Argonaute proteins are the key effectors of post-transcriptional gene silencing. Despite the essential role for Argonaute2 in mammalian gene silencing, the mechanisms regulating Ago2 protein expression are poorly understood. Using mouse embryonic stem cells (ESCs) deficient for components of the small RNA biogenesis pathway, the authors show that Ago2 expression is post-transcriptionally controlled by miRNAs, and illuminate a feedback mechanism that couples Ago2 protein levels with small RNA abundance. Their findings also support a role for the Hsp90 chaperone machinery in Ago2 loading and for the cellular autophagy pathway in the degradation of unloaded Ago2.
Keywords: Ago2, microRNA (miRNA), Dicer, DGCR8, Hsc70, Hsp90, autophagy
Abstract
Argonaute proteins are essential components of microRNA (miRNA)- and small interfering (siRNA)-mediated post-transcriptional gene-silencing pathways. In mammals, Argonaute2 (Ago2) is the catalytic center of the RNA-induced silencing complex (RISC) that recognizes and endonucleolytically cleaves messenger RNAs of complementary sequence. Although Ago2 is essential for RISC activity, the mechanisms regulating Argonaute protein expression are largely unknown. Here we report that Ago2 expression is dependent on miRNA abundance and that unloaded Ago2 protein is unstable. We observed a low level of Ago2 protein in Dicer- or DGCR8-deficent mouse embryonic stem cells (ESCs) that could be rescued by reintroduction of the respective cDNAs or by transfection of miRNAs or siRNAs. We found expression of Ago2 protein from a transgene to be similarly regulated, further supporting a post-transcriptional control mechanism. Inhibition of Hsc70/Hsp90 led to decreased Ago2 expression consistent with the reported role of this chaperone complex in RISC assembly. We furthermore found that the degradation of Ago2 was specifically blocked by inhibition of the lysosome, but not the proteasome. Our results illuminate a novel feedback mechanism that post-transcriptionally couples Ago2 protein levels with small RNA abundance with implications for RNA-interference (RNAi) and miRNA function.
INTRODUCTION
Gene silencing pathways triggered by small interfering RNAs (siRNAs) and microRNAs (miRNAs) function in a sequence-specific manner to regulate mRNA stability and protein translation, and in some cases, heterochromatin formation in diverse organisms (Ketting 2011). To do so, these regulatory RNAs associate with Argonaute (Ago) proteins as a part of a RNA-induced silencing complex (RISC). Ago proteins bind short duplex RNAs of ∼22 bp and subsequently one strand (guide strand) is stably incorporated while the other strand (passenger strand) gets degraded (Kawamata and Tomari 2010). Loaded Ago serves then as a guide to specifically recognize target mRNAs to inhibit their translation and/or induce their cleavage. Of the four Ago proteins (Ago1–4), Ago2 is the only mammalian Ago protein with intrinsic endonuclease activity, encoded by its PIWI domain (Hammond et al. 2001; Liu et al. 2004; Rivas et al. 2005). Moreover, the endonucleolytic activity of Ago2 has also been implicated in miRNA biogenesis (Diederichs and Haber 2007; O’Carroll et al. 2007; Cheloufi et al. 2010).
Numerous reports have identified Hsp90 (as well as other heat shock proteins) as an Ago-interacting protein (Tahbaz et al. 2001; Maniataki and Mourelatos 2005; Hock et al. 2007; Landthaler et al. 2008). Hsp90 is part of a multiprotein chaperone complex that functions to regulate the folding, turnover, or activity of a small set of “client” proteins (Pratt et al. 2008). Illustrative of this function is the example of steroid receptors (Pratt and Toft 2003). The Hsp90 chaperone complex regulates steroid signaling by opening the ligand-binding clefts of steroid receptors to allow the access of the steroid ligand (Pratt et al. 2008). Hsp90 action requires ATP hydrolysis, and in the presence of specific inhibitors that mimic ATP-binding such as geldanamycin or its analog 17-AAG, client proteins become unstable and are quickly degraded (Pratt et al. 2008). Indeed, Hsp90 inhibitors were shown to significantly reduce the levels of human Ago proteins as well as their recruitment to Processing bodies (P-bodies), discrete cytoplasmic speckles involved in miRNA-mediated repression (Pare et al. 2009; Johnston et al. 2010). In the report by Johnston et al. (2010), the action of Hsp90 inhibitors on Ago2 levels was found to be counteracted by the proteasome inhibitor MG132, and it was concluded that Hsp90 functions to protect unloaded Ago proteins from proteasomal degradation. Two recent in vitro studies indicate that Hsp90 is specifically required for loading small RNA duplexes into fly-Ago1, fly-Ago2, and hAgo2, but not for subsequent duplex unwinding or target cleavage (Iwasaki et al. 2010; Miyoshi et al. 2010). Altogether, these reports propose that Hsp90 functions to change the conformation of Ago proteins, likely by hydrolyzing ATP, enabling it to accommodate the small RNA “ligands” (Iwasaki et al. 2010; Johnston et al. 2010; Miyoshi et al. 2010).
Here we report that miRNA ligands post-transcriptionally regulate the levels of Ago2 and that unloaded Ago2 protein is unstable. Specifically, we show that Dicer and DGCR8-deficient mouse embryonic stem cells (ESCs) display reduced Ago2 protein, but not mRNA levels, compared with their wild-type counterparts. This effect could be rescued by restoring miRNA levels, either by expressing their respective cDNAs or by transfection of small RNAs. We found that expression of Ago2 protein from a transgene was similarly regulated, supporting a post-transcriptional mechanism. We found that inhibition of the Hsc70/Hsp90 chaperone machinery led to decreased Ago2 levels, consistent with the reported role of the chaperones in the loading of Ago2. Furthermore, we found that the degradation of Ago2 was specifically blocked by inhibition of autophagy, but not the proteasome. Given that levels of Ago proteins within the cell reportedly influence global miRNA levels, our results illuminate a novel feedback mechanism that post-transcriptionally couples Ago2 protein levels with miRNA/siRNA abundance to form stoichiometric and functional RISCs.
RESULTS AND DISCUSSION
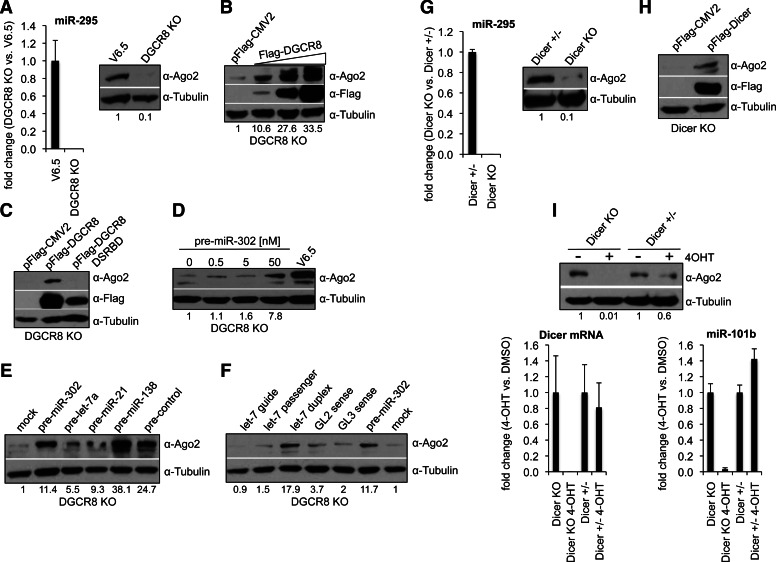
To investigate the role of miRNA ligands in Ago2 protein stability, we assessed Ago2 protein levels in mouse embryonic stem cells (ESCs) that lack DGCR8, a key component of the Microprocessor required for canonical miRNA biogenesis (Gregory et al. 2004; Wang et al. 2007). DGCR8 KO ESCs, hence, lack mature canonical miRNAs (Fig. 1A). DGCR8 KO ESCs displayed dramatically reduced Ago2 protein levels compared with their wild-type counterpart (V6.5) (Fig. 1A). Importantly, Ago2 protein levels were increased in DGCR8 KO cells rescued with a pFlag-DGCR8 expressing construct, but not a characterized DGCR8 mutant construct impaired for RNA binding (pFlag-DGCR8-DSRB) (Fig. 1B,C; Yeom et al. 2006). To rule out the possibility that the DGCR8 protein is itself required to maintain high Ago2 protein levels (e.g., via protein–protein interactions or other unknown mechanisms) and to explore whether particular miRNAs are required for Ago2 stability, we transfected DGCR8 KO ESCs with pre-miRNAs or with small (∼22 nt) RNAs which are either direct or downstream products of Microprocessor activity, respectively. Introduction of pre-miR-302, a miRNA normally expressed in ESCs, resulted in increased Ago2 protein levels (Fig. 1D). This increase was not specific to pre-miR-302, since other miRNA precursors were also able to up-regulate Ago2 (Fig. 1E). Notably, transfection of a control pre-miRNA, whose sequence is not encoded in the genome, also increases the Ago2 protein levels of DGCR8 KO cells. Ago2 also increased upon transfection of small RNA duplexes, but to a lesser extent by the transfection of single-stranded small RNAs (Fig. 1F). These results indicate that small RNA ligands are required to maintain high Ago2 protein levels. Mouse Dicer KO ESCs also lack mature miRNAs as well as other types of small endogenous RNAs (Babiarz et al. 2008). In agreement with our previous findings, the levels of Ago2 protein were reduced in Dicer KO compared with heterozygous Dicer+/− ESCs (Fig. 1G). Ago2 protein levels were increased in Dicer KO ESCs rescued with a pFlag-Dicer expressing construct (Fig. 1H). Furthermore, we investigated Ago2 protein levels following a global shutdown of miRNA processing. Specifically, we utilized tamoxifen inducible Dicer-null mouse embryonic fibroblasts (MEFs) derived from mouse embryos that are either heterozygous (Dicer+/−) or homozygous (Dicer KO) for a targeted allele of Dicer (Tan et al. 2009; Gantier et al. 2011). Treatment with 4-hydroxytamoxifen (4-OHT) induced deletion of Dicer and a concomitant reduction in miRNA levels (Fig. 1I). Supporting our previous findings, cells treated with 4-OHT also displayed reduced Ago2 protein levels, indicating that the observed effects are not ESC specific and are also seen in differentiated cells (Fig. 1I).
FIGURE 1.
miRNAs are required to maintain high levels of Ago2 expression. (A, left) Analysis of miR-295 levels by TaqMan qRT-PCR in DGCR8 KO vs. wild-type V6.5 ESCs. snoRNA142 was used for normalization control. Error bars represent mean ± SD with n = 3. (Right) Analysis of Ago2 protein levels in total cell extracts by Western blot. (B) DGCR8 KO ESCs were transfected with either empty vector (pFlag-CMV2) or increasing amounts of a pFlag-DGCR8 expressing construct. Ago2 and Flag-DGCR8 levels in total cell extracts were analyzed 48 h post-transfection (p-t) by Western blot. (C) DGCR8 KO ESCs were transfected with either empty vector (pFlag-CMV2), a pFlag-DGCR8, or a pFlag-DGCR8 DSRBD (mutated in the double-stranded RNA-binding domain) expressing construct. (D–F) DGCR8 KO ESCs were transfected with the indicated miRNA precursor, single- or double-stranded RNA at 30 nM final for D or 25 nM final for E and F. Ago2 levels in total cell extracts were analyzed 48 h p-t by Western blot. (G, left) Analysis of miR-295 levels by TaqMan qRT-PCR in Dicer KO vs. Dicer+/− ESCs. snoRNA142 was used for normalization control. Error bars represent mean ± SD with n = 3. (Right) Analysis of Ago2 protein levels in total cell extracts by Western blot. (H) Dicer KO ESCs were transfected with either empty vector (pFlag-CMV2) or a pFlag-Dicer expressing construct. Ago2 and Flag-Dicer levels in total cell extracts were analyzed 48 h p-t by Western blot. (I) Dicer KO and Dicer+/− MEFs were treated with 4-OHT or DMSO (see Materials and Methods). (Top) Ago2 protein levels were analyzed in total cell extracts by Western blot. (Left) Total RNA was collected for analysis of Dicer mRNA and miR-101b levels by qRT-PCR and TaqMan qRT-PCR, respectively. Actin and snoRNA142 were used for normalization controls, respectively. Error bars represent mean ± SD with n = 3. (A–I) Tubulin serves as loading control. The numbers on the bottom of Western blots indicate the relative Ago2 abundance (normalized to loading control).
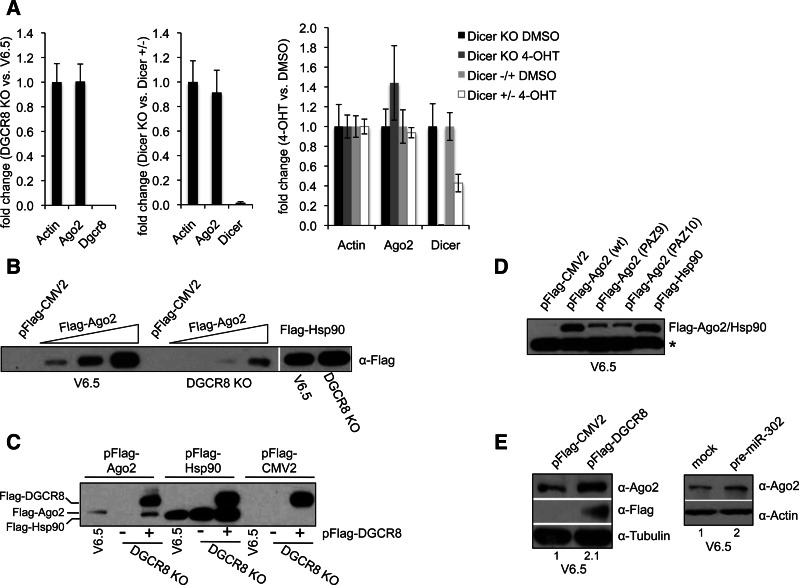
To determine whether the observed decrease in Ago2 levels in cells with no miRNAs (or reduced levels of miRNAs) is a post-transcriptional event, we measured Ago2 transcript levels by quantitative Real-Time PCR (qRT-PCR). We found no significant change in Ago2 mRNA levels between miRNA-deficient cells and their wild-type counterpart, indicating that miRNAs are able to stabilize Ago2 expression post-transcriptionally (Fig. 2A). To confirm the observed post-transcriptional regulation, we reasoned that transfection of a construct expressing the Ago2 cDNA under the control of an exogenous promoter would be expressed at lower levels in miRNA-deficient than in wild-type ESCs. To test this, we transfected a plasmid expressing pFlag-Ago2 or control pFlag-Hsp90 under the CMV promoter into V6.5 and DGCR8 KO cells. Indeed, we detected Flag-Ago2 protein at higher levels in wild-type than in DGCR8 KO cells (Fig. 2B). In contrast, control Flag-Hsp90 was detected at comparable levels in both cell types. Upon cotransfection of a pFlag-DGCR8 expressing construct into DGCR8 KO cells, we were able to detect Flag-Ago2 at comparable levels to wild-type cells (Fig. 2C). These findings strongly indicate that Ago2 is post-transcriptionally regulated by miRNAs, and in the absence of miRNAs Ago2 protein is unstable and degraded. We hypothesized that Ago2 that is impaired in small RNA binding would be expressed at lower levels than wild-type Ago2. Mutations in the PAZ domain of Ago2 were shown to decrease small RNA-binding activity (Liu et al. 2005). Consistent with our model, when transfected into mouse ESCs, two characterized Ago2 mutants containing nine or 10 mutations in the PAZ domain are expressed at lower levels than wild-type Ago2 (Fig. 2D). Finally, in support of our model, we found that increasing miRNA levels in wild-type ESCs by overexpressing of pFlag-DGCR8 or pre-miR-302 results in a twofold increase in Ago2 protein levels (Fig. 2E).
FIGURE 2.
Ago2 is post-transcriptionally stabilized by miRNAs. (A) Total RNA from DGCR8 KO and V6.5 ESCs (left); Dicer KO and Dicer+/− ESCs (middle); and Dicer KO and Dicer+/− MEFs treated with 4-OHT or DMSO (see Materials and Methods) (right) was analyzed by qRT-PCR for the indicated transcripts. Actin was used as normalization control. Error bars represent mean ± SD with n = 3. (B) V6.5 and DGCR8 KO ESCs were transfected with either empty vector (pFlag-CMV2), control pFlag-Hsp90, or increasing amounts of pFlag-Ago2 as indicated. Total cell extracts were collected after 48 h p-t and equal amounts of extracts were subjected to Flag IP and Western blot with a-Flag antibody. (C) V6.5 and DGCR8 KO ESCs were cotransfected with pFlag-Ago2, pFlag-Hsp90, or pFlag-CMV2, and either pFlag-DGCR8 or empty vector as indicated. Total cell extracts were collected after 48 h p-t and equal amounts of extracts were subjected to Flag IP and Western blot with a-Flag antibody. (D) V6.5 cells were transfected with empty vector, control pFlag-Hsp90, wild-type, or PAZ mutant Flag-Ago2 constructs. Total cell extracts were collected after 48 h p-t and equal amounts of extracts were subjected to Flag IP and Western blot with a-Flag antibody. Asterisk indicates a the presence of a contaminating band that serves as a loading control for the IPs (E, left) V6.5 cells were transfected with empty vector or pFlag-DGCR8, and Ago2 levels in total cell extracts were analyzed 48 h p-t by Western blot. (Right) V6.5 cells were either mock transfected or transfected with (20 nM final) pre-miR-302. Ago2 levels in total cell extracts were analyzed 48 h p-t by Western blot. Tubulin or Actin serves as loading control. The numbers on the bottom of Western blots indicate the relative Ago2 abundance (normalized to loading control).
Cross-regulation between components of the small RNA biogenesis machinery has been widely observed. For instance, the main components of the Microprocessor responsible for processing primary miRNA transcripts into miRNA precursors (pre-miRNAs), Drosha and DGCR8, have been shown to post-transcriptionally regulate each other. The Drosha–DGCR8 complex cleaves the hairpins embedded in the 5′ end of the DGCR8 mRNA, thereby destabilizing the mRNA and DGCR8 and, in turn, stabilizes the Drosha protein via protein–protein interactions (Han et al. 2009; Triboulet et al. 2009). Nuclear pre-miRNAs are recognized by exportin5 (Xpo5) and transported to the cytoplasm, where the RNase III Dicer in collaboration with the RNA-binding proteins Trbp and Pact generates the short RNA duplexes of ∼22 bp. Xpo5 was shown to post-transcriptionally regulate Dicer expression by exporting the Dicer mRNA out of the nucleus (Bennasser et al. 2011). The protein–protein association of Pact and Trbp with Dicer reportedly enhances Dicer stability and processing activity (Chendrimada et al. 2005; Haase et al. 2005). Finally, levels of Ago proteins within the cell have been shown to influence global miRNA levels. While ectopic expression of Ago proteins increases mature miRNAs, MEFs and hematopoietic cells lacking the Ago2 gene exhibit reduced levels of mature miRNAs (Diederichs and Haber 2007; O’Carroll et al. 2007). These changes in mature miRNA levels mediated by increased Ago expression suggest that Ago proteins assist to stabilize miRNAs likely by protecting bound miRNAs from the action of exoribonucleases (Winter and Diederichs 2011). Our findings indicate that miRNAs, in turn, serve to stabilize Ago2 protein levels and uncover a novel feedback mechanism to titrate the levels of Ago2 protein with the availability of mature miRNAs to form stoichiometric and functional RISCs.
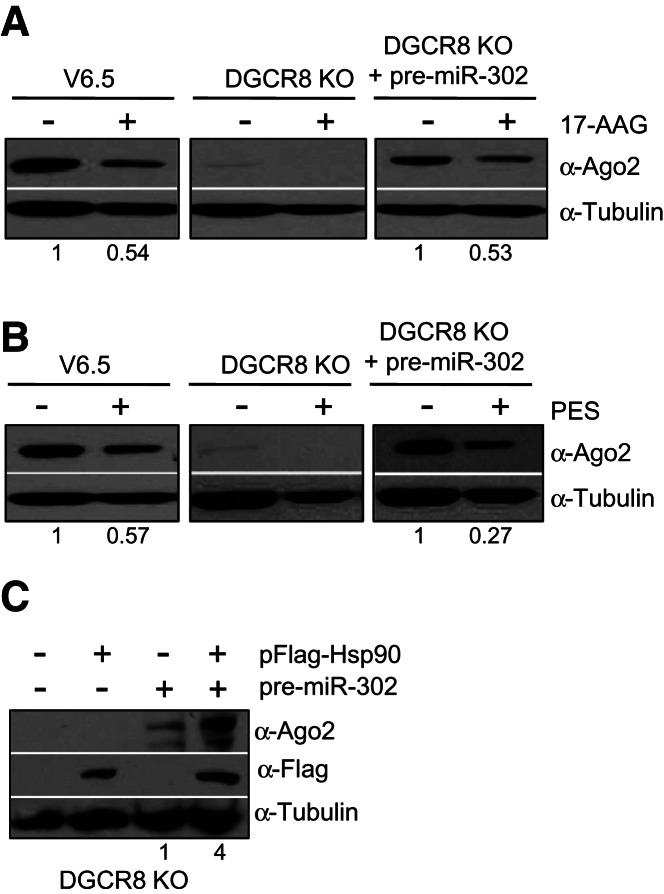
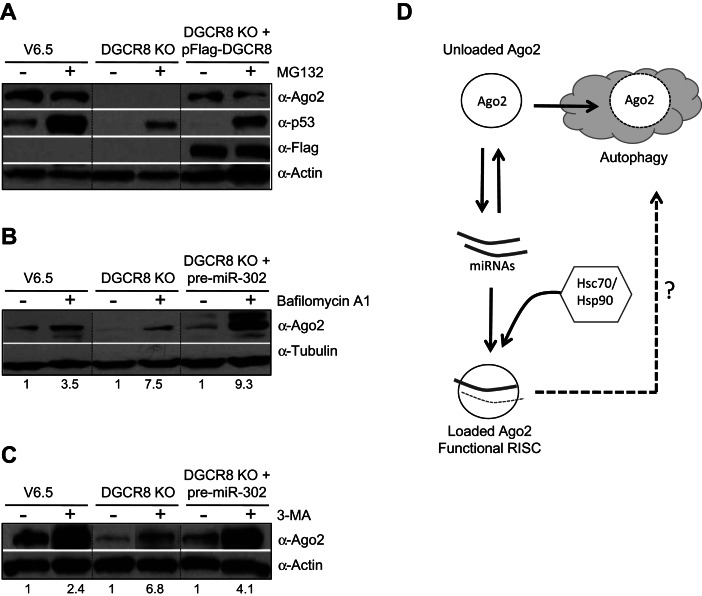
It has been proposed that Hsp90 functions to change the conformation of Ago proteins, likely by hydrolyzing ATP, enabling it to accommodate the small RNA ligands (Iwasaki et al. 2010; Johnston et al. 2010; Miyoshi et al. 2010). Mammalian cells treated with geldanamycin (or its analog 17-AAG) to specifically inhibit the function of Hsp90, exhibit reduced levels of Ago proteins (Johnston et al. 2010). However, in Drosophila cell extracts the stability of Ago proteins does not change upon inhibition of the Hsp90/Hsc70 chaperone complexes (Iwasaki et al. 2010; Miyoshi et al. 2010). Therefore, to test whether the heat-shock chaperones play a role in Ago2 stability in mouse ESCs, we treated cells with 17-AAG and PES (2-phenylethynesulfonamide) to specifically inhibit Hsp90 and Hsc70, respectively. Both inhibitors significantly reduced Ago2 levels in wild-type (V6.5) as well as miRNA-deficient ESCs previously transfected with pre-miR-302 (Fig. 3A,B). In addition, we performed gain-of-function experiments by overexpressing Hsp90 in DGCR8 KO ESCs. While transfection of a pFlag-Hsp90-expressing construct failed to increase Ago2 protein levels in the absence of miRNAs, Hsp90 overexpression was able to increase Ago2 levels of DGCR8 KO ESCs previously transfected with a miRNA precursor (Fig. 3C). Altogether, these results indicate that Hsp90 plays a role in stabilizing loaded Ago2 and highlights the importance of miRNA ligands in Ago2 protein levels. The report by Johnston et al. (2010) indicates that the effect of geldanamycin treatment could be abrogated by addition of the proteasome inhibitor MG132, suggesting that unloaded Ago2 is degraded via the proteasome. We reasoned that in miRNA-deficient cells, where Ago2 is unloaded, inhibition of the proteasome would increase Ago2 protein levels. However, MG132 treatment failed to up-regulate Ago2 levels in DGCR8 KO ESCs (Fig. 4A). Importantly, we detected increased p53 levels, indicating a successful inhibition of the proteasome. Moreover, we did not detect any increase in levels of Ago2 protein even in wild-type ESCs treated with MG132, where the stem-cell factor Trim71 has been reported to function as an E3 Ubiquitin ligase targeting Ago2 for proteasomal degradation (Rybak et al. 2009). However, other recent studies found Ago2 protein levels to be unchanged in Trim71-depleted ESCs and neural progenitors, raising doubts about the relevance of Trim71-mediated control of Ago2 ubiquitination and stability (Chang et al. 2012; Chen et al. 2012). Ago2 was initially identified as a membrane-bound protein and later showed to associate with multivesicular bodies for secretion and/or lysosomal degradation (Cikaluk et al. 1999; Tahbaz et al. 2001; Gibbings et al. 2009; Lee et al. 2009). We hypothesized that inhibition of autophagy may increase unloaded Ago2 levels. To test this idea, we treated DGCR8 KO ESCs with Bafilomycin A1, which has been shown to inhibit acidification and protein degradation in the lysosome (Yoshimori et al. 1991). Indeed, we found that Bafilomycin A1 treatment increased Ago2 protein levels in miRNA-deficient as well as wild-type ESCs (Fig. 4B). Treatment with a different autophagy inhibitor, 3-Methyladenine (3-MA), which has been shown to inhibit autophagosome formation by inhibiting type III phosphatidylinositol 3-kinases (PI-3K) (Blommaart et al. 1997), also increased Ago2 protein levels (Fig. 4C). These results indicate that the lysosome likely plays a role in the degradation of unloaded and perhaps also loaded Ago2. While this work was under review, Gibbings et al. (2012) also reported the involvement of selective autophagy in the degradation of unloaded Ago2 in HeLa cells, which is in accordance with our findings in mouse ESCs.
FIGURE 3.
Hsp90 plays a role in the stabilization of loaded Ago2. (A,B) V6.5, DGCR8 KO, and DGCR8 KO ESCs previously transfected with pre-miR-302 (30 nM final) for 24 h were treated with 17-AAG (A) or PES (B) (see Materials and Methods). Ago2 levels in total cell extracts were analyzed by Western blot. (C) DGCR8 KO ESCs were cotransfected with 20 nM final of pre-miR-302 and 0.5 ng/μL final of pFlag-Hsp90 (or empty pFlag-CMV2) construct as indicated. Ago2 and Flag-Hsp90 levels in total cell extracts were analyzed by Western blot. (A–C) Tubulin serves as loading control. The numbers on the bottom of Western blots indicate the relative Ago2 abundance (normalized to loading control). For each cell type the relative Ago2 levels in the untreated lane is set to 1.0.
FIGURE 4.
Inhibition of the autophagy stabilizes unloaded Ago2 protein. (A) V6.5, DGCR8 KO, and DGCR8 KO ESCs previously transfected with pFlag-DGCR8 for 24 h were treated with MG132 (see Materials and Methods). Ago2, Flag-DGCR8, and p53 levels in total cell extracts were analyzed by Western blot. Actin serves as loading control. (B,C) V6.5 and DGCR8 KO ESCs were treated with the lysosomal inhibitor Bafilomycin A1 (B) and the inhibitor of autophagosome formation 3-MA (C) (see Materials and Methods). Ago2 levels in total cell extracts were analyzed by Western blot. Tubulin or Actin serves as loading control. The numbers on the bottom of Western blots indicate the relative Ago2 abundance (normalized to loading control). For each cell-type the relative Ago2 levels in the untreated lane is set to 1.0. (D) Model: Ago2 protein levels are post-transcriptionally coupled to miRNA levels. The Hsc70/Hsp90 chaperone complex aids in the loading of miRNAs into Ago2. Unloaded Ago2 is degraded—a process that involves cellular autophagy. The involvement of autophagy and/or other pathways in the turnover of loaded Ago2 still remains unclear.
In vitro studies indicate that the Hsp90 (and Hsc70) chaperone machinery is required for loading of small RNA duplexes into Ago proteins, but not for subsequent strand separation or target cleavage (Iwasaki et al. 2010; Miyoshi et al. 2010). These studies propose that the chaperone machinery utilizes ATP to mediate a conformational opening of Ago proteins so that they can receive bulky small RNA duplexes. Interestingly, a recent report showed that purified recombinant Ago2 that was not loaded with a small RNA is more susceptible to digestion by thermolysin than loaded Ago2, suggesting that, at least in vitro, small RNA loading confers a conformational change that protects Ago2 from degradation by proteases (Elkayam et al. 2012). Our study demonstrates that Ago2 levels are tightly coupled to miRNA levels in vivo (Fig. 4D). This novel feedback mechanism therefore ensures stoichiometric RISC assembly and could potentially prevent the accumulation of unloaded Ago2 protein or Ago2 loaded with “nonspecific ligands,” which might otherwise have deleterious consequences to the cell transcriptome.
Our findings also support a role for the Hsc70/Hsp90 chaperone machinery in the stabilization of loaded Ago2 and implicate the autophagy pathway in the destabilization of unloaded Ago2 (Fig. 4D). Given the physical association of Ago2 with Hsc70, one possibility is that unloaded Ago2 is subject to a specialized form of autophagy called chaperone-mediated autophagy (CMA) (Kaushik and Cuervo 2012). In this model the Hsc70/Hsp90 complex would play a dual role in both RISC assembly and in cases where miRNA levels are limiting, and may also be responsible for initiating degradation of unloaded Ago2. However, the autophagy inhibitor 3-MA has been shown to selectively inhibit autophagy but not CMA, since CMA does not require formation of autophagosomes (Finn et al. 2005). We find that 3-MA is indeed a potent up-regulator of Ago2 levels, indicating that CMA does not likely play a role in Ago2 stability. In support of this idea, Gibbings et al. (2012) recently reported that the autophagy receptor NDP52 is responsible to target unloaded Ago2 for degradation. However, it remains possible that CMA may also contribute to the regulation of Ago2. Another aspect of Ago2 regulation that still remains unclear is what factors/processes are required for the turnover of loaded Ago2 and whether autophagy may also be involved.
MATERIALS AND METHODS
Cell culture, cell lines, and inhibitors
Dicer inducible null MEFs (Dicer+/− and Dicer KO) were maintained in DMEM (GIBCO, #11965) and ESCs (V6.5, DGCR8 KO, Dicer+/− and Dicer KO) in DMEM with ESGRO (1000 units/mL, Millipore, ESG1107). All media were supplemented with antibiotics (GIBCO, 15140) and with either 10% (v/v) fetal bovine serum (GIBCO, 26140) for MEFs or 15% (v/v) Stasis Stem Cell fetal bovine serum (GEMINI, 100-125) for ESCs. To induce deletion of the Dicer allele, Dicer inducible null MEFs were treated with 100 nM 4-OHT (Sigma, H7904) for 4 d (split 1:5 every other day) and cultured for an additional 4 d before RNA and protein extraction (Gantier et al. 2011; Tan et al. 2009). MG132 (Z-Leu-Leu-Leu-al; Sigma, C2211) and Bafilomycin A1 (Sigma, B1793) were dissolved in DMSO and added to cells for 5 h to a final concentration of 20 μM and 0.1 μM, respectively. 3-MA (3-Methyladenine; Sigma, M9281) was dissolved in water and added to cells at a final concentration of 5 mM for 24 h. PES (2-Phenylethynesulfonamide; Sigma, P0122) and 17-AAG (17-demethoxy-17-allylaminogeldanamycin; LC LABS, A-6880) were dissolved in DMSO and added to cells to a final concentration of 20 µM for 18 and 24 h, respectively.
DNA constructs and RNA
The cDNA of human Hsp90AA1 was cloned into the XbaI and SmaI sites of pFlag-CMV2 using the primers Fw: 5′-AAAAATCTAGAATGCCTGAGGAAACCCAGAC-3′ and Rv: 5′-AAAACCCGGGTTAGTCTACTTCTTCCATGCGTGATGTGTCG-3′. Human Flag-DGCR8 wt and DSRB mutant were previously described and were generated by site-directed mutagenesis (Yeom et al. 2006). Human Ago2 (pIRESneo-Flag/HA-Ago2) and human Dicer (pFRT/TO/Flag/HA-DEST-Dicer) expressing constructs were obtained from Addgene (plasmids 10822 and 19881, respectively). For Figure 2D, the cDNA of human wt, PAZ9, and PAZ10 Ago2 mutants were PCR amplified from the Myc-Ago2, PAZ9, and PAZ10 constructs (Liu et al. 2005) and subcloned into the EcoRI/XbaI sites of pFlag-CMV2. Precursor miRNAs for negative control (pre-control, AM17110) and mmu-miR-302a were purchased from Ambion. Pre-mmu-let-7a1, pre-mmu-miR-21, and pre-mmu-miR-138 were purchased from Dharmacon. The sequence for hsa-let-7 guide and passenger strands are as follows: guide 5′-UGAGGUAGUAGUUUGUAC-3′; passenger: 5′-CUGUACAGGCCACUGCCU-3′. GL2 and GL3 sense sequences have been previously described (Elbashir et al. 2001).
Cell transfections
For transfection of DNA and RNA, Lipofectamine 2000 (Invitrogen, 11668-019) was used according to the manufacturer’s instructions. Otherwise noted, cells were collected 48 h after transfection.
Cell extracts, Flag IP, and Western blotting
Cells were harvested in lysis buffer (20 mM Tris-HCl at pH 8.0, 137 mM NaCl, 1 mM EDTA, 1% (v/v) Triton X-100, 10% [v/v] Glycerol, 1.5 mM MgCl2, 1 mM DTT, 0.2 mM PMSF). For Flag-IP, Flag-tagged proteins were affinity-purified using α-Flag M2 agarose beads (Sigma) and washed 6× with BC-100 buffer (20 mM Tris at pH 7.6, 100 mM KCl, 10 mM β-Mercaptoethanol, 10% [v/v] Glycerol, 0.2% [v/v] Nonidet P-40, 0.2 mM PMSF) before elution with 0.5 mg/mL Flag peptide. Cell extracts and affinity eluates were analyzed by SDS-PAGE and Western blotting. Antibodies used in this study are listed as follows: α-Tubulin, (Abcam, ab6046), α-Ago2 (Cell Signaling, C34C6), α-Flag (Sigma, A8592), α-p53 (Leica Microsystems, NCL-p53-CM5p), and α-Actin (Sigma, A2066). All Western blots were quantified using the ImageJ software from the National Institutes of Health (Bethesda, MD).
qRT-PCR and miRNA quantification
Total RNA samples were isolated using TRIzol reagent (Invitrogen, 15596-018). For mRNA measurement, 100 ng–1 μg of total RNA was reverse transcribed using random hexamers and SuperScript III reverse transcriptase (Invitrogen, 18080). Quantitative PCR was performed using iQ SYBR Green Supermix (Bio-Rad, 170-8880). Mouse β-Actin was used for normalization control. Primers for qRT-PCR are listed as follows: Ago2 forward: 5′-TGCCATGGTACGAGAGTTGCTCAT-3′, reverse: 5′-ACGATGAACGTGATTCCTGGCTGA-3′; b-Actin forward: 5′-CAGAAGGAGATTACTGCTCTGGCT-3′, reverse: 5′-TACTCCTGCTTGCTGATCCACATC-3′; mDGCR8 forward: 5′-TATGGCGGAGACAGTGATCATCCA-3′, reverse: 5′-TGCAGGTATACAGGGACTCCAGAA-3′; mDicer forward: 5′-ACATGGCCTCAGAATTTGCCTTGG-3′ and reverse: 5′-AGAAGGCTGCAGAGTGTGATGTGA-3′. To confirm the deletion of Dicer alleles in Dicer inducible null MEFs (Dicer+/− and Dicer KO) we used the previously described forward (5′-TCTGCAGGCTTTTACACACG-3′) and reverse (5′-CCAATGATGCAAAGATGGTG-3′) primers (Gantier et al. 2011). For miRNA quantification, 5 ng of total RNA was reverse transcribed using gene-specific stem–loop RT primers and Multiscribe reverse transcriptase (Applied Biosystems, Part #4311235). The resulting cDNAs were mixed with TaqMan Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems, Part #4324018). Mouse snoRNA-142 was used as normalization control. Quantitative PCR was then performed using iCycler iQ Multicolor Real-Time PCR Detection System (Bio-Rad).
ACKNOWLEDGMENTS
We thank Robert Blelloch (UCSF) for DGCR8 KO ES cells, Klaus Rajewsky (Harvard) for Dicer+/− and KO ESCs, and Michael McManus (UCSF) for inducible Dicer-null MEFs, and Jidong Liu (MSKCC) for Myc-Ago2 PAZ mutant constructs. Thanks to Robert LaPierre for the generation of pFlag-DGCR8 DSRB mutant plasmid. R.I.G. was supported by a grant from the US National Institute of General Medical Sciences (NIGMS) (R01GM086386). N.J.M. is supported by NIGMS (1F32GM095102).
REFERENCES
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R 2008. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev 22: 2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennasser Y, Chable-Bessia C, Triboulet R, Gibbings D, Gwizdek C, Dargemont C, Kremer EJ, Voinnet O, Benkirane M 2011. Competition for XPO5 binding between Dicer mRNA, pre-miRNA and viral RNA regulates human Dicer levels. Nat Struct Mol Biol 18: 323–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ 1997. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem 243: 240–246 [DOI] [PubMed] [Google Scholar]
- Chang HM, Martinez NJ, Thornton JE, Hagan JP, Nguyen KD, Gregory RI 2012. Trim71 cooperates with microRNAs to repress Cdkn1a expression and promote embryonic stem cell proliferation. Nat Commun 3: 923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ 2010. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 465: 584–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lai F, Niswander L 2012. The ubiquitin ligase mLin41 temporally promotes neural progenitor cell maintenance through FGF signaling. Genes Dev 26: 803–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cikaluk DE, Tahbaz N, Hendricks LC, DiMattia GE, Hansen D, Pilgrim D, Hobman TC 1999. GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol Biol Cell 10: 3357–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs S, Haber DA 2007. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 131: 1097–1108 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L 2012. The structure of human argonaute-2 in complex with miR-20a. Cell 150: 100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn PF, Mesires NT, Vine M, Dice JF 2005. Effects of small molecules on chaperone-mediated autophagy. Autophagy 1: 141–145 [DOI] [PubMed] [Google Scholar]
- Gantier MP, McCoy CE, Rusinova I, Saulep D, Wang D, Xu D, Irving AT, Behlke MA, Hertzog PJ, Mackay F, et al. 2011. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res 39: 5692–5703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O 2009. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol 11: 1143–1149 [DOI] [PubMed] [Google Scholar]
- Gibbings D, Mostowy S, Jay F, Schwab Y, Cossart P, Voinnet O 2012. Selective autophagy degrades DICER and AGO2 and regulates miRNA activity. Nat Cell Biol 14: 1314–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Laine S, Sack R, Gatignol A, Filipowicz W 2005. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep 6: 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150 [DOI] [PubMed] [Google Scholar]
- Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN 2009. Posttranscriptional cross-regulation between Drosha and DGCR8. Cell 136: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G 2007. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep 8: 1052–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y 2010. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell 39: 292–299 [DOI] [PubMed] [Google Scholar]
- Johnston M, Geoffroy MC, Sobala A, Hay R, Hutvagner G 2010. HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. Mol Biol Cell 21: 1462–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik S, Cuervo AM 2012. Chaperone-mediated autophagy: A unique way to enter the lysosome world. Trends Cell Biol 22: 407–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Tomari Y 2010. Making RISC. Trends Biochem Sci 35: 368–376 [DOI] [PubMed] [Google Scholar]
- Ketting RF 2011. The many faces of RNAi. Dev Cell 20: 148–161 [DOI] [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen PY, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T 2008. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA 14: 2580–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Pressman S, Andress AP, Kim K, White JL, Cassidy JJ, Li X, Lubell K, Lim do H, Cho IS, et al. 2009. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol 11: 1150–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol 7: 719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z 2005. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev 19: 2979–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Takeuchi A, Siomi H, Siomi MC 2010. A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat Struct Mol Biol 17: 1024–1026 [DOI] [PubMed] [Google Scholar]
- O’Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, Miska EA, Tarakhovsky A 2007. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev 21: 1999–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare JM, Tahbaz N, Lopez-Orozco J, LaPointe P, Lasko P, Hobman TC 2009. Hsp90 regulates the function of argonaute 2 and its recruitment to stress granules and P-bodies. Mol Biol Cell 20: 3273–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt WB, Toft DO 2003. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood) 228: 111–133 [DOI] [PubMed] [Google Scholar]
- Pratt WB, Morishima Y, Osawa Y 2008. The Hsp90 chaperone machinery regulates signaling by modulating ligand binding clefts. J Biol Chem 283: 22885–22889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L 2005. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 12: 340–349 [DOI] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Hadian K, Smirnova L, Wulczyn EA, Michel G, Nitsch R, Krappmann D, Wulczyn FG 2009. The let-7 target gene mouse lin-41 is a stem cell specific E3 ubiquitin ligase for the miRNA pathway protein Ago2. Nat Cell Biol 11: 1411–1420 [DOI] [PubMed] [Google Scholar]
- Tahbaz N, Carmichael JB, Hobman TC 2001. GERp95 belongs to a family of signal-transducing proteins and requires Hsp90 activity for stability and Golgi localization. J Biol Chem 276: 43294–43299 [DOI] [PubMed] [Google Scholar]
- Tan GS, Garchow BG, Liu X, Yeung J, Morris JP, Cuellar TL, McManus MT, Kiriakidou M 2009. Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res 37: 7533–7545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triboulet R, Chang HM, Lapierre RJ, Gregory RI 2009. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA 15: 1005–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R 2007. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39: 380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Diederichs S 2011. Argonaute proteins regulate microRNA stability: Inc reased microRNA abundance by Argonaute proteins is due to microRNA stabilization. RNA Biol 8: 1149–1157 [DOI] [PubMed] [Google Scholar]
- Yeom KH, Lee Y, Han J, Suh MR, Kim VN 2006. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res 34: 4622–4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y 1991. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)- ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem 266: 17707–17712 [PubMed] [Google Scholar]