Loading of microRNAs by Argonaute2 is strand-specific and depends upon their PACT and TRBP, each of which is a double-stranded RNA-binding protein. This study reveals surprising differences between PACT and TRBP in their ability to enhance strand selection and loading.
Keywords: RNAi, RISC, Dicer, TRBP, PACT, strand selection
Abstract
Small RNAs guide RNA-induced silencing complexes (RISCs) to bind to cognate mRNA transcripts and trigger silencing of protein expression during RNA interference (RNAi) in eukaryotes. A fundamental aspect of this process is the asymmetric loading of one strand of a short interfering RNA (siRNA) or microRNA (miRNA) duplex onto RISCs for correct target recognition. Here, we use a reconstituted system to determine the extent to which the core components of the human RNAi machinery contribute to RNA guide strand selection. We show that Argonaute2 (Ago2), the endonuclease that binds directly to siRNAs and miRNAs within RISC, has intrinsic but substrate-dependent RNA strand selection capability. This activity can be enhanced substantially when Ago2 is in complex with the endonuclease Dicer and the double-stranded RNA-binding proteins (dsRBPs)—trans-activation response (TAR) RNA-binding protein (TRBP) or protein activator of PKR (PACT). The extent to which human Dicer/dsRBP complexes contribute to strand selection is dictated by specific duplex parameters such as thermodynamics, 5′ nucleotide identity, and structure. Surprisingly, our results also suggest that strand selection for some miRNAs is enhanced by PACT-containing complexes but not by those containing TRBP. Furthermore, overall mRNA targeting by miRNAs is disfavored for complexes containing TRBP but not PACT. These findings demonstrate that multiple proteins collaborate to ensure optimal strand selection in humans and reveal the possibility of delineating RNAi pathways based on the presence of TRBP or PACT.
INTRODUCTION
Short double-stranded RNAs (dsRNAs) are capable of directing sequence-specific post-transcriptional gene silencing as part of evolutionarily conserved RNA interference (RNAi) pathways. RNAi is initiated in the cytoplasm by long dsRNAs (pre-siRNAs) or hairpin RNAs (pre-miRNAs), which are processed by Dicer to yield short, 21- to 23-nt duplex RNAs termed short interfering RNAs (siRNAs) and microRNAs (miRNAs), respectively (Bernstein et al. 2001; Elbashir et al. 2001a,b; Hutvagner et al. 2001; Jinek and Doudna 2009). In Drosophila, which expresses two separate Dicer enzymes, Dicer-2 generates siRNAs, whereas Dicer-1 generates miRNAs (Lee et al. 2004). This distinction is less clear in mammals, which have only one Dicer enzyme that is capable of processing both pre-siRNAs and pre-miRNAs (Provost et al. 2002; Zhang et al. 2002). Human Dicer has two known double-stranded RNA-binding protein (dsRBP) partners, trans-activation response (TAR) RNA-binding protein (TRBP) and protein activator of PKR (PACT). Both proteins form heterodimers with Dicer and are important for the processing of certain RNAs (Chendrimada et al. 2005; Haase et al. 2005; Lee et al. 2006; Kok et al. 2007). An early report indicated that siRNA-initiated gene silencing may be more profoundly affected by an RNAi-induced knockdown of TRBP than by knockdown of PACT (Lee et al. 2006), but conflicting results were later obtained, using similar methodologies, which found a similar effect for the knockdown of either protein (Kok et al. 2007). A specific delineation of the roles of these two dsRBPs has thus not yet been achieved.
Following dicing, duplex RNAs are loaded onto Argonaute2 (Ago2), the catalytic component of the RNA-induced silencing complex (RISC) (Liu et al. 2004; Rivas et al. 2005). Loading of these RNAs is enhanced by the Hsc70/Hsp90 chaperone machinery, and in Drosophila requires Dicer/dsRBP heterodimers (Liu et al. 2003; Pare et al. 2009; Iki et al. 2010; Iwasaki et al. 2010; Johnston et al. 2010; Miyoshi et al. 2010). Upon loading, Ago2 cleaves and/or removes one strand of the duplex (the passenger strand), driving unwinding with its N-terminal domain and the aid of an endoribonuclease protein complex called C3PO (Matranga et al. 2005; Rand et al. 2005; Leuschner et al. 2006; Kim et al. 2007; Liu et al. 2009; Tian et al. 2011; Ye et al. 2011; Kwak and Tomari 2012). The remaining RNA strand (the guide strand) base-pairs with complementary sequences typically located in the 3′ untranslated regions of mRNAs, leading to their Ago2-mediated cleavage or translational repression.
Interestingly, a Drosophila RISC loading complex (RLC) composed of Ago2, Dicer-2, and R2D2 promotes the loading of siRNAs, but selects against miRNAs. Conversely, miRNAs are preferentially loaded onto Ago1 by a poorly understood mechanism (Tomari et al. 2007). In mammals, siRNAs and miRNAs do not appear to be differentially sorted into the four Ago proteins (Ago1–Ago4), although Ago2 is the most highly expressed and is the only human Ago that exhibits strand-specific endonuclease activity (Liu et al. 2004; Meister et al. 2004; Wang et al. 2009, 2012; Dueck et al. 2012). Since Ago2 binds to both siRNAs and miRNAs, the delineation of these pathways at this level in humans remains unclear.
Guide strand selection in Drosophila is based primarily on the detection of RNA duplex thermodynamics by the RLC. Thermodynamically asymmetric siRNAs are preoriented by Dicer-2 and R2D2 such that Dicer-2 binds to the less stable end and R2D2 binds to the more stable end prior to loading onto Ago2 (Khvorova et al. 2003; Schwarz et al. 2003; Tomari et al. 2004). Analogous human complexes containing either TRBP or PACT function as siRNA thermodynamic sensors as well, but the ultimate importance of these complexes for strand-selective RISC loading has yet to be fully demonstrated (Noland et al. 2011). Although Ago2/Dicer/dsRBP complexes have been shown to be the primary complexes involved in RISC loading in humans (Liu et al. 2012), for some siRNAs and miRNAs, Ago2 is sufficient for loading and accurate guide strand selection in the absence of Dicer (Kanellopoulou et al. 2005; Murchison et al. 2005; Betancur and Tomari 2012). The extent to which human complexes containing Dicer and either of its associated dsRBPs are true RLCs and contribute to strand selection is, therefore, a topic of debate. Furthermore, it is unknown how competing parameters such as duplex thermodynamics, 5′ nucleotide identity, and other structural features may affect the relative contributions of these different RNAi components to overall guide strand selection efficiency in humans.
Here, we show that recombinant human Ago2 alone can acquire a guide RNA strand by first associating with duplex siRNAs or miRNAs, leading to targeted cleavage of transcripts in vitro. Furthermore, we demonstrate that, although Ago2 alone is sufficient for strand selection for particular siRNAs and miRNAs, this is not universally the case. Dicer and TRBP or PACT are often essential for achieving maximal levels of strand selection. For some substrates, the level of strand selection depends on the particular dsRBP present in the complex. Interestingly, complexes containing TRBP are less efficient than those containing PACT at RNA targeting using miRNAs. These findings demonstrate that specific RNA duplex features dictate the relative contributions of human RNAi components to overall guide strand selection and suggest that TRBP and PACT may play distinct roles in this process.
RESULTS
Recombinant Ago2 cleaves target RNAs in vitro using duplex siRNAs and miRNAs
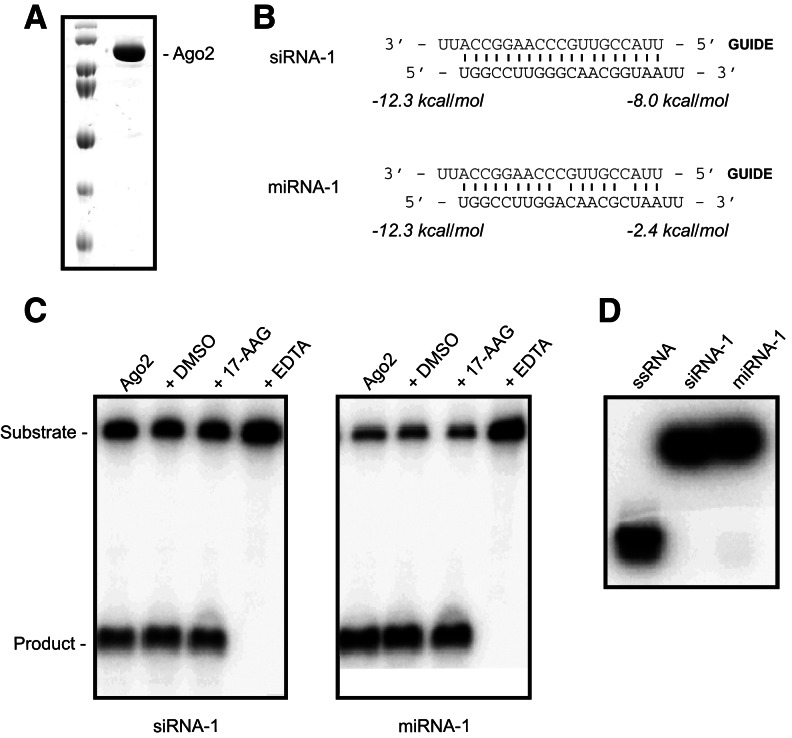
We wanted to establish a minimal in vitro system for assessing the individual contributions of different RNAi proteins to strand-selective target cleavage. As a starting point, we attempted to load purified, recombinant human Ago2 with a thermodynamically asymmetric duplex siRNA or miRNA to test for conditions that would allow for slicing activity (Fig. 1A,B). Importantly, these duplex RNAs were gel-purified by 15% native PAGE to ensure that no single-stranded RNAs (ssRNAs) were present. Surprisingly, although recombinant human Ago2 alone had previously been shown to be unable to bind duplex siRNAs (Rivas et al. 2005; Ye et al. 2011), we found that Ago2 was able to cleave target RNAs using a duplex siRNA or miRNA without the need for either ATP or accessory proteins (Fig. 1C). A native gel analysis performed at 37°C showed that the duplex RNAs remained double-stranded after 30 min, ruling out the possibility that strand dissociation and subsequent loading of ssRNAs accounted for the observed slicing activity (Fig. 1D).
FIGURE 1.
Human Ago2 efficiently cleaves target RNAs in vitro using duplex siRNAs and miRNAs. (A) Coomassie Blue-stained SDS-PAGE gel of recombinantly purified Ago2. (B) Sequences and thermodynamic analysis of siRNA-1 and miRNA-1. Thermodynamics were calculated for the four terminal base pairs and 2-nt 3′ overhangs at each end of the duplex. (C) Recombinant human Ago2 is active for target cleavage using either a duplex siRNA (siRNA-1, left panel) or duplex miRNA (miRNA-1, right panel) as a guide. Neither DMSO nor the Hsp90 inhibitor 17-AAG inhibit the activity of human Ago2, whereas EDTA does inhibit activity. (D) siRNA-1 and miRNA-1 remain double-stranded at 37°C, as demonstrated by 15% native PAGE analysis.
Heat shock proteins have been shown to enhance Ago2 loading in both Drosophila and humans (Iwasaki et al. 2010; Miyoshi et al. 2010), and it was, therefore, possible that these results were simply due to trace amounts of insect Hsp90 that may have copurified with Ago2. To determine if this was the case, we performed RNA cleavage assays in the presence of 1 mM of the Hsp90 inhibitor 17-(allylamino)-17-demethoxygeldanamycin (17-AAG). In the presence of this inhibitor, we saw no decrease in cleavage activity for either the siRNA or miRNA, whereas EDTA (a metal-chelating agent that prevents Ago2 from binding catalytically important magnesium ions) strongly inhibited cleavage activity (Fig. 1C). These findings support the conclusion that Ago2 alone can bind to duplex RNAs and select one strand as a guide for target recognition.
Dicer and dsRBPs are required for efficient strand selection with some siRNAs
We next tested the extent to which Dicer and TRBP or PACT contribute to strand-selective RISC loading in humans. For this experiment, we used siRNA-1, which is thermodynamically asymmetric and has a 5′ uridine on both duplex strands (Fig. 1B, siRNA-1). We designed a pair of 41-nt target RNAs that were perfectly complementary to one or the other strand of the duplex (except for mismatches corresponding to the last 4 nt at the 3′ end of either strand) and carried out target cleavage assays to determine the extent to which each duplex strand is used for target RNA cleavage.
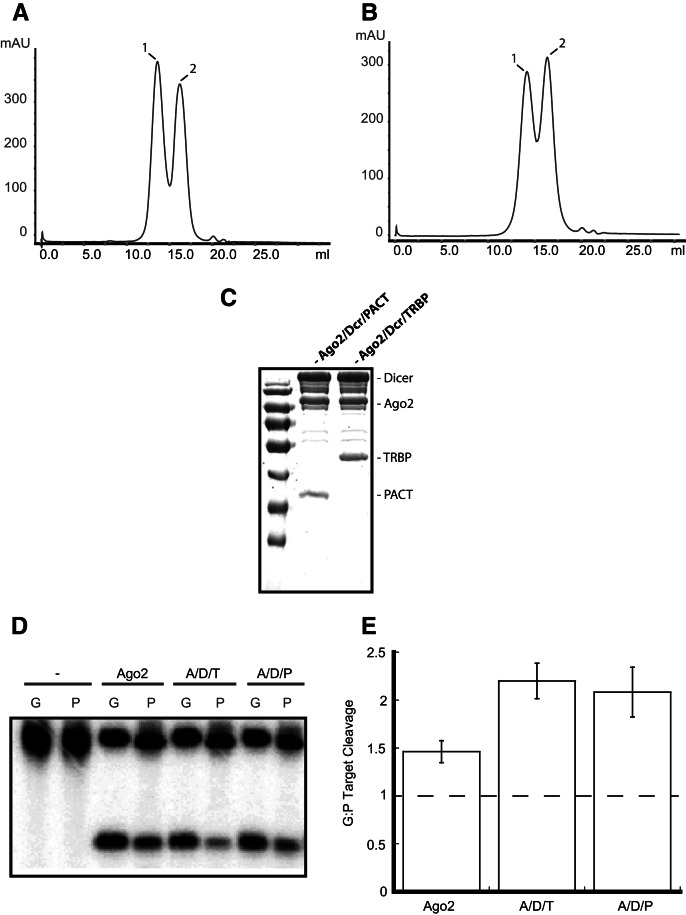
Ago2 alone or in complex with Dicer and either TRBP or PACT (Fig. 2A–C) was preincubated with siRNA-1 on ice for 30 min. Following this initial RISC loading reaction, radiolabeled target RNAs were added and incubated for 30 min at 37°C. We found that under these conditions, Ago2 used both the predicted guide strand and passenger strands to nearly equal levels. Both Dicer/dsRBP-containing complexes, however, preferentially used the predicted guide strand for targeting (Fig. 2D,E). This finding demonstrates that, for a siRNA that has the same 5′ nucleotide on both strands, preformed Ago2/Dicer/dsRBP complexes are essential for achieving optimal strand-selective RISC loading and thus target specificity. In this respect, human Dicer/dsRBP-containing complexes have the ability to function similarly to the Drosophila RLC.
FIGURE 2.
Dicer and dsRBPs are required for efficient strand selection with some siRNAs. (A) Superose 6 elution profile for reconstituted Ago2/Dicer/PACT. Peak 1 contains the full complex and Peak 2 contains free Ago2 and free PACT. (B) Superose 6 elution profile for reconstituted Ago2/Dicer/TRBP. Peak 1 contains the full complex and Peak 2 contains free Ago2 and free TRBP. (C) Coomassie Blue-stained SDS-PAGE gel showing purified Ago2/Dicer/PACT and Ago2/Dicer/TRBP. (D) Ago2 alone exhibits minimal levels of strand selection when loaded with siRNA-1. Complexes containing Dicer and either TRBP or PACT enhance strand selection. Lanes labeled “G” contain a target corresponding to the guide strand, and lanes labeled “P” contain a target that is complementary to the passenger strand. (E) Quantification of the ratio of target cleavage mediated by the guide strand compared to that mediated by the passenger strand. Data shown are means (±SD) from three separate experimental replicates.
Ago2/Dicer/dsRBP complexes are sensitive to changes in duplex thermodynamics
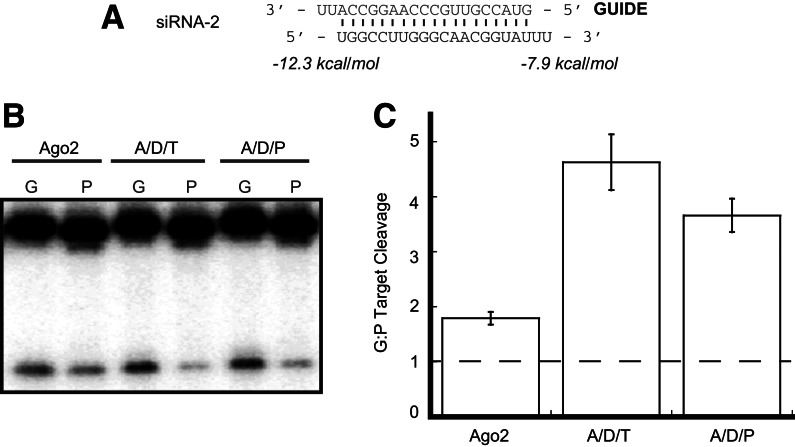
We were interested to know if a wider window of siRNA thermodynamic asymmetry would allow for strand selection by Ago2 alone or result in further enhancement of strand selection by Dicer/dsRBP-containing complexes. To this end, we altered siRNA-1 by changing the terminal A:U base pair at the 5′ end of the guide strand to a G:U wobble pair (Fig. 3A, siRNA-2). This change subtly decreases the thermodynamic stability of that end of the duplex and has a structural effect on the base pair itself. With this RNA, we again found that Ago2 exhibited only low levels of strand-selective target cleavage. As expected, strand selection was enhanced by both Ago2/Dicer/dsRBP complexes to a higher degree than was observed for siRNA-1 (Fig. 3B,C). This finding indicates that human Ago2/Dicer/dsRBP complexes are more sensitive to siRNA thermodynamics (and potentially to structural features of the terminal base pair) than Ago2 alone.
FIGURE 3.
Ago2/Dicer/dsRBP complexes are sensitive to changes in duplex thermodynamics. (A) Sequence and thermodynamic analysis of siRNA-2. Thermodynamics were calculated for the four terminal base pairs and 2-nt 3′ overhangs at each end of the duplex. (B) Ago2 alone exhibits minimal levels of strand selection when loaded with siRNA-2. Complexes containing Dicer and either TRBP or PACT enhance strand selection. Lanes labeled “G” contain a target corresponding to the guide strand, and lanes labeled “P” contain a target that is complementary to the passenger strand. (C) Quantification of the ratio of target cleavage mediated by the guide strand compared to that mediated by the passenger strand. Data shown are means (±SD) from three separate experimental replicates.
5′ nucleotide identity affects strand selection by human Ago2
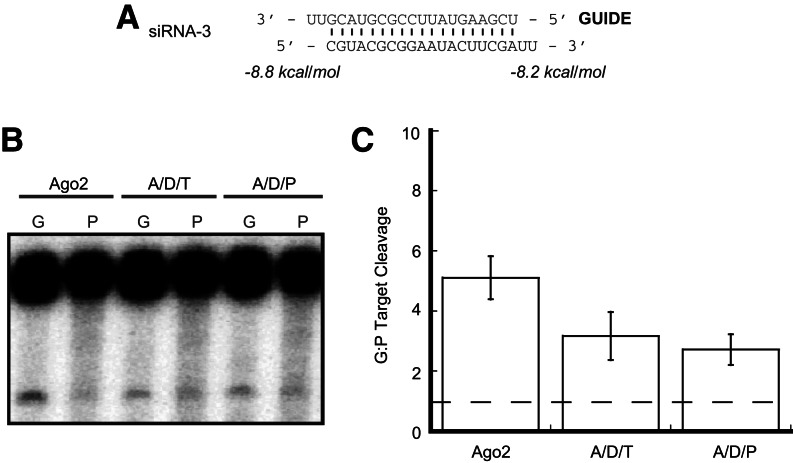
In addition to purely thermodynamic considerations, Ago2 is known to have a binding preference for small RNAs with 5′ uridine and adenosine nucleotides, while 5′ cytidines and guanosines are selected against during RISC loading. This is due in part to a conserved selectivity loop in the Ago2 Mid domain (Hu et al. 2009; Frank et al. 2010). In light of this 5′ nucleotide specificity, we were intrigued by the finding that Ago2/Dicer/dsRBP complexes achieved stronger strand-selective target cleavage with siRNA-2 despite the fact that it contained a guanosine nucleotide at the 5′ end of the guide strand. This result demonstrates that the thermodynamics of the duplex are more important than 5′ nucleotides in strand selection by human Ago2/Dicer/dsRBP complexes but does not rule out the possibility that 5′ nucleotides might play some role. In particular, if an unfavorable nucleotide were present at the 5′ end of the passenger strand, the combined thermodynamic and 5′ nucleotide effects may allow for Ago2 alone to achieve strand-selective target cleavage. In a previous study using a siRNA that had a 5′ C on the passenger strand of a siRNA duplex, target RNAs were cleaved strand-specifically by Ago2 in the absence of Dicer (Betancur and Tomari 2012). We used the same siRNA—which has a small window of thermodynamic asymmetry—in our strand selective RNA cleavage assay to further test the effect that 5′ nucleotide preference may have on strand selection (Fig. 4A, siRNA-3). For unknown reasons, the target cleavage activity when this siRNA was used was quite low. Despite this lower overall activity, in support of the previous results (Betancur and Tomari 2012), Ago2 was able to cleave targets strand-selectively on its own using this duplex (Fig. 4B,C). Interestingly, in our hands, Ago2 alone exhibited a higher degree of strand selection than either Dicer/dsRBP-containing complex in this case, indicating that these complexes may predetermine the guide strand based on thermodynamics such that unfavorable 5′ nucleotides on the passenger strand do not have a stimulatory effect. Nonetheless, these findings indicate that an unfavorable 5′ nucleotide on the passenger strand potentially facilitates strand-selectivity for human Ago2 in the absence of Dicer or dsRBPs.
FIGURE 4.
5′ nucleotide identity affects strand selection by Ago2. (A) Sequence and thermodynamic analysis of siRNA-3. Thermodynamics were calculated for the four terminal base pairs and 2-nt 3′ overhangs at each end of the duplex. (B) Ago2 alone cleaves target RNAs strand-selectively using siRNA-3. This level of strand selection is slightly attenuated by complexes containing Dicer and either TRBP or PACT. Lanes labeled “G” contain a target corresponding to the guide strand, and lanes labeled “P” contain a target that is complementary to the passenger strand. (C) Quantification of the ratio of target cleavage mediated by the guide strand compared to that mediated by the passenger strand. Data shown are means (±SD) from three separate experimental replicates.
RNA duplex terminal base-pairing affects strand selection by human Ago2
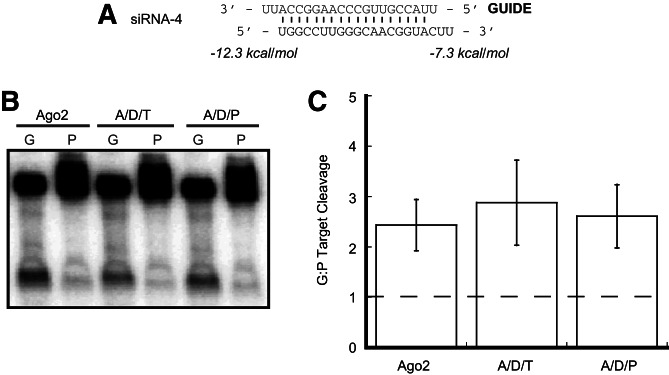
Prokaryotic Ago proteins bind to duplex nucleic acids such that the 5′ nucleotide of the guide strand is splayed apart from the corresponding base of the target—a property that appears to be conserved in human Ago2 (Wang et al. 2008; Elkayam et al. 2012). We were interested to know if this binding geometry might allow Ago2 to preferentially select the guide strand in the absence of Dicer. To this end, we modified siRNA-1 by introducing a mismatch at the 5′ end of the guide strand (Fig. 5A, siRNA-4). With a similar RNA, it was recently shown that endogenous mammalian Ago2 alone can be asymmetrically loaded as efficiently as when it is supplemented with recombinant Dicer (Betancur and Tomari 2012). In support of those data, we found that with this siRNA, Ago2 was again able to cleave its target in a strand-selective manner with no further enhancement in the presence of Dicer and TRBP or PACT (Fig. 5B,C). Thus, in addition to the specific identity of the 5′ nucleotide of the passenger strand, certain structural features of duplex ends likely provide a preferred binding geometry that allows Ago2 to selectively cleave the guide strand target in the absence of Dicer.
FIGURE 5.
RNA duplex terminal base-pairing affects strand selection by Ago2. (A) Sequence and thermodynamic analysis of siRNA-4. Thermodynamics were calculated for the four terminal base pairs and 2-nt 3′ overhangs at each end of the duplex. (B) Ago2 alone cleaves targets strand-selectively using siRNA-4. Complexes containing Dicer and either TRBP or PACT do not enhance this strand selection. Lanes labeled “G” contain a target corresponding to the guide strand, and lanes labeled “P” contain a target that is complementary to the passenger strand. (C) Quantification of the ratio of target cleavage mediated by the guide strand compared to that mediated by the passenger strand. Data shown are means (±SD) from three separate experimental replicates.
Ago2 alone cleaves target RNAs strand-selectively using a miRNA
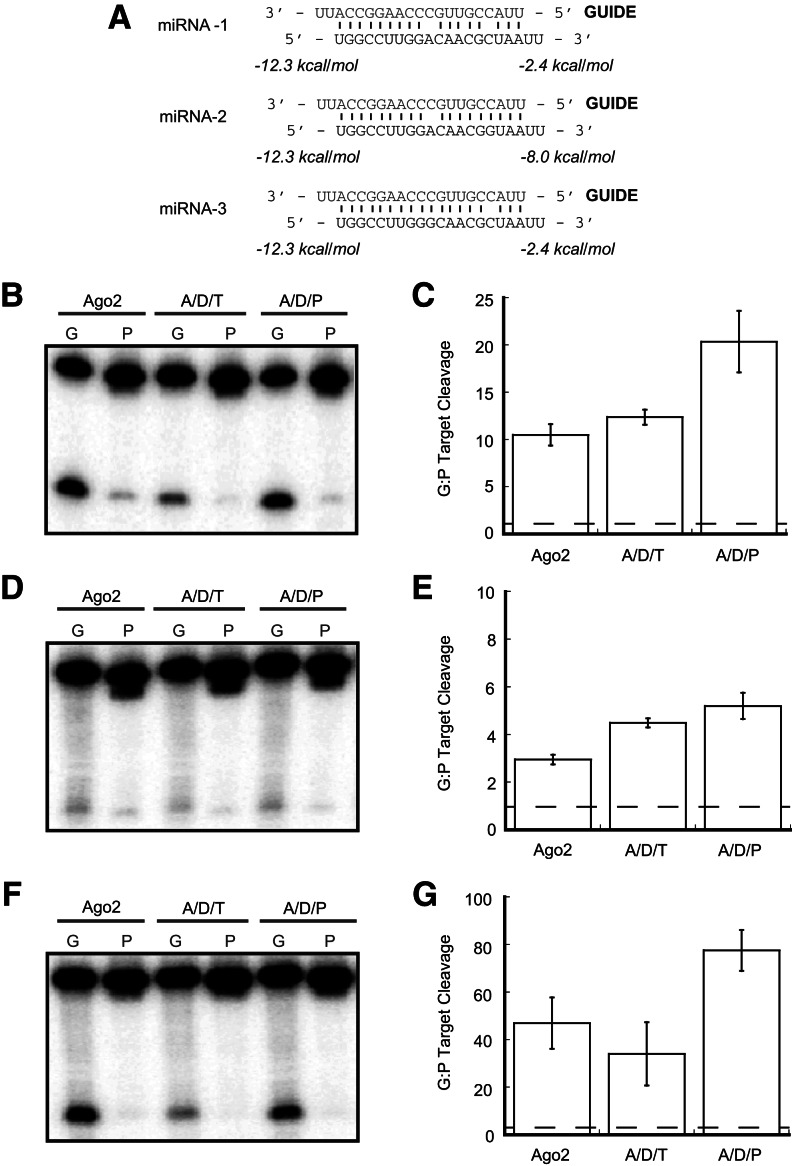
We were curious to know how internal duplex structural features such as those present in natural miRNAs might affect the ability of Ago2 alone to achieve strand-selective target cleavage. Such structural features are known to affect asymmetric RISC loading in Drosophila (Czech et al. 2009; Okamura et al. 2009). Additionally, miRNAs often have much wider windows of thermodynamic asymmetry compared with siRNAs, and it is possible that such a large thermodynamic difference between the duplex ends would be beneficial for strand-selective Ago2 loading in a manner similar to that of a terminal mismatch. We, therefore, created a model miRNA by introducing a central mismatch at position 10 of the guide strand of siRNA-1 as well as a seed mismatch at position 4 of the guide strand (Fig. 6A, miRNA-1; note that the synthetic miRNA naming system used here should not be confused with the nomenclature generally used for natural miRNAs). Target RNAs were again complementary to either strand to allow for their endonucleolytic cleavage. Ago2 alone exhibited strong strand selection with this miRNA (Fig. 6B,C), indicating that certain combinations of internal duplex structure and thermodynamics greatly enhance strand selection by human Ago2.
FIGURE 6.
Complexes containing PACT but not TRBP enhance miRNA strand selection. (A) Sequence and thermodynamic analysis of miRNA-1, miRNA-2, and miRNA-3. Thermodynamics were calculated for the four terminal base pairs and 2-nt 3′ overhangs at each end of the duplex. (B) Ago2 alone cleaves target RNAs strand-selectively using miRNA-1. This strand selection is not enhanced by the presence of Dicer and TRBP but is strongly enhanced by the presence of Dicer and PACT. Lanes labeled “G” contain a target corresponding to the guide strand, and lanes labeled “P” contain a target that is complementary to the passenger strand. (C) Quantification of the ratio of target cleavage mediated by the guide strand compared to that mediated by the passenger strand. Data shown are means (±SD) from three separate experimental replicates. (D) Ago2 alone exhibits low levels of strand-selectivity using miRNA-2. Complexes containing Dicer and either TRBP or PACT enhance this strand-selectivity. (E) Quantification of the ratio of target cleavage mediated by the guide strand compared to that mediated by the passenger strand. Data shown are means (±SD) from three separate experimental replicates. (F) Ago2 alone exhibits strong strand-selectivity using miRNA-3. This strand selection is not enhanced by the presence of Dicer and TRBP but is strongly enhanced by the presence of Dicer and PACT. (G) Quantification of the ratio of target cleavage mediated by the guide strand compared to that mediated by the passenger strand. Data shown are means (±SD) from three separate experimental replicates.
miRNA strand selection by Ago2 is affected to differing degrees by TRBP and PACT
With the siRNA duplexes that we tested, Ago2/Dicer/dsRBP complexes generally exhibited similar levels of strand-selection enhancement. We were surprised to find that with miRNA-1, strand selection by Ago2/Dicer/TRBP complexes was not enhanced above that observed for Ago2 alone, whereas a complex containing PACT enhanced strand-selective target cleavage (Fig. 6B,C). Additionally, the overall cleavage efficiency was substantially lower for the complex containing TRBP, indicating that such complexes may disfavor RNAs with internal duplex structures such as those found in most natural miRNAs (Fig. 6B). Together, these findings point to the possibility of delineating specific roles for TRBP and PACT in the fine-tuning of target specificity in RNAi.
To determine the specific elements of miRNA-1 that are important for this distinction (and for the ability of Ago2 alone to select the guide strand), we designed two miRNAs with a single mismatch either at position 10 (Fig. 6A, miRNA-2) or position 4 (Fig. 6A, miRNA-3) of the guide strand. Targets were designed to match each miRNA strand. The overall target cleavage activity for miRNA-2 was much lower than for miRNA-1 (Fig. 6D), which can be explained by previous reports that central miRNA mismatches promote Ago loading but not unwinding in both humans and Drosophila (Kawamata et al. 2009; Yoda et al. 2010). Although this low overall activity makes the interpretation of the results for this miRNA difficult, it appears that Ago2 alone was again able to cleave target RNAs strand-specifically in this case, indicating that internal duplex structure may play a role in dictating Ago2’s ability to select the guide strand, as in Drosophila (Okamura et al. 2009). Interestingly, both Ago2/Dicer/dsRBP complexes enhanced Ago2’s basal strand selection to similar degrees, indicating that the mismatch at position 10 is likely not responsible for the disproportionate enhancement of strand selection that was noted for the Ago2/Dicer/PACT complex with miRNA-1 (Fig. 6D,E).
The overall activities of each protein when programmed with miRNA-3 were similar to those observed with miRNA-1 (Fig. 6B,F). Again in this case, Ago2 exhibited strong strand selection on its own, indicating that seed mismatches and/or high levels of duplex thermodynamic asymmetry influence strand selection in the absence of Dicer (Fig. 6F,G). The abilities of Ago2/Dicer/dsRBP complexes to enhance this strand selection was also similar for miRNA-3 and miRNA-1, such that TRBP did not enhance Ago2’s strand-selectivity, whereas PACT did enhance strand selection (Fig. 6F,G). Thus, complexes containing PACT are potentially more sensitive to duplex structure in the seed region than those containing TRBP when it comes to strand selection. Furthermore, complexes containing TRBP are likely generally less equipped to tolerate internal mismatches such as those present in many natural miRNAs.
DISCUSSION
This study has revealed the nuanced roles of the human RNAi machinery in strand-selective target cleavage and lays the groundwork for the further delineation of those roles. Our results indicate that recombinant human Ago2 expressed in insect cells is fully competent to bind duplex siRNAs and miRNAs and to select one strand as a guide for target cleavage. This finding is contrary to previous reports that recombinant Ago2 alone is only active when programmed with ssRNAs (Rivas et al. 2005; Ye et al. 2011). A separate study showed that, while bacterially expressed human Ago2 binds to duplex siRNAs and miRNAs, with siRNAs it could not release cleaved passenger strands (Wang et al. 2009). It is unclear why such discrepancies in the field have arisen. One possibility is that the purification of N-terminal GST-fusion proteins from bacterial sources may lead to a less active protein than a wild-type protein from insect cells, given that human Ago2’s N-terminal domain has been implicated in duplex unwinding (Kwak and Tomari 2012). Similarly, we have found that a number of purification strategies for Ago2 lead to less active or inactive protein (data not shown). Another possible reason for these discrepancies is that some experimental procedures involve annealing duplex RNAs by adding an excess of passenger strand, which may compete with duplex RNAs for Ago2 binding, leading to the conclusion that the recombinant protein is not active with dsRNAs (Ye et al. 2011). Here, we have gel-purified each duplex RNA to ensure that no ssRNAs are present in our reactions (Fig. 1D).
Heat shock proteins are known to facilitate Ago2 loading, but the fact that our reactions do not contain ATP and that the Hsp90 inhibitor 17-AAG does not inhibit Ago2 indicates that the activity we observe is not likely the result of chaperone activity. With respect to passenger strand removal, we cannot rule out the possibility that insect C3PO has copurified with Ago2, although the protein appears to be pure by SDS-PAGE (Fig. 1A). Based on previous studies, it is likely that the addition of either C3PO or heat shock proteins to our assays would further enhance the activity of Ago2 as well as each Ago2/Dicer/dsRBP complex (Liu et al. 2009; Iwasaki et al. 2010; Miyoshi et al. 2010; Ye et al. 2011).
The results of our strand-selective target cleavage assays support and expand upon previous findings that, in certain cases, human Ago2 alone is able to cleave targets strand-specifically (Betancur and Tomari 2012). It was previously concluded that Dicer is dispensable for asymmetric RISC loading, but our expanded set of duplex RNAs has demonstrated that this conclusion is true only for certain types of duplex RNAs such as those with unfavorable 5′ nucleotides on the passenger strand or with terminal mismatches. Furthermore, even when Ago2 alone is capable of robust strand selection, Dicer and TRBP or PACT, in some cases, enhance that selection, as is the case with the miRNAs tested here. It should also be noted that despite the ability of Ago2 to bind certain duplex RNAs asymmetrically in the absence of Dicer, Ago2/Dicer/dsRBP complexes were recently shown to be the primary machinery responsible for miRNA loading to Ago2 in mammals (Liu et al. 2012). In light of this finding and the data presented here, it seems clear that rather than functioning exclusively in human strand selection, Ago2’s binding preferences serve instead as a secondary RISC-loading checkpoint that acts in concert with Dicer/dsRBP asymmetry sensors to ensure proper strand selection.
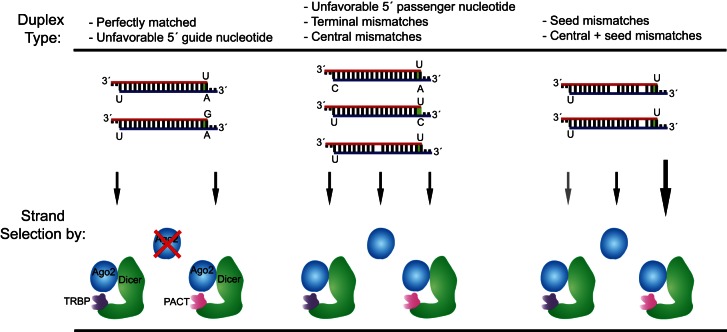
The data presented here indicate the presence of a hierarchy of parameters that dictate the necessity of human Dicer and its associated dsRBPs for strand-selection enhancement that considers thermodynamic, 5′ nucleotide, and other structural contributions (Fig. 7). For perfectly duplexed siRNAs with a 5′ uridine on both strands, Dicer/dsRBP-containing complexes are required for optimal strand selection. This remains true when unfavorable 5′ nucleotides are present on the guide strand. When present on the passenger strand, however, unfavorable 5′ nucleotides are able to drive strand selection by Ago2 alone. Similarly, terminal mismatches, which increase the thermodynamic asymmetry of the duplex and likely provide a favorable binding geometry for Ago2, allow for strand selection by that protein alone. Central and seed mismatches also allow robust strand-selective target cleavage by Ago2. In the case of miRNAs containing only a central mismatch, complexes containing Dicer and either TRBP or PACT enhance this strand selection. In the case of miRNAs containing seed mismatches, Ago2’s strand-selectivity is significantly enhanced in the presence of Dicer and PACT but not TRBP, demonstrating that these dsRBPs play differing roles in strand selection that are likely governed by internal duplex structural elements. Many more natural miRNAs will need to be tested to fully define the duplex features that dictate these roles.
FIGURE 7.
Several duplex parameters dictate the relative contributions of human RNAi components to guide strand selection. Left: Only Ago2/Dicer/dsRBP complexes are able to efficiently direct strand selection with perfectly matched siRNAs containing identical 5′ nucleotides on both strands or a disfavored 5′ nucleotide on the guide strand. Center: Ago2 alone as well as Dicer/dsRBP-containing complexes are capable of strand selection with duplex RNAs harboring a disfavored 5′ nucleotide on the passenger strand, terminal mismatches, or central mismatches. In certain cases (as with siRNA-3), strand selection by Ago2 may be enhanced above that of Dicer/dsRBP-containing complexes. Right: Ago2 and an Ago2/Dicer/TRBP complexes exhibit robust strand selection with duplex RNAs containing seed mismatches or combined seed and central mismatches, although overall activity is diminished with a TRBP-containing complex (denoted by gray arrow). Strand selection with these RNAs is enhanced by Ago2/Dicer/PACT complexes (denoted by large arrow). Red strand denotes the guide strand. Green highlights the less stable duplex ends.
We also found that the presence of TRBP substantially attenuates overall RISC activity when programmed with a miRNA containing a seed sequence mismatch. This finding is reminiscent of small RNA sorting by the Drosophila RLC, where Dicer-2/R2D2 heterodimers disfavor miRNA loading onto Ago2, and a separate mechanism supports preferential loading of miRNAs onto Ago1 (Tomari et al. 2007). Our study indicates that in humans the Dicer/TRBP heterodimer may be analogous to the Dicer-2/R2D2 heterodimer in its relative selection against miRNAs. Interestingly, while we found that Ago2/Dicer/TRBP complexes disfavored miRNA processing at the level of mRNA targeting, separate studies have found that Ago2/Dicer/PACT complexes disfavor siRNA processing at the level of dicing and that TRBP enhances pre-siRNA processing by Dicer (Chakravarthy et al. 2010; HY Lee, K Zhou, A Smith, CL Noland, JA Doudna, in prep.). These studies complement the present work to provide a picture of human siRNA and miRNA pathway delineation wherein Ago2/Dicer/TRBP complexes seem optimized for the siRNA pathway, whereas Ago2/Dicer/PACT complex are optimized for the miRNA pathway.
MATERIALS AND METHODS
Expression and purification of RNAi proteins
Purification of TRBP and PACT
TRBP was purified as described (Noland et al. 2011). PACT was purified similarly, but with the exception that following cleavage by tobacco etch virus (TEV) protease, the sample was dialyzed against Buffer A (20 mM HEPES-KOH, pH 7.5; 150 mM KCl; 5% glycerol; and 1 mM Tris-[2-carboxyethyl]phosphene [TCEP]) for 2 h, replacing the buffer with fresh Buffer A after 1 h. The dialyzed sample was then bound to a 5-mL HiTrap S (GE) column that had been pre-equilibrated with Buffer A. PACT was eluted from the column using a 100-mL linear gradient of KCl (150 mM to 1 M). Fractions containing PACT were pooled and applied to a HiLoad 16/60 Superdex 200 prep grade gel filtration column (GE) that had been pre-equilibrated with gel filtration buffer (50 mM HEPES-KOH, pH 8.0; 300 mM KCl; 10% glycerol; 1 mM TCEP).
Purification of Dicer
Dicer was purified as described (Macrae et al. 2008) with the exception that expression was carried out in High5 cells. Additionally, prior to gel filtration, flow-through from the HisTrap column (GE) was dialyzed against Buffer A and applied to 5-mL HiTrap S and HiTrap Q columns (GE) connected in tandem that had been pre-equilibrated with Buffer A. Dicer flowed through the HiTrap S column and bound to the HiTrap Q column. Dicer was then eluted from the column using a 100-mL linear gradient of KCl (150 mM to 1 M). Fractions containing Dicer were pooled and applied to a HiLoad 16/60 Superdex 200 prep grade gel filtration column that had been pre-equilibrated with gel filtration buffer.
Purification of Ago2
Ago2 was purified as described (Macrae et al. 2008) with the exception that, prior to gel filtration, flow-through from the HisTrap column was concentrated to 5 mL and dialyzed against Ago2 Buffer A (10 mM KH2PO4, pH 7.4; 150 mM KCl; 1 mM TCEP). This sample was then applied to a pre-equilibrated 5-mL Bio-Scale CHT5 Ceramic Hydroxyapatite column (Type I; Bio-Rad), and Ago2 was eluted from the column using a 100-mL linear gradient of KH2PO4 (10 mM to 500 mM). Fractions containing Ago2 were pooled and applied to a HiLoad 16/60 Superdex 200 prep grade gel filtration column that had been pre-equilibrated with gel filtration buffer.
In vitro reconstitution of Ago2/Dicer/dsRBP complexes
Binding reactions were carried out in a final volume of 500 μL gel filtration buffer containing a 1:2:4 molar ratio of Dicer:Ago2:dsRBP (assuming an initial dsRBP dimer). Reactions were incubated at 4°C for 30 min and then applied to a Superose 6 10/300 GL column (GE) that had been pre-equilibrated with gel filtration buffer. Fractionation occurred at 4°C, and fractions containing the desired complex were pooled conservatively to ensure that no free Ago2 or dsRBPs were present. All complexes were concentrated to 5 μM and aliquots stored at −80°C.
RNA preparation
All ssRNAs were chemically synthesized by Integrated DNA Technologies. ssRNAs to be used for siRNA and miRNAs were 5′-phosphorylated. All target RNAs were 41 nt long and were perfectly complementary to their respective guide strands except for mismatches with the last four bases at the 3′ end of the guide strand to promote turnover (Haley and Zamore 2004; Liu et al. 2011). All ssRNAs were gel-purified on a 15% denaturing PAGE gel. Eluted RNAs were washed thoroughly to remove contaminating urea. dsRNAs were prepared by incubating equimolar amounts of each ssRNA together in annealing buffer (100 mM HEPES, pH 7.5; 30 mM KCl; 3 mM MgCl2) at 65°C for 10 min. Annealing reactions were then slow-cooled by removing the heat block and placing it on the benchtop to equilibrate to room temperature. Annealed dsRNAs were further gel-purified on a 15% native gel to remove any remaining ssRNAs.
Nearest-neighbor RNA duplex thermodynamics analysis
Thermodynamic stabilities of the RNA duplex ends were calculated using nearest-neighbor methods for the terminal 4 bp and 2-nt 3′ overhangs at each end of the duplex (Tinoco et al. 1973; Borer et al. 1974; Freier et al. 1986; Xia et al. 1998; Mathews et al. 1999; O’Toole et al. 2005).
Slicing assays
For slicing assays using siRNAs, 50 nM Ago2 or either complex was preincubated on ice for 30 min with 50 nM siRNA or miRNA in 18 μL reaction buffer (20 mM Tris-HCl, pH 6.5; 75 mM KCl; 1 mM MgCl2; 2.5% glycerol; 0.1 mg/mL BSA; 2.5 mM TCEP). Following an incubation on ice for 30 min, these RISC-loading reactions were then divided into two 9-μL slicing reactions and 1 μL (1000 cpm, ∼0.5 nM) 5′ 32P-radiolabeled target RNA corresponding to the guide strand was added to one reaction and 1 μL (1000 cpm, ∼0.5 nM) 5′ 32P-radiolabeled target RNA corresponding to the passenger strand was added to the other. Slicing reactions were carried out for 30 min at 37°C and were stopped by phenol:chloroform extraction and ethanol precipitation of the RNAs. RNA pellets were resuspended in 2× formamide RNA loading dye (95% formamide; 18 mM EDTA; 0.025% SDS; 0.1% xylene cyanol; 0.1% bromophenol blue). Product RNAs were resolved by 12% denaturing PAGE and visualized by phosphorimaging.
Slicing assays using miRNAs were identical to those using siRNAs with the exception that two separate 9-μL reactions were set up for each protein. One of these reactions contained a 5′ biotinylated DNA capture oligonucleotide with complementarity to the passenger strand and the other contained a capture oligonucleotide complementary to the guide strand (this is to prevent reloading of unwound strands, since Ago2 binds efficiently to ssRNAs). Following an initial 30-min incubation on ice, 1 μL (1000 cpm, ∼0.5 nM) 5′ 32P-radiolabeled target RNA corresponding to the guide strand was added to the reaction containing the passenger strand capture oligo. A radiolabeled target corresponding to the passenger strand was added to the reaction containing the guide strand capture oligo. Slicing reactions were then carried out as above.
ACKNOWLEDGMENTS
We thank members of the Doudna lab for valuable discussions, A. Fisher for tissue culture assistance, and the Keck MacroLab for the use of their resources. This work was supported in part by a grant from the US National Institutes of Health (J.A.D.). J.A.D. is a Howard Hughes Medical Institute Investigator.
REFERENCES
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Betancur JG, Tomari Y 2012. Dicer is dispensable for asymmetric RISC loading in mammals. RNA 18: 24–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borer PN, Dengler B, Tinoco I, Uhlenbeck OC 1974. Stability of ribonucleic acid double-stranded helices. J Mol Biol 86: 843–853 [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Sternberg SH, Kellenberger CA, Doudna JA 2010. Substrate-specific kinetics of Dicer-catalyzed RNA processing. J Mol Biol 404: 392–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R 2005. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436: 740–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Zhou R, Erlich Y, Brennecke J, Binari R, Villalta C, Gordon A, Perrimon N, Hannon GJ 2009. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell 36: 445–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueck A, Ziegler C, Eichner A, Berezikov E, Meister G 2012. microRNAs associated with the different human Argonaute proteins. Nucleic Acids Res 40: 9850–9862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T 2001a. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T 2001b. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15: 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkayam E, Kuhn C-D, Tocilj A, Haase AD, Greene EM, Hannon GJ, Joshua-Tor L 2012. The structure of human argonaute-2 in complex with miR-20a. Cell 150: 100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F, Sonenberg N, Nagar B 2010. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465: 818–822 [DOI] [PubMed] [Google Scholar]
- Freier SM, Kierzek R, Jaeger JA, Sugimoto N, Caruthers MH, Neilson T, Turner DH 1986. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci 83: 9373–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase AD, Jaskiewicz L, Zhang H, Lainé S, Sack R, Gatignol A, Filipowicz W 2005. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with Dicer and functions in RNA silencing. EMBO Rep 6: 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley B, Zamore PD 2004. Kinetic analysis of the RNAi enzyme complex. Nat Struct Mol Biol 11: 599–606 [DOI] [PubMed] [Google Scholar]
- Hu HY, Yan Z, Xu Y, Hu H, Menzel C, Zhou YH, Chen W, Khaitovich P 2009. Sequence features associated with microRNA strand selection in humans and flies. BMC Genomics 10: 413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838 [DOI] [PubMed] [Google Scholar]
- Iki T, Yoshikawa M, Nishikiori M, Jaudal MC, Matsumoto-Yokoyama E, Mitsuhara I, Meshi T, Ishikawa M 2010. In vitro assembly of plant RNA-induced silencing complexes facilitated by molecular chaperone HSP90. Mol Cell 39: 282–291 [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y 2010. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell 39: 292–299 [DOI] [PubMed] [Google Scholar]
- Jinek M, Doudna JA 2009. A three-dimensional view of the molecular machinery of RNA interference. Nature 457: 405–412 [DOI] [PubMed] [Google Scholar]
- Johnston M, Geoffroy M-C, Sobala A, Hay R, Hutvagner G 2010. HSP90 protein stabilizes unloaded Argonaute complexes and microscopic P-bodies in human cells. Mol Biol Cell 21: 1462–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19: 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamata T, Seitz H, Tomari Y 2009. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol 16: 953–960 [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Kim K, Lee YS, Carthew RW 2007. Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes. RNA 13: 22–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok KH, Ng M-HJ, Ching Y-P, Jin D-Y 2007. Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J Biol Chem 282: 17649–17657 [DOI] [PubMed] [Google Scholar]
- Kwak PB, Tomari Y 2012. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat Struct Mol Biol 19: 145–151 [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117: 69–81 [DOI] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park S-Y, Kim Y-K, Suh MR, Kim VN 2006. The role of PACT in the RNA silencing pathway. EMBO J 25: 522–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuschner PJF, Ameres SL, Kueng S, Martinez J 2006. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep 7: 314–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim H-E, Smith DP, Wang X 2003. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921–1925 [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song J-J, Hammond SM, Joshua-Tor L, Hannon GJ 2004. Argonaute2 is the catalytic engine of mammalian RNAi. Science 305: 1437–1441 [DOI] [PubMed] [Google Scholar]
- Liu Y, Ye X, Jiang F, Liang C, Chen D, Peng J, Kinch LN, Grishin NV, Liu Q 2009. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science 325: 750–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Tan H, Tian H, Liang C, Chen S, Liu Q 2011. Autoantigen La promotes efficient RNAi, antiviral response, and transposon silencing by facilitating multiple-turnover RISC catalysis. Mol Cell 44: 502–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jin D-Y, McManus MT, Mourelatos Z 2012. Precursor microRNA-programmed silencing complex assembly pathways in mammals. Mol Cell 46: 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae IJ, Ma E, Zhou M, Robinson CV, Doudna JA 2008. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci 105: 512–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol 288: 911–940 [DOI] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD 2005. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123: 607–620 [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T 2004. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell 15: 185–197 [DOI] [PubMed] [Google Scholar]
- Miyoshi T, Takeuchi A, Siomi H, Siomi MC 2010. A direct role for Hsp90 in pre-RISC formation in Drosophila. Nat Struct Mol Biol 17: 1024–1026 [DOI] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ 2005. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci 102: 12135–12140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noland CL, Ma E, Doudna JA 2011. siRNA repositioning for guide strand selection by human Dicer complexes. Mol Cell 43: 110–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Liu N, Lai EC 2009. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell 36: 431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole AS, Miller S, Serra MJ 2005. Stability of 3′ double nucleotide overhangs that model the 3′ ends of siRNA. RNA 11: 512–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare JM, Tahbaz N, López-Orozco J, LaPointe P, Lasko P, Hobman TC 2009. Hsp90 regulates the function of Argonaute 2 and its recruitment to stress granules and P-bodies. Mol Biol Cell 20: 3273–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Rådmark O 2002. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J 21: 5864–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X 2005. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123: 621–629 [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song J-J, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L 2005. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol 12: 340–349 [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Tian Y, Simanshu DK, Ascano M, Diaz-Avalos R, Park AY, Juranek SA, Rice WJ, Yin Q, Robinson CV, Tuschl T, et al. 2011. Multimeric assembly and biochemical characterization of the Trax-translin endonuclease complex. Nat Struct Mol Biol 18: 658–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I, Borer PN, Dengler B, Levin MD, Uhlenbeck OC, Crothers DM, Bralla J 1973. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol 246: 40–41 [DOI] [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD 2004. A protein sensor for siRNA asymmetry. Science 306: 1377–1380 [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD 2007. Sorting of Drosophila small silencing RNAs. Cell 130: 299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ 2008. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456: 921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Li S, Qi HH, Chowdhury D, Shi Y, Novina CD 2009. Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nat Struct Mol Biol 16: 1259–1266 [DOI] [PubMed] [Google Scholar]
- Wang D, Zhang Z, O’Loughlin E, Lee T, Houel S, O’Carroll D, Tarakhovsky A, Ahn NG, Yi R 2012. Quantitative functions of Argonaute proteins in mammalian development. Genes Dev 26: 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T, SantaLucia J, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH 1998. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry 37: 14719–14735 [DOI] [PubMed] [Google Scholar]
- Ye X, Huang N, Liu Y, Paroo Z, Huerta C, Li P, Chen S, Liu Q, Zhang H 2011. Structure of C3PO and mechanism of human RISC activation. Nat Struct Mol Biol 18: 650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, Tomari Y 2010. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol 17: 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W 2002. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J 21: 5875–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]