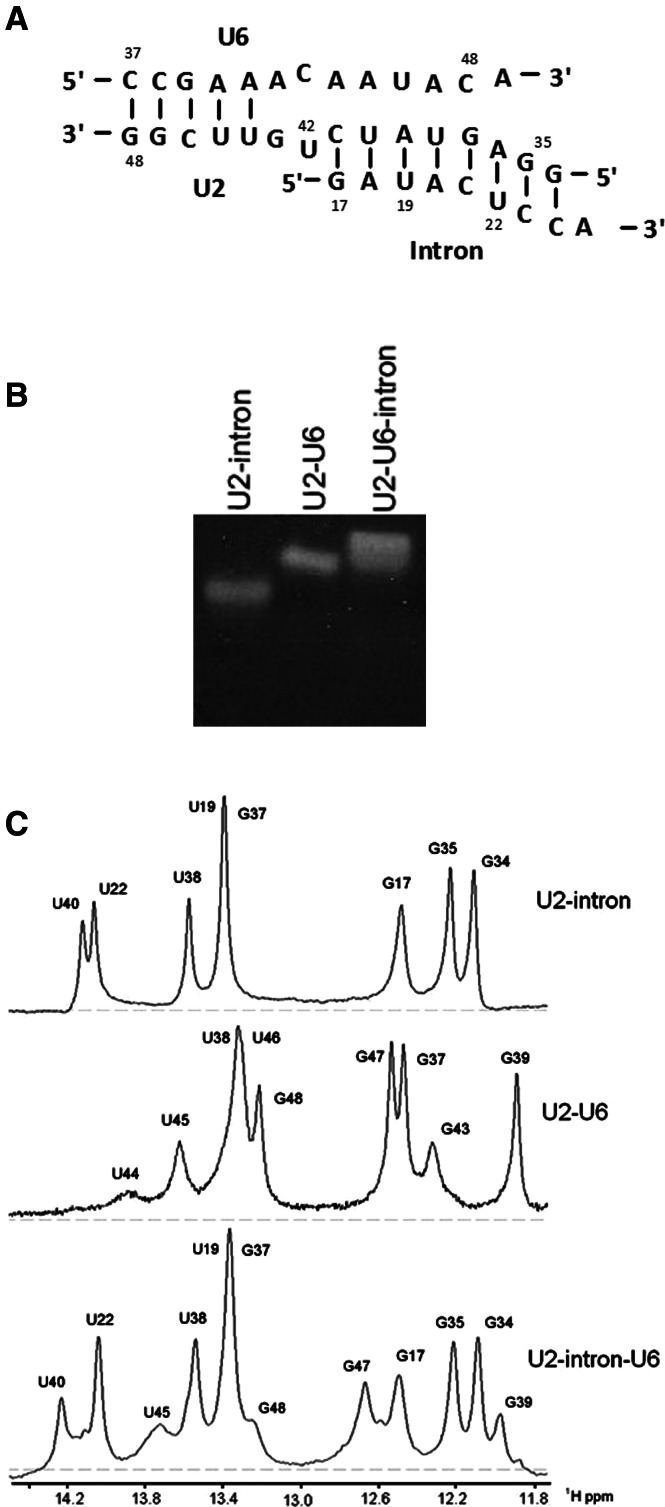
The segment of U2 snRNA proposed to pair with U6 snRNA to form Helix III includes 3 nt that are also complementary to those of the intron. We used nondenaturing polyacrylamide gels and solution NMR spectroscopy to help distinguish whether this region of U2 snRNA pairs with U6 snRNA, the intron, or both via formation of a triple helix. RNA oligomers whose interaction was tested include (1) a 13-nt U6 snRNA strand including ACA of the ACAGAGA loop and 10 nt 5′ to it, (2) a segment of U2 snRNA (15 nt) including nucleotides pairing with the intron’s branch site sequence and nucleotides 3′ to it, and (3) a 9-nt intron strand that included a UAC sequence that is identical in the U6 strand. Two Gs were added to both ends of the U2 strand and the corresponding Cs at the 5′ and the 3′ end of U6 and intron strands, respectively, to promote pairing (Fig. 4A). The branch site residues were not included in the intron strand, and the native segment of the strand ends with U22 (Fig. 4A).
FIGURE 4.
(A) Pairing of fragments of U2 and U6 snRNAs and an intron oligomer representing the proposed Helix III interaction. Results of NMR studies (panel C of this figure) indicate that part of the U2-U6 Helix III dissociates in the presence of the intron, and the U2-intron pairing predominates in the three-strand complex. (B) Nondenaturing gel demonstrating the pairing of U2, U6 snRNA, and intron oligomers representing the branch site helix and Helix III. Components electrophoresed in each lane are labeled above the gel. (C) Imino 1H region of one-dimensional spectra of exchangeable protons of the strands depicted in panel A to study the (1) U2-intron duplex (Newby and Greenbaum 2001), (2) U2-U6 duplex, and (3) U2-intron-U6 complex. Eight imino resonances observed in the spectra of the U2-intron are also observed in the spectrum of the U2-intron-U6, consistent with unperturbed U2-intron pairing in the complex. Four imino resonances attributed to the U2-U6 duplex are observed in the spectra of the U2-intron-U6 complex; three terminal G-C base pairs that were added to stabilize U2-U6 interaction and the adjacent A-U base pair are detected in the spectra of the three-strand complex. These data suggest that U6 does not interact with the U2-intron in the vicinity of the proposed Helix III.
A 1:1 ratio of RNA oligomers was combined, and their relative mobility on nondenaturing polyacrylamide gels was compared with that of individual strands. An equimolar mixture of the U2 snRNA and intron strands, and of the U2 and U6 snRNA strands, each formed a single band migrating at a slower rate, consistent with duplex formation; the slowest migrating band was observed in the lane containing a stoichiometric mixture of all three strands, indicating formation of a U2-U6-intron interaction (Fig. 4B).
One-dimensional spectra of the U2-U6-intron three-strand complex exhibited resonances (these data were corroborated by data from NOESY spectra; data not shown) consistent with formation of 12 base pairs, eight of which were attributed to the U2-intron duplex (Newby and Greenbaum 2001) and four attributed to a U2-U6 pairing. The NMR data were consistent with formation of a short U2-U6 stem involving the 5′ end of U6, i.e., residues G48, G47, and U45 of U2 and G39 of U6 (Fig. 4C). No interactions were detected between the U6 strand and the U2-intron duplex, nor did we observe any evidence for formation of a triple helix or Hoogsteen interactions. As a result, the ACAGAGA loop extends to include at least residues 46–48 of the yeast U6 snRNA, leaving them available to pair with the segment of the intron adjacent the 5′ splice site.
DISCUSSION
Results of enzymatic structure probing and 19F NMR reported here demonstrate formation of U2 snRNA Stem I in the predominant lowest energy conformation of the human U2-U6 snRNA complex, consistent with formation of a four-helix junction by the protein-free RNA complex. This conformation agrees with the functional model proposed by Sun and Manley (1995) based on results of genetic experiments in human cells and is analogous to the conclusion reached by Sashital et al. (2004) from NMR studies of protein-free yeast RNA fragments. Our results also suggest that the predominant four-helix junction is in equilibrium with the three-helix conformation; features of this model have been proposed for a protein-free yeast complex in vitro (Burke et al. 2012) and during the splicing process of yeast in situ (Madhani and Guthrie 1992; Hilliker and Staley 2004; Mefford and Staley 2009).
Consistent with these different models, it is possible that the spliceosome exists in at least two distinct conformational states during the course of the splicing activity. Opposite stereochemistry and different substrates associated with the two splicing reactions suggest the possibility of different catalytic centers for each of the two cleavage steps (Moore and Sharp 1993), which, in turn, may require conformational rearrangement within the U2-U6 snRNA complex. Accordingly, Query and Konarska proposed a “two-state” model for the two steps of splicing in which dual conformations are in equilibrium (Query and Konarska 2004; Liu et al. 2007).
In addition, computational studies by Cao and Chen (2006) demonstrated a propensity for conformational heterogeneity of protein-free U2-U6 snRNA complexes from both yeast and human sequences with a truncated Helix I/III. They observed a different distribution for the yeast and human sequences, which they have attributed to different sequences near the junction region. The appearance of multiple low-energy folds may reflect the potential for rearrangement of the junction region during splicing activity. Quantification of conformational heterogeneity of cell-free U2-U6 snRNA complexes, in which coaxial stacking patterns of RNA helices are not modulated by proteins, provides an advantage in characterization of conformational change associated with metal ions and spliceosomal proteins.
Structural rearrangement of individual protein-free yeast U2-U6 snRNA complexes into at least three distinct states in the presence of Mg2+ was shown by fluorescence resonance energy transfer experiments (Guo et al. 2009), suggesting the propensity for ion-dependent conformational change.Specifically, two major conformations were attributed to four-helix and three-helix models, respectively, with an obligatory intermediate. The equilibrium distribution they found was very different from that predicted by Cao and Chen (2006) using computational analysis. This observation may be the result of differences in the lengths and sequences of the stems the two teams examined or the result of differences in data obtained from experimental and statistical approaches, respectively.
The computational study also found a different distribution of folds for the human complex than for that of yeast, specifically favoring formation of a four-helix conformer, which they attributed to the sequence differences in the human complex junction (Cao and Chen 2006). We note that several minor conformations identified by their calculations were associated with the presence of highly truncated Helix I/III in the human U2 and U6 snRNA sequences tested.
Our first approach to probing the human U2 and U6 snRNA complex in vitro was to employ enzymatic structure mapping techniques, which are useful in identifying single- and double-stranded regions of folded RNA molecules (Ehresmann et al. 1987) on sequences including more complete Helix I and III stems. Results from this approach were most consistent with formation of a four-helix junction. However, these data are not absolute in their nucleotide specificity and cannot reliably differentiate minor populations of alternative conformations from nonspecific reaction. For example, RNase V1 cleaves not only after base-paired nucleotides but also stacked single-stranded regions, and enzymes specific for single-strand regions may not have access to certain nucleotides (Lowman and Draper 1986). In particular, our data do not exclude the possibility of equilibrium between the major fold and a minor coexisting conformation.
In addition to inclusion of more complete stem lengths for Helix III and Helix I, we evaluated changes in the base-pairing patterns of the U2-U6 snRNA complex on the Helix III side upon binding of an intron strand. A segment of the conserved intron sequence is identical to the UAC sequence of U6 snRNA in the region immediately 5′ of the ACAGAGA loop that pairs with U2 snRNA in the absence of the intron. Our NMR investigation of a protein-free complex showed that in the presence of the intron strand, the U2-U6 snRNA pairing opens up so that the 3 nt of U2 complementary to both U6 and the intron pair with the intron, leaving that region of U6 snRNA unpaired. We observed no evidence of higher-order structure such as a triple helix. Opening of this segment of Helix III, which creates a larger ACAGAGA loop, frees these nucleotides to interact with their respective pre-mRNA targets (Parker et al. 1987; Wu and Manley 1989; Zhuang et al. 1989; Sawa and Abelson 1992; Sawa and Shimura 1992) and proteins (Yan and Ares 1996). These findings are also consistent with mutational and photo-cross-linking data in a yeast system (Ryan and Abelson 2002; Ryan et al. 2004).
We then exploited different chemical shifts of 5-19F-incorporated pyrimidine nucleotide, in either a single- or double-stranded region of RNA, which has previously been used to probe the secondary structure of RNA (Horowitz et al. 1977; Marshall and Smith 1977; Gollnick et al. 1986; Chu et al. 1992; Kanyo et al. 1996; Sahasrabudhe and Gmeiner 1997; Arnold and Fisher 2000; Hammann et al. 2001; Olejniczak et al. 2002; Hennig et al. 2007; Puffer et al. 2009) to demonstrate the presence of an alternative conformation of U2-U6 snRNA complex conclusively. The advantage of observing the 19F nucleus by NMR is the broader range and the greater sensitivity of fluorine chemical shifts in response to the local environment as compared to those of hydrogen because the fluorine nucleus is surrounded by nine electrons vs. a single electron in hydrogen. We specifically targeted C13 of U2 snRNA, which resides within the helix of Stem I in the four-helix junction model but would be single-stranded otherwise. Although this is also an ensemble approach, it allowed us to quantify distribution between populations for a specific state.
Identification of a very dominant resonance peak at the chemical shift value corresponding to 5-19F-cytidine in a double-stranded Stem I reinforces results of the enzymatic probing experiments, specifically inclusion of U2 snRNA Stem I in the lowest energy state conformation. However, the presence of the lesser peak corresponding to the single-stranded 5-19F-cytidine suggests that there are alternative conformations present in the complex, which may be in equilibrium. NMR data verifying exchange between conformations in which the 5-19F-cytidine is located in different environments strongly suggest a time scale far slower than that expected for transient opening and closing of the base pairs (C Zhao and NL Greenbaum, unpubl.). 19F NMR data for the third mutant sequence, designed to favor formation of a three-helix model (Fig. 1E), indicated that the human U2-U6 snRNA complex is, indeed, capable of forming Helix Ib. Therefore, while it is likely that part of the broad peak observed for the original sequence corresponds to “breathing,” our data are consistent with the ability to adopt the three-helix conformer. The low energy barrier between conformations suggests that the human U2-U6 snRNA complex adopts alternative structures under different conditions, perhaps stabilized by specific contacts favoring the formation or presentation of different active sites associated with the two steps of splicing.
Several lines of investigation support conformational rearrangement between the two steps of splicing experimentally. For example, Tseng and Cheng (2008) demonstrated that both catalytic steps of splicing are reversible, which suggests the possibility of conformational rearrangement (Tseng and Cheng 2008). Also, Prp8 assists substrate repositioning by altering the equilibrium between the two steps (Query and Konarska 2004; Liu et al. 2007). Prp16p-dependent opening and closing of Helix I was demonstrated (Mefford and Staley 2009). In agreement with their conclusions, results of site-directed hydroxyl radical cleavage have shown an alteration in the spatial relationship between U6 snRNA ISL and the ACAGAGA loop between the two steps of splicing (Rhode et al. 2006).
In vitro experiments also provide evidence for conformational rearrangement upon addition of Mg2+. Butcher and coworkers observed an ∼9% decrease in the radius of gyration at 2 mM Mg2+ for the yeast complex assayed by small angle X-ray scattering (Burke et al. 2012). However, a notably greater change was reported from single-molecule FRET studies (Guo et al. 2009), in which a large shift in the fraction of the three-helix conformation from the four-helix junction conformation was observed in the presence of 10 mM Mg2+. We also investigated whether Mg2+ induced conformational change in the human sequence as was found by Guo et al. (2009) in the yeast sequence. 19F NMR experiments indicated a small increase in the three-helix structure, and sedimentation velocity data suggested a small amount of compaction of the three-dimensional structure in the presence of Mg2+; these changes may occur in the protein-free system as a result of altered patterns of coaxial stacking of stems upon interaction with the metal ion. The single symmetrical peak we observed in analytical ultracentrifugation experiments suggests that the two conformations measured by 19F NMR either have similar sedimentation velocities or produced an average behavior.
Based upon the relative population of the two conformers obtained from the distribution of 19F NMR resonance peaks at 25°C, we calculated a ΔG of −4.6 kJ/mol for formation of the four-helix structure from the three-helix structure in the absence of Mg2+ and −4.0 kJ/mol in the presence of 5 mM Mg2+, consistent with a very low inter-conversion barrier that would favor the changes necessary to form different active sites for each of the two steps in the context of the active spliceosome. Since it is likely that some component of the broader peak is the result of “breathing,” that is, transient opening and closing of the Stem I helix, assignment of the entire broad peak, which represents ∼14% of the total, to the alternative conformation may be an overestimate. If, in fact, the alternative conformation only reflects a lesser population, then the actual ΔG value would be somewhat greater.
In summary, our data show that the human U2-U6 snRNA complex adopts two conformations with an apparent low energy barrier between conformations. Such a finding is energetically consistent with inter-conversion alternative structures under different conditions, perhaps stabilized by specific contacts favoring the formation or presentation of different active sites that facilitate the two steps of splicing.
MATERIALS AND METHODS
Models for folding of the U2-U6 snRNA complex
Based upon proposed models for secondary structural folds of the yeast (Madhani and Guthrie 1992; Sashital et al. 2004; Burke et al. 2012) and the human U2-U6 snRNA complexes (Sun and Manley 1995), we constructed two alternative folds of the human U2-U6 snRNA complex: one including a four-helix junction in which the AGC triad is paired with opposing nucleotides at the base of the U6 ISL, and the other describing a three-helix structure in which the AGC sequence pairs intermolecularly with U2 snRNA to form Helix Ib (Fig. 1A,B, respectively).
Formation of the U2-U6 snRNA complex
RNA fragments representing the regions of human U2 and U6 snRNA sequences were designed with several modifications to minimize formation of undesirable self-paired complexes. Specifically, we replaced the hairpin loop of U6 snRNA ISL, GCGCA, with the yeast sequence GCAUA, changed the U9 of the U2 strand to A9 to form a complementary pair with U89 of the U6 strand, and truncated the 3′ and 5′ sequences of U6 and U2, respectively, so that helix III comprised 9 bp (Fig. 1A). NMR evidence of formation of a U-U pair in a complex containing the native sequence in this region confirmed the formation of Helix II in either case. In addition, two guanosines were added to the 5′ end of each strand for efficient in vitro transcription, as well as cytidines on the 3′ termini. None of these changes were predicted by m-Fold to induce any conformational change from the native fold (Zuker 2003).
Design of the mutants
To validate the results obtained with the wild-type U2-U6 snRNA complex, we have designed mutants that will either stabilize/destabilize the formation of Stem I, thus favoring the formation of four-helix or three-helix structures respectively. In total, we have designed three mutants: (1) extension of U2 Stem I mutant by addition of two extra G-C/C-G base pairs to increase thermal stability of the stem (Fig. 1C); (2) substitution of the UUUU tetra-loop sequence of Stem I to the hyperstable sequence UUCG, and mutation of the top pair of Stem I from G-C to C-G (which would inhibit the formation of Helix Ib) (Fig. 1D). Both of these mutations favor the formation of Stem I and the four-helix structure; and (3) mutation of U2 snRNA G12 and G19 to A12/19 to disfavor the formation of Stem I, along with mutation of U6 snRNA G80C and C81G to disfavor the extended ISL and C61U to disfavor the potential interactions between U6 ISL and the single-stranded loop in U2 snRNA. This latter set of mutations was designed to favor formation of the putative three-helix conformation (Fig. 1E). In mutations 1 and 2, a 5-19F-C was introduced at U2 snRNA position 14 and13; in mutation 3, separate samples were created with 5-19F-C in U2 snRNA positions 13 and 21, respectively (Fig. 1C–F, red nucleotide).
Both hU6 and hU2 RNAs, as well as the mutant sequences without 19F-substituted nucleotides, were transcribed from synthetic double-stranded DNA templates (Integrated DNA Technologies) using T7 RNA polymerase expressed and purified in the laboratory. Transcribed RNA was PAGE-purified, eluted using an electroeluter, precipitated, washed with a suitable buffer using a Centricon filter, dried, and resuspended to the final concentration.
In order to characterize intermolecular pairing and exclude significant contributions of self-paired U2 or U6 snRNA, we measured migration of individual and paired strands on a nondenaturing gel. Equal amounts of purified U2 and U6 snRNA strands were heated to 70°C for 3 min in a buffer containing 50 mM Tris, 100 mM NaCl, pH 7.5, and cooled at room temperature for 30–45 minutes. Samples of U2 and U6 strands alone were treated equivalently and observed as controls. The reaction mixtures were loaded onto a 12% nondenaturing gel and electrophoresed at 100 mV for 4 h at 4°C. Following staining by Nuclistain, we observed a single band in the lane with combined strands, which migrated more slowly than individual U6 or U2 strands, and which we attributed to the U2-U6 snRNA complex (Fig. 1G). The pairing experiment was repeated in various buffers that were used for the enzymatic structure probing and spectroscopic experiments, with the same result. In all cases, we saw essentially complete pairing of the U2 and U6 snRNA strands, with no evidence of unpaired or self-paired U2 or U6 snRNA strands. For the mutant sequences, we verified pairing of the two strands by the method outlined above.
Enzymatic structural probing
Individual U6 and U2 snRNA fragments were dephosphorylated at the 5′ terminus using Antarctic phosphatase (New England Biolabs) and labeled with γ-32P-ATP using T4 polynucleotide kinase (New England Biolabs). Labeled RNA was purified on a 12% denaturing gel, eluted by a crush-and-soak method, precipitated by ethanol or isopropanol, dried, and resuspended in 50 mM Tris, 100 mM NaCl pH 7.5 for all the RNase A and RNase T1, with 1 mM MgCl2 for assays with RNase V1 (minimal [MgCl2] requirement for enzyme activity). Each labeled strand was paired with a 1.5× excess of unlabeled partner strand using the above-mentioned protocol. To assess pairing, an aliquot of paired samples (with nanomolar concentrations of either labeled U2 or U6 strand) was electrophoresed on a nondenaturing gel against labeled individual strands. From quantification of paired vs. unpaired strands, we observed that effectively 100% of each labeled strand was in a paired form. We have repeated this experiment with a constant amount of labeled strand with an increase in concentrations of the unlabeled counter strand; we observed essentially complete pairing at all the concentrations. This suggests that, at the experimental conditions that we are using, we are looking at ∼100% complex formation.
For reactions at high [Mg2+], MgCl2 was added to a final concentration of 100 mM to the folded RNA and equilibrated at room temperature for 30 min. The resulting complex was subjected to partial cleavage by ribonucleases RNase A, T1 (both from Ambion, Inc.), and RNase V1 (Pierce Milwaukee). For each experiment, reference guanosine ladders were prepared by incubation with RNase T1 (Fermentas, Inc.) under semidenaturing conditions, as were alkaline hydrolysis ladders.
Enzymatic probing assays were performed essentially by protocols accompanying the enzymes, using tRNA as carrier and identifying conditions that resulted in less than one “hit” per labeled molecule, and were electrophoresed on a denaturing gel against T1 and alkaline hydrolysis ladders. The gel was exposed to a phosphor screen for 12–16 h (overnight) at 4°C and scanned by a phosphorimager (STORM scanner from GE Health Care). The resulting picture was analyzed with SAFA (Laederach et al. 2008), which calculates the density of each band. The band densities were normalized, and the resulting values were then plotted against the sequence of the RNA to give the cleavage profiles for various enzymes. Cleavage intensities were mapped onto the two possible secondary structural folds of the complex (Fig. 2B–E).
Interaction of an intron strand with U2-U6 snRNA
RNA oligomers used for 1H NMR studies of U2-U6 Helix III were purchased from Dharmacon and were purified using an anion-exchange resin (DEAE Sephadex). Sequences of oligomers were U2-GGAGUAUCUGUUCGG (U2 strand), CCGAAACAAUACA (U6 strand), and intron-GAUACUCCA (intron strand). Integrity of oligomers was verified by denaturing PAGE.
Pairing of RNAs was tested by nondenaturing PAGE and visualized following staining with SYBR Gold. A single band observed in the lane of the stoichiometrically paired U2-U6, as compared to more rapidly migrating bands of individual U2 and U6 oligomers in separate lanes, indicated that the U2 and U6 snRNA strands paired at a stoichiometric ratio and formed a thermally stable duplex.
1H-NMR spectra
1H-NMR spectra were acquired on a 720-MHz Varian Unity Plus spectrometer (National High Magnetic Field Laboratory, Tallahassee, FL) for testing formation of Helix III, and on a Bruker Avance III 600 MHz spectrometer equipped with a cryoprobe (Hunter College, New York, NY) for analysis of the base-pairing patterns of 19F samples. Quadrature detection was achieved using the States-TPPI method (Marion et al. 1989). Spectra were processed and assigned using Varian VNMRJ/Bruker Topspin and NMRPipe software (Delaglio et al. 1995). One-dimensional experiments were acquired with 3-9-19 watergate pulse sequence for water suppression. Two-dimensional spectra were apodized using a Gaussian function, and zero filling was performed in both dimensions.
19F NMR studies
RNA sequences for 19F NMR experiments were the same as those used for enzymatic structure probing, with the exception that the U2 sequence contained a single 5-fluoro-cytidine (5-19F-C) residue at position C13 (Fig. 1C, red nucleotide). This site was chosen because it would reside within a double-stranded stem (the middle residue of Stem I) in the four-helix junction model or single-stranded in the three-helix structure. For the mutants, the 5-19F-C substitutions were located either at position C14 for mutation 1, C13 for mutation 2 (Fig. 1C,D, red nucleotide), or at position C13 (region of Stem I) or C21 (putative Helix Ib) for the two forms of mutation 3, respectively (Fig. 1E,F, red nucleotides).
Two short oligomers containing 5-19F-C in place of cytidine, 5′-UG5FCCUUUU-3′ and 5′-GG5FCCUUUUGGCCG-3′, were designed as controls in which the 5-19F-C was predicted to be in a single- or double-stranded environment, respectively. The 5-19F-C oligomers were purchased from Dharmacon and deprotected according to their protocols. Unmodified sequences were prepared by in vitro transcription using synthetic DNA templates and purified by gel electrophoresis. The modified U2 snRNA was paired with a small excess (1:1.1) of the transcribed U6 snRNA by heating both strands to 70°C and cooling slowly to room temperature as described above. The paired complex was then dried and resuspended in 95% H2O/5% 2H2O (Cambridge Isotope Laboratories) for NMR experiments. Pairing of the two strands was verified on a nondenaturing gel. NMR samples had a concentration of ∼0.35 mM RNA in 5 mM NaPi, pH 6.5, 50 mM NaCl, and 0.1 mM EDTA.
19F NMR spectra with 1H-coupling were acquired on a Varian INOVA 500 MHz spectrometer equipped with a broadband probe. Acquisition parameters of 19F NMR experiments were as follows: spectrometer frequency 470.220 MHz, spectral width 61633.3 Hz, 19F excitation pulse length 15 µsec, number of scans 12,000–24,000, acquisition time 1.063 sec, and relaxation delay 1.5 sec. Data were processed by Varian VNMRJ software with a line broadening factor of 30 Hz. Spectra were referenced by external neat trifluoroacetic acid (−78.5 ppm).
Analytical ultracentrifugation
Equimolar amounts of U2 and U6 snRNA strands were resuspended in a buffer containing 50 mM Tris-HCl, 100 mM NaCl at pH 7.5, to a final absorbance value ∼1 OD, heated to 70°C, and cooled for 15–30 min. MgCl2 was added to a final concentration of 5 mM or 100 mM at least 30 min before loading the samples into the centrifuge cells. Sedimentation velocity experiments were performed using a Beckman XL-I analytical ultracentrifuge at 20°C in double-sector cells loaded into a Ti-60 rotor and centrifuged at 30,000 rpm as described (Mitra 2009). Sedimentation boundaries were fit using DCDT+ (Philo 2000, 2006) to determine the sedimentation and diffusion coefficients, and the values were normalized to standard conditions using the buffer density calculated using SEDNTERP (http://www.jphilo.mailway.com/download.htm) with hydration and partial specific volume values of 0.59 and 0.53 cm3/g, respectively (Mitra 2009). The Stokes radius and axial ratio were calculated using SEDNTERP as described (Mitra 2009).
ACKNOWLEDGMENTS
We thank Dr. Louis Levinger, York College of CUNY, for the gift of V1 RNase, the NMR Facility, Hunter College of CUNY, NMR Facility at Florida State University, and the National High Magnetic Field Laboratory (Tallahassee, FL). This investigation was supported by NSF grant MCB 0929394 (to N.L.G.), and NIH grant 1RO1-GM085130 (to M.B.). The project described was supported by Grant Number RR003037 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH); its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH.
REFERENCES
- Arnold JR, Fisher J 2000. Structural equilibria in RNA as revealed by 19F NMR. J Biomol Struct Dyn 17: 843–856 [DOI] [PubMed] [Google Scholar]
- Black DL, Chabot B, Steitz JA 1985. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell 42: 737–750 [DOI] [PubMed] [Google Scholar]
- Burke JE, Sashital DG, Zuo X, Wang YX, Butcher SE 2012. Structure of the yeast U2/U6 snRNA complex. RNA 18: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Chen SJ 2006. Free energy landscapes of RNA/RNA complexes: With applications to snRNA complexes in spliceosomes. J Mol Biol 357: 292–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WC, Feiz V, Derrick WB, Horowitz J 1992. Fluorine-19 nuclear magnetic resonance as a probe of the solution structure of mutants of 5-fluorouracil-substituted Escherichia coli valine tRNA. J Mol Biol 227: 1164–1172 [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A 1995. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6: 277–293 [DOI] [PubMed] [Google Scholar]
- Ehresmann C, Baudin F, Mougel M, Romby P, Ebel JP, Ehresmann B 1987. Probing the structure of RNAs in solution. Nucleic Acids Res 15: 9109–9128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Abelson J 1990. Two domains of yeast U6 small nuclear RNA required for both steps of nuclear precursor messenger RNA splicing. Science 250: 404–409 [DOI] [PubMed] [Google Scholar]
- Gollnick P, Hardin CC, Horowitz J 1986. Fluorine-19 nuclear magnetic resonance study of codon–anticodon interaction in 5-fluorouracil-substituted E. coli transfer RNAs. Nucleic Acids Res 14: 4659–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon PM, Sontheimer EJ, Piccirilli JA 2000. Metal ion catalysis during the exon-ligation step of nuclear pre-mRNA splicing: Extending the parallels between the spliceosome and group II introns. RNA 6: 199–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Karunatilaka KS, Rueda D 2009. Single-molecule analysis of protein-free U2–U6 snRNAs. Nat Struct Mol Biol 16: 1154–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammann C, Norman DG, Lilley DM 2001. Dissection of the ion-induced folding of the hammerhead ribozyme using 19F NMR. Proc Natl Acad Sci 98: 5503–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig M, Scott LG, Sperling E, Bermel W, Williamson JR 2007. Synthesis of 5-fluoropyrimidine nucleotides as sensitive NMR probes of RNA structure. J Am Chem Soc 129: 14911–14921 [DOI] [PubMed] [Google Scholar]
- Hilliker AK, Staley JP 2004. Multiple functions for the invariant AGC triad of U6 snRNA. RNA 10: 921–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker AK, Mefford MA, Staley JP 2007. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev 21: 821–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz J, Ofengand J, Daniel WE Jr, Cohn M 1977. 19F nuclear magnetic resonance of 5-fluorouridine-substituted tRNA1Val from Escherichia coli. J Biol Chem 252: 4418–4420 [PubMed] [Google Scholar]
- Huppler A, Nikstad LJ, Allmann AM, Brow DA, Butcher SE 2002. Metal binding and base ionization in the U6 RNA intramolecular stem-loop structure. Nat Struct Biol 9: 431–435 [DOI] [PubMed] [Google Scholar]
- Kanyo JE, Duhamel J, Lu P 1996. Secondary structure of the r(CUUCGG) tetraloop. Nucleic Acids Res 24: 4015–4022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating KS, Toor N, Perlman PS, Pyle AM 2010. A structural analysis of the group II intron active site and implications for the spliceosome. RNA 16: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laederach A, Das R, Vicens Q, Pearlman SM, Brenowitz M, Herschlag D, Altman RB 2008. Semiautomated and rapid quantification of nucleic acid footprinting and structure mapping experiments. Nat Protoc 3: 1395–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser CF, Guthrie C 1993. Mutations in U6 snRNA that alter splice-site specificity: Implications for the active-site. Science 262: 1982–1988 [DOI] [PubMed] [Google Scholar]
- Liu L, Query CC, Konarska MM 2007. Opposing classes of prp8 alleles modulate the transition between the catalytic steps of pre-mRNA splicing. Nat Struct Mol Biol 14: 519–526 [DOI] [PubMed] [Google Scholar]
- Lowman HB, Draper DE 1986. On the recognition of helical RNA by cobra venom-V1 nuclease. J Biol Chem 261: 5396–5403 [PubMed] [Google Scholar]
- Madhani HD, Guthrie C 1992. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71: 803–817 [DOI] [PubMed] [Google Scholar]
- Marion D, Driscoll PC, Kay LE, Wingfield PT, Bax A, Gronenborn AM, Clore GM 1989. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H–15N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: Application to interleukin 1β. Biochemistry 28: 6150–6156 [DOI] [PubMed] [Google Scholar]
- Marshall AG, Smith JL 1977. Nuclear-spin-labeled nucleic acids. 1. 19F nuclear magnetic resonance of Escherchia coli 5-fluorouracil-5S-RNA. J Am Chem Soc 99: 635–636 [DOI] [PubMed] [Google Scholar]
- McPheeters DS, Abelson J 1992. Mutational analysis of the yeast U2 snRNA suggests a structural similarity to the catalytic core of group I introns. Cell 71: 819–831 [DOI] [PubMed] [Google Scholar]
- Mefford MA, Staley JP 2009. Evidence that U2/U6 helix I promotes both catalytic steps of pre-mRNA splicing and rearranges in between these steps. RNA 15: 1386–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S 2009. Using analytical ultracentrifugation (AUC) to measure global conformational changes accompanying equilibrium tertiary folding of RNA molecules. Methods Enzymol 469: 209–236 [DOI] [PubMed] [Google Scholar]
- Moore MJ, Sharp PA 1993. Evidence for two active-sites in the spliceosome provided by stereochemistry of premessenger RNA splicing. Nature 365: 364–368 [DOI] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL 2001. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. RNA 7: 833–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olejniczak M, Gdaniec Z, Fischer A, Grabarkiewicz T, Bielecki L, Adamiak RW 2002. The bulge region of HIV-1 TAR RNA binds metal ions in solution. Nucleic Acids Res 30: 4241–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R, Siliciano PG, Guthrie C 1987. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell 49: 229–239 [DOI] [PubMed] [Google Scholar]
- Pena V, Rozov A, Fabrizio P, Luhrmann R, Wahl MC 2008. Structure and function of an RNase H domain at the heart of the spliceosome. EMBO J 27: 2929–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philo JS 2000. A method for directly fitting the time derivative of sedimentation velocity data and an alternative algorithm for calculating sedimentation coefficient distribution functions. Anal Biochem 279: 151–163 [DOI] [PubMed] [Google Scholar]
- Philo JS 2006. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem 354: 238–246 [DOI] [PubMed] [Google Scholar]
- Puffer B, Kreutz C, Rieder U, Ebert MO, Konrat R, Micura R 2009. 5-Fluoro pyrimidines: Labels to probe DNA and RNA secondary structures by 1D 19F NMR spectroscopy. Nucleic Acids Res 37: 7728–7740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Query CC, Konarska MM 2004. Suppression of multiple substrate mutations by spliceosomal prp8 alleles suggests functional correlations with ribosomal ambiguity mutants. Mol Cell 14: 343–354 [DOI] [PubMed] [Google Scholar]
- Rhode BM, Hartmuth K, Westhof E, Luhrmann R 2006. Proximity of conserved U6 and U2 snRNA elements to the 5′ splice site region in activated spliceosomes. EMBO J 25: 2475–2486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie DB, Schellenberg MJ, Gesner EM, Raithatha SA, Stuart DT, Macmillan AM 2008. Structural elucidation of a PRP8 core domain from the heart of the spliceosome. Nat Struct Mol Biol 15: 1199–1205 [DOI] [PubMed] [Google Scholar]
- Ryan DE, Abelson J 2002. The conserved central domain of yeast U6 snRNA: Importance of U2-U6 helix Ia in spliceosome assembly. RNA 8: 997–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DE, Kim CH, Murray JB, Adams CJ, Stockley PG, Abelson J 2004. New tertiary constraints between the RNA components of active yeast spliceosomes: A photo-crosslinking study. RNA 10: 1251–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahasrabudhe PV, Gmeiner WH 1997. Solution structures of 5-fluorouracil-substituted RNA duplexes containing G-U wobble base pairs. Biochemistry 36: 5981–5991 [DOI] [PubMed] [Google Scholar]
- Sashital DG, Cornilescu G, McManus CJ, Brow DA, Butcher SE 2004. U2–U6 RNA folding reveals a group II intron-like domain and a four-helix junction. Nat Struct Mol Biol 11: 1237–1242 [DOI] [PubMed] [Google Scholar]
- Sawa H, Abelson J 1992. Evidence for a base-pairing interaction between U6 small nuclear RNA and 5′ splice site during the splicing reaction in yeast. Proc Natl Acad Sci 89: 11269–11273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa H, Shimura Y 1992. Association of U6 snRNA with the 5′-splice site region of pre-mRNA in the spliceosome. Genes Dev 6: 244–254 [DOI] [PubMed] [Google Scholar]
- Sontheimer EJ, Sun S, Piccirilli JA 1997. Metal ion catalysis during splicing of premessenger RNA. Nature 388: 801–805 [DOI] [PubMed] [Google Scholar]
- Sontheimer EJ, Gordon PM, Piccirilli JA 1999. Metal ion catalysis during group II intron self-splicing: Parallels with the spliceosome. Genes Dev 13: 1729–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H, Luhrmann R 2006. Cryo-electron microscopy of spliceosomal components. Annu Rev Bioph Biom 35: 435–457 [DOI] [PubMed] [Google Scholar]
- Steitz TA, Steitz JA 1993. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci 90: 6498–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JS, Manley JL 1995. A novel U2–U6 snRNA structure is necessary for mammalian mRNA splicing. Genes Dev 9: 843–854 [DOI] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM 2008. Crystal structure of a self-spliced group II intron. Science 320: 77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CK, Cheng SC 2008. Both catalytic steps of nuclear pre-mRNA splicing are reversible. Science 320: 1782–1784 [DOI] [PubMed] [Google Scholar]
- Valadkhan S, Manley JL 2001. Splicing-related catalysis by protein-free snRNAs. Nature 413: 701–707 [DOI] [PubMed] [Google Scholar]
- Valadkhan S, Manley JL 2002. Intrinsic metal binding by a spliceosomal RNA. Nat Struct Biol 9: 498–499 [DOI] [PubMed] [Google Scholar]
- Valadkhan S, Mohammadi A, Wachtel C, Manley JL 2007. Protein-free spliceosomal snRNAs catalyze a reaction that resembles the first step of splicing. RNA 13: 2300–2311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valadkhan S, Mohammadi A, Jaladat Y, Geisler S 2009. Protein-free small nuclear RNAs catalyze a two-step splicing reaction. Proc Natl Acad Sci 106: 11901–11906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner AM 1993. mRNA splicing and autocatalytic introns: Distant cousins or the products of chemical determinism? Cell 72: 161–164 [DOI] [PubMed] [Google Scholar]
- Wu J, Manley JL 1989. Mammalian pre-mRNA branch site selection by U2 snRNP involves base pairing. Genes Dev 3: 1553–1561 [DOI] [PubMed] [Google Scholar]
- Yan D, Ares M Jr 1996. Invariant U2 RNA sequences bordering the branchpoint recognition region are essential for interaction with yeast SF3a and SF3b subunits. Mol Cell Biol 16: 818–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K, Zhang L, Xu T, Heroux A, Zhao R 2008. Crystal structure of the β-finger domain of Prp8 reveals analogy to ribosomal proteins. Proc Natl Acad Sci 105: 13817–13822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yean SL, Wuenschell G, Termini J, Lin RJ 2000. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature 408: 881–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F, Griffin L, Phelps L, Buschmann V, Weston K, Greenbaum NL 2007. Use of a novel Förster resonance energy transfer method to identify locations of site-bound metal ions in the U2–U6 snRNA complex. Nucleic Acids Res 35: 2833–2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang YA, Goldstein AM, Weiner AM 1989. UACUAAC is the preferred branch site for mammalian mRNA splicing. Proc Natl Acad Sci 86: 2752–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]