This study presents a detailed analysis of a plant RNA ligase. The results highlight how its RNA repair activity is affected by active-site mutations and how mutations are bypassed by RNA alterations.
Keywords: RNA ligase, RNA quality control, cyclic phosphodiesterase, polynucleotide kinase, tRNA splicing
Abstract
tRNA ligases are essential components of informational and stress-response pathways entailing repair of RNA breaks with 2′,3′-cyclic phosphate and 5′-OH ends. Plant and fungal tRNA ligases comprise three catalytic domains. Phosphodiesterase and kinase modules heal the broken ends to generate the 3′-OH, 2′-PO4, and 5′-PO4 required for sealing by the ligase. We exploit RNA substrates with different termini to define rates of individual steps or subsets of steps along the repair pathway of plant ligase AtRNL. The results highlight rate-limiting transactions, how repair is affected by active-site mutations, and how mutations are bypassed by RNA alterations. We gain insights to 2′-PO4 specificity by showing that AtRNL is deficient in transferring AMP to pRNAOH to form AppRNAOH but proficient at sealing pre-adenylylated AppRNAOH. This strategy for discriminating 2′-PO4 versus 2′-OH ends provides a quality-control checkpoint to ensure that only purposeful RNA breaks are sealed and to avoid nonspecific “capping” of 5′-PO4 ends.
INTRODUCTION
Intron-containing tRNAs are widespread in eukarya; the intron is usually located in the anticodon loop of the pre-tRNA and must be removed precisely for the tRNA to function in protein synthesis. tRNA splicing occurs in two stages: intron excision and joining of the broken tRNA halves (Popow et al. 2012). The intron removal phase of tRNA splicing requires two incisions of the pre-tRNA at the exon–intron borders (Fig. 1A). The breakage reactions are catalyzed by a tRNA splicing endonuclease that recognizes the fold of the pre-tRNA and yields 2′,3′-cyclic phosphate and 5′-OH termini in the cleaved tRNA and the excised linear intron.
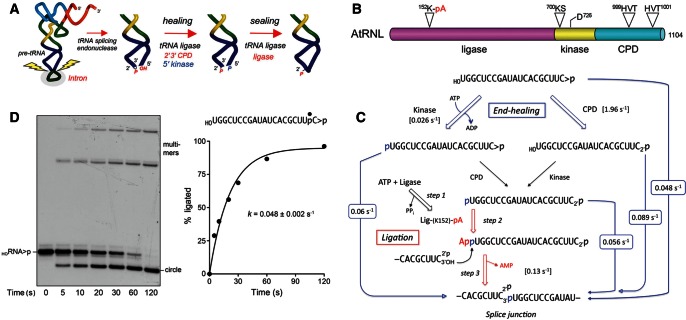
FIGURE 1.
Pathway and kinetics of end healing and sealing by tRNA ligase. (A) Splicing pathway. The pre-tRNA is cleaved at the exon–intron junctions in the anticodon loop by a tRNA splicing endonuclease, which leaves a 2′,3′-cyclic phosphate end on the proximal half-molecule and a 5′-OH on the distal half-molecule. In yeast and plants, the ends are healed and sealed by tRNA ligase, a multifunctional enzyme with 2′,3′-cyclic phosphodiesterase (CPD), 5′-OH polynucleotide kinase, and ATP-dependent ligase activities. The residual 2′-PO4 at the splice junction is eventually removed by the NAD+-dependent 2′-phosphotransferase Tpt1. (B) Arabidopsis tRNA ligase is composed of an N-terminal ligase module, a central 5′ kinase module, and a C-terminal CPD module. The site of covalent adenylylation at the ligase active site, the P-loop motif and aspartate general base at the kinase active site, and the two HxT motifs that comprise the CPD active site are highlighted. (C) The chemical steps of AtRNL-mediated healing and sealing of a HORNA>p substrate are shown. The rate constants for the individual steps or subsets of steps (denoted by arrows between input substrate and product) that were measured under single-turnover conditions in the present study are indicated. (D) Single-turnover OHRNA>p ligation. Reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 10 mM MgCl2, 0.1 mM ATP, 2 mM DTT, 20 nM HORNA>p substrate (shown at top right with the 32P label denoted by •), and 1 µM AtRNL were incubated at 22°C. Aliquots (40 µL) were removed at the times specified and quenched immediately with an equal volume of 90% formamide, 30 mM EDTA. The products were resolved by electrophoresis through a 20% polyacrylamide gel containing 7 M urea in TBE. An autoradiogram of the gel is shown in the left panel. The positions of the input substrate and the ligated circle and multimer products are indicated. The extent of ligation ([circle + multimers]/total RNA) is plotted as a function of time in the right panel. Each datum is the average of three separate time-course experiments ±SEM. A nonlinear regression curve fit of the data to a single exponential is shown.
The joining phase of the tRNA splicing pathway has been characterized best in yeast and plants, where a multifunctional tRNA ligase enzyme (Trl1 in Saccharomyces cerevisiae; AtRNL in Arabidopsis thaliana) repairs the ends of the broken tRNA half-molecules (Xu et al. 1990; Apostol et al. 1991; Sawaya et al. 2003; Englert and Beier 2005). Trl1 and AtRNL perform three reactions: (1) the 2′,3′-cyclic phosphate (>p) of the proximal tRNA half-molecule is hydrolyzed to a 3′-OH, 2′-PO4 by a cyclic phosphodiesterase (CPD); (2) the 5′-OH of the distal half-molecule is phosphorylated by a polynucleotide kinase; and (3) the 3′-OH, 2′-PO4, and 5′-PO4 ends are sealed by an ATP-dependent RNA ligase to form a tRNA containing an unconventional 2′-PO4, 3′–5′ phosphodiester at the splice junction. Reactions 1 and 2 are collectively referred to as “end-healing” (Fig. 1A; Schwer et al. 2004). The healing reactions provide the proper termini for end sealing, which is dependent on the 2′-PO4 moiety. The final step of removing the 2′-PO4 at the splice junction is performed by a separate enzyme, Tpt1 (Spinelli et al. 1997).
Yeast and plant tRNA ligases are homologous proteins composed of separable healing and sealing domains (Fig. 1B; Apostol et al. 1991; Sawaya et al. 2003; Nandakumar et al. 2008). AtRNL is an ortholog of Trl1 that provides all the essential tRNA splicing functions in vivo when expressed in yeast trl1Δ cells (Wang et al. 2006). Each of the component catalytic modules of tRNA ligase is essential for yeast viability, and missense mutations in the active sites of the CPD, kinase, and ligase domains are lethal in vivo (Sawaya et al. 2003; Wang et al. 2006). The C-terminal CPD module belongs to the “2H” phosphoesterase superfamily, named for the pair of HxT peptides that comprise the CPD active site (Fig. 1B; Mazumder et al. 2002). The central kinase module is a member of the P-loop phosphotransferase enzyme superfamily, named for the signature GxGK(S/T) motif that binds the NTP phosphate donor (Fig. 1B). The N-terminal ligase domain belongs to the covalent nucleotidyltransferase enzyme superfamily that includes RNA and DNA ligases and mRNA capping enzymes (Shuman and Lima 2004). Ligation requires ATP and proceeds via three adenylate transfer steps. First, the ligase domain reacts with ATP to form a covalent ligase-(lysyl-Nζ)-AMP intermediate and displace pyrophosphate. Adenylylation occurs at a conserved KxxG motif that defines the enzyme superfamily (Fig. 1B). Second, ligase transfers AMP to the 5′-PO4 RNA terminus to form an RNA-adenylate intermediate (AppRNA). Third, ligase directs the attack of the 3′-OH on AppRNA to form the splice junction and displace AMP.
The plant and yeast tRNA ligases evince no specificity for the native tRNA fold during RNA sealing in vitro, i.e., deleting the TψC loop or the D loop plus the TψC loop did not affect the efficiency of ligation (Nandakumar et al. 2008). Rather, their specificity derives from the fact that their sealing activity is restricted to breaks in which one partner has a 3′-OH, 2′-PO4 terminus. Because there is no enzymatic pathway known in which an RNA 3′-OH, 2′-OH could be directly phosphorylated to a 3′-OH, 2′-PO4, this means that eukaryal tRNA ligases are unable to catalyze sealing events at the unprocessed 3′ ends of native RNAs. The route to a sealable substrate for plant/yeast ligases is via hydrolysis of an RNA>p with scission of the O3′—P bond. This is exactly what is accomplished when the CPD domain of the tRNA ligases acts at an RNA>p end. Consequently, the yeast and plant tRNA repair systems are hard-wired so that only the purposefully broken RNAs (which have >p ends) will be joined.
A manifestation of the dominance of end specificity in the joining reaction of plant tRNA ligase is that, when it is presented with a mixture of cleaved pre-tRNA and excised intron generated by splicing endonuclease, it catalyzes both tRNA splicing and intron circularization, without preference for the broken tRNA versus the intron (Nandakumar et al. 2008). Moreover, when plant tRNA ligase substitutes for Trl1 in yeast in vivo, it generates intron circles when splicing HAC1 mRNA during the unfolded protein response (Mori et al. 2010).
Despite the centrality of tRNA splicing to eukaryal gene expression, very little is known about the kinetic mechanisms of the several RNA repair reactions catalyzed by tRNA ligase, and virtually nothing is known about how the specificity for sealing 2′-PO4 ends is enforced. Here we address these knowledge gaps by studying the kinetics of end healing and sealing by plant tRNA ligase under single-turnover conditions. We exploit a series of linear RNA substrates with different terminal structures (HORNA>p, HORNA2′p, pRNA>p, pRNA2′p, pRNAOH, AppRNA>p, AppRNA2′p, AppRNAOH) and a collection of AtRNL mutants to dissect individual steps or subsets of steps along the RNA repair pathway. The results of this inquiry illuminate rate-limiting transactions and how they are affected and/or bypassed by changes in the enzyme or the RNA substrate. We pinpoint the RNA 5′-adenylylation step as the source of the 2′-PO4 requirement.
RESULTS
Kinetics of HORNA>p ligation
To assay the composite healing and sealing pathway of AtRNL, we used a 20-mer RNA with 5′-OH and 2′,3′-cyclic phosphate (>p) ends and a single radiolabel between the 3′-terminal and penultimate nucleosides. This linear molecule is analogous to an excised 20-mer tRNA intron that is efficiently sealed by AtRNL in vitro (Nandakumar et al. 2008). To prepare this substrate, we used T4 RNA ligase 1 to transfer [5′-32P]pCp to the 3′ end of a 19-mer RNA oligonucleotide to generate a 20-mer strand with 5′-OH and 3′-PO4 ends. We then used Escherichia coli RNA cyclase RtcA to convert the 3′-PO4 end of the labeled 20-mer RNA into a 2′,3′-cyclic phosphodiester. The HORNA>p substrate (20 nM) was reacted with a 50-fold molar excess of recombinant wild-type AtRNL in the presence of 10 mM Mg2+ (an essential cofactor for the ligase and kinase reactions) and 0.1 mM ATP (an adenylyltransferase substrate for the sealing reaction and a phosphate donor for the kinase step). The reactions were quenched with EDTA at 5, 10, 20, 30, 60, and 120 sec, and the products were analyzed by urea-PAGE (Fig. 1D). AtRNL effected a time-dependent conversion of the HORNA>p substrate into a circular intramolecular ligation product that migrated ahead of the substrate strand. AtRNL also generated more slowly migrating multimers via intermolecular end joining. A plot of the percent ligation versus time is shown in Figure 1D, with each datum being the average of three separate experiments ±SEM. This regime of AtRNL excess sufficed to attain pseudo-first-order kinetics, i.e., the rate of the reaction did not change when the AtRNL concentration was either halved or doubled (data not shown). A nonlinear regression fit of data in Figure 1D to a single exponential yielded an apparent rate constant of 0.048 ± 0.002 sec−1 for the complete ligation pathway. This entails at least five chemical steps, as depicted in Figure 1C.
Kinetics of the 2′,3′-cyclic phosphodiesterase reaction
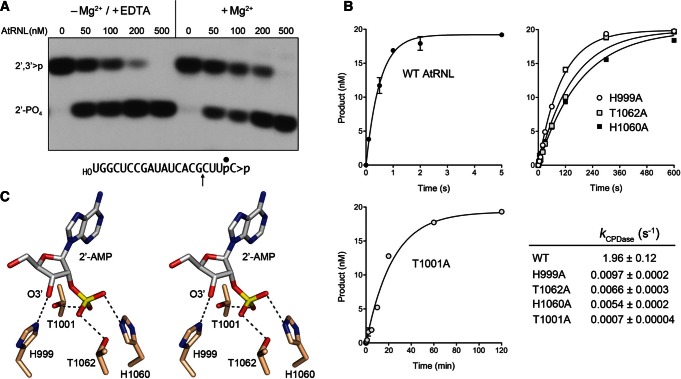
The CPD domain of AtRNL is a 2H phosphotransferase (Mazumder et al. 2002). Although 2H enzymes are posited to catalyze phosphoryl transfer via a general acid–base mechanism, it is not clear whether the CPD reaction of AtRNL requires or is affected by the presence of a divalent cation. Beyond mechanistic interest, this issue is pertinent to the implementation of an effective quench protocol to study the kinetics of the CPD step. A pilot experiment was performed in which AtRNL was reacted with 32P-labeled HORNA>p substrate in the absence of ATP (to preclude the kinase reaction); the reaction mixtures contained either 10 mM Mg2+ or 10 mM EDTA (and no added Mg2+). To enable resolution of the reactants and products with different 3′ ends, the mixtures were digested with RNase T1 prior to analysis by denaturing PAGE (Fig. 2A). RNase T1 incised the substrate 3′ of the most distal guanosine to yield the 32P-labeled tetranucleotide HOCUUpC>p (Fig. 2A, lanes 0). Reaction of increasing concentrations of AtRNL with the RNA>p substrate depleted the HOCUUpC>p T1 fragment and generated a more rapidly migrating T1 fragment corresponding to the CPD product, HOCUUpC2′p (Fig. 2A). The CPD reaction was independent of a divalent cation and indifferent to the presence of EDTA.
FIGURE 2.
Kinetics and mechanism of the CPD reaction. (A) Metal independence. Reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 2 mM DTT, 20 nM HORNA>p substrate (shown at bottom with the position of the 32P-label denoted by • and the site of RNase T1 scission indicated by ↑), increasing concentrations of AtRNL as specified, and either 10 mM EDTA or 10 mM MgCl2 were incubated for 1 min at 22°C. The products were digested with RNase T1 and then analyzed by urea-PAGE. An autoradiogram of the gel is shown. The phosphorylation states of the terminal RNase T1 fragments HOCUUpC>p (2′,3′>p) or HOCUUpC2′p (2′-PO4) are indicated at left. (B) Single-turnover kinetics. Reaction mixtures contained 20 nM HORNA>p substrate and 1 µM wild-type or mutant AtRNL as specified. The extents of cyclic phosphate hydrolysis (HOCUUpC2′p/[HOCUUpC>p + HOCUUpC2′p]) normalized to input RNA concentration are plotted as a function of reaction time. Each datum in the graphs is the average of three separate experiments ±SEM. Nonlinear regression curve fits of the data to a single exponential are shown. The CPD rate constants for the wild-type and mutant enzymes are tabulated. (C) Stereo view of the active site of mammalian 2′,3′-cyclic phosphodiesterase in complex with the reaction product 2′-AMP (from pdb 2YDD). The histidine and threonine residues are numbered according to their counterparts in AtRNL. Hydrogen bonds to the 2′-PO4 and 3′-OH are indicated by dashed lines.
Accordingly, we relied on SDS to quench the CPD reaction and recovered the RNAs (free of SDS) by phenol-CHCl3 and CHCl3 extraction prior to digestion with RNase T1 and analysis by PAGE. The single-turnover CPD reaction of 1 µM wild-type (WT) AtRNL with 20 nM HORNA>p substrate was sufficiently brisk to require the use of a rapid mix-quench apparatus to access the kinetic profile, which is depicted in Figure 2B. The data fit well to a single exponential with an apparent rate constant of 1.96 ± 0.12 sec−1. The finding that the rate of the CPD reaction was 40-fold faster than the rate of the composite HORNA>p ligation pathway signified that CPD chemistry is not rate limiting.
Catalytic contributions of the CPD HxT motifs
The HxT motifs of yeast and plant tRNA ligases are necessary for their function in vivo (Sawaya et al. 2003; Wang et al. 2006). To gauge the impact of mutations in the motifs on CPD activity in vitro, we analyzed the kinetics of cyclic phosphate hydrolysis by AtRNL mutants H999A, T1001A, H1060A, and T1062A. The rate constants for the single-turnover CPD reactions of H999A (0.0097 sec−1), T1062A (0.0066 sec−1), and H1060A (0.0054 sec−1) were several hundredfold slower than wild-type AtRNL (Fig. 2B). The T1001A variant (k = 0.0007 sec−1) was several thousandfold slower than wild-type CPD (Fig. 2B), consistent with the unconditional lethality of this mutation in vivo (Wang et al. 2006). The mutational effects may be interpreted in light of the structure of mammalian 2′,3′-cyclic nucleotide phosphodiesterase (CNPase) in complex with the reaction product 2′-AMP (Myllykoski et al. 2012). CNPase catalyzes the same chemical reaction as the tRNA ligase CPD module and is a functional ortholog of CPD, insofar as CNPase can substitute for the tRNA ligase CPD to perform the 3′ end-healing step of yeast tRNA splicing in vivo (Schwer et al. 2008). The active site of the CNPase•2′-AMP product complex is shown in Figure 2C, with the HxT residues labeled according to their equivalents in AtRNL. The His999 Nϵ makes a hydrogen bond to the ribose O3′ atom, the leaving group in the CPD reaction, thereby implicating His999 as a general acid catalyst. Thr1001 and Thr1062 donate hydrogen bonds to the 2′-PO4 oxygens, interactions that presumably stabilize the transition state. His1060 donates a hydrogen bond to the 2′-PO4 oxygen of the product complex that is proposed to derive from the water nucleophile that attacks the cyclic phosphate substrate (Myllykoski et al. 2012), indicative of its imputed function as a general base catalyst. The catalytic defects we see for the alanine mutants are consistent with these functional assignments in the CPD mechanism.
Kinetics and specificity of the 5′-OH kinase reaction
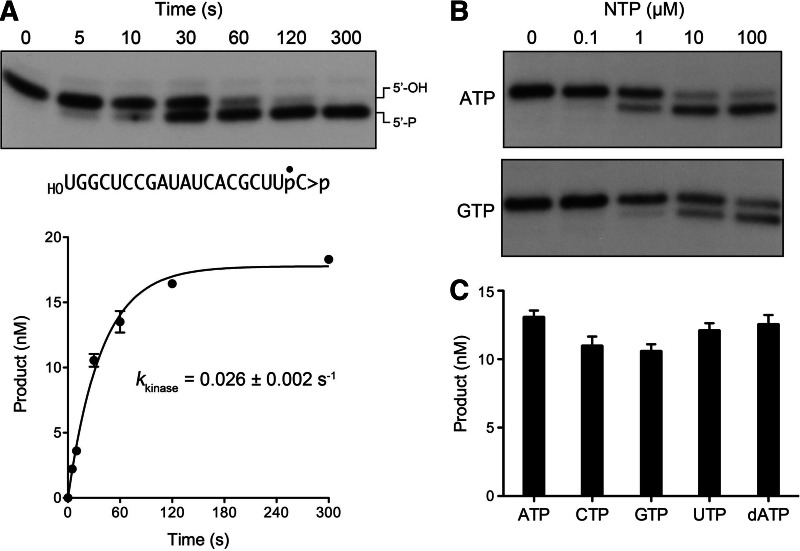
We exploited the CPD-defective T1001A variant to study the 5′-phosphorylation reaction of AtRNL with the HORNA>p substrate in the presence of magnesium and 0.1 mM ATP, it being necessary to suppress the CPD step so as to prevent ligation of the 5′-PO4 kinase reaction product. The reactions were quenched with EDTA, and the products were analyzed by denaturing PAGE, which revealed a time-dependent conversion of the HORNA>p strand to a slightly faster migrating pRNA>p product (Fig. 3A, top panel) without generation of ligated circles (which migrate ahead of pRNA>p) or ligated multimers during the time frame of the experiment. The extent of phosphorylation is plotted as a function of time in Figure 3A, bottom panel. The data fit well to a single exponential with a rate constant of 0.026 ± 0.002 sec−1. The rate of the isolated HORNA>p kinase reaction of T1001A was within a factor of two of the composite HORNA>p ligation reaction of AtRNL and was 75-fold slower than the isolated CPD reaction with the same substrate (Fig. 1C). We surmise that the kinase step is rate limiting during the end-healing phase of the tRNA ligase pathway.
FIGURE 3.
Kinetics and NTP specificity of the 5′ kinase reaction. (A) Reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 10 mM MgCl2, 0.1 mM ATP, 2 mM DTT, 20 nM HORNA>p substrate, and 1 µM AtRNL-T1001A were incubated at 22°C. Aliquots (40 µL) were removed at the times specified and quenched immediately with an equal volume of 90% formamide, 30 mM EDTA. The products were resolved by urea-PAGE. An autoradiogram of the gel is shown in the top panel. Product formation, (pRNA>p/[pRNA>p + HORNA>p]) normalized to input RNA concentration is plotted as a function of reaction time. Each datum is the average of four separate experiments ±SEM. A nonlinear regression curve fit of the data to a single exponential is shown. (B) NTP dependence. Kinase reaction mixtures containing 20 nM HORNA>p substrate, 1 µM AtRNL-T1001A, and ATP or GTP as specified were incubated for 1 min at 22°C. The products were resolved by urea-PAGE. (C) NTP specificity. Reaction mixtures containing 20 nM HORNA>p substrate, 1 µM AtRNL-T1001A, and 0.1 mM ATP, CTP, GTP, UTP, or dATP as specified were incubated for 1 min at 22°C. The extents of product formation are shown; each datum in the graph is the average of three separate experiments ±SEM.
It was of interest to evaluate the NTP donor specificity of AtRNL in light of biochemical studies showing that yeast Trl1 preferentially uses GTP as the phosphate donor for its kinase reaction and genetic evidence that GTP is the physiological substrate for Trl1 in vivo (Westaway et al. 1993; Sawaya et al. 2003). Greer and colleagues discovered that the Trl1 kinase is 100-fold more efficient with GTP versus ATP, as reflected in the yield of pRNA product as a function of NTP concentration (Westaway et al. 1993). Indeed, GTP has routinely been included in the reaction mixtures in prior studies of tRNA splicing by AtRNL (Englert and Beier 2005). Here we compared the extents of phosphorylation of 20 nM HORNA>p strand by 1 µM AtRNL-T1001A during a 1 min reaction in the presence of increasing concentrations of ATP or GTP (Fig. 3B). Omission of exogenous NTP resulted in no 5′-phosphorylation by AtRNL. Whereas little or no product was formed with 0.1 µM ATP, the substrate was partially phosphorylated at 1 µM ATP. The extent of product formation increased at 10 µM ATP and was not further enhanced at 100 µM ATP. GTP titration showed it to be less effective than ATP at limiting concentrations (1–10 µM), though additional product was formed as GTP was increased to 100 µM (Fig. 3B). Thus, AtRNL kinase does not display a GTP preference akin to yeast Trl1. To build on this finding, we surveyed more broadly the ability of various NTPs at 100 µM concentration to support AtRNL-T1001A-mediated phosphorylation of the HORNA>p strand. The kinase readily used ATP, GTP, CTP, UTP, or dATP as the phosphate donor (Fig. 3C). Thus, the AtRNL kinase is not fastidious with respect to the nucleobase or the pentose sugar of the NTP substrate.
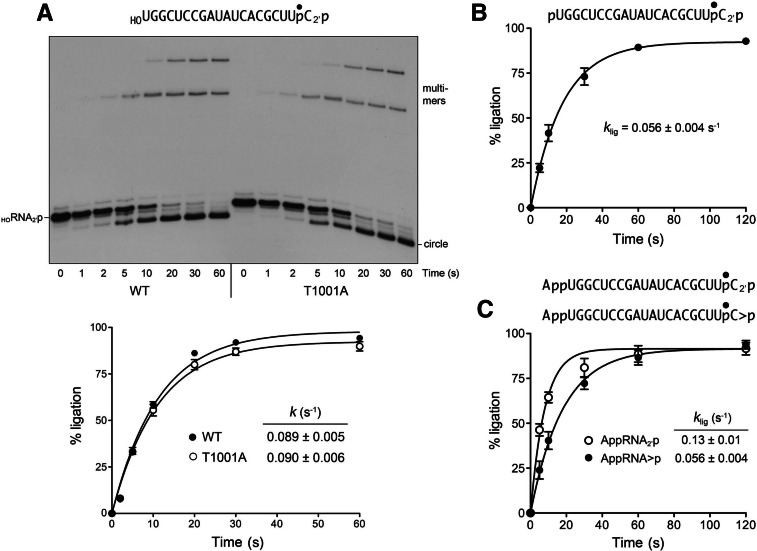
Kinetics of HORNA2′p ligation and bypass of the T1001A CPD mutation
To bypass the biochemical requirement for 2′,3′-cyclic phosphate hydrolysis in the RNA splicing pathway, we converted the 32P-labeled HORNA>p strand to HORNA2′p by incubation with AtRNL in the absence of Mg2+ and ATP. The HORNA2′p strand was isolated and the 2′-PO4 terminal structure was verified by RNase T1 digestion. Reaction of the HORNA2′p substrate (20 nM) with wild-type AtRNL (1 µM) in the presence of Mg2+ and ATP elicited its conversion to circle and multimer ligation products (Fig. 4A). The reaction was complete in 30–60 sec. The extent of ligation is plotted as a function of time in Figure 4A. The data fit to a single exponential with a rate constant of 0.089 ± 0.005 sec−1. This value is similar to the rate of single-turnover ligation of HORNA> p (0.048 sec−1). An instructive finding was that the ligation defect of the AtRNL-T1001A mutant was bypassed when the mutant was reacted with the HORNA2′p substrate (Fig. 4A). The rate of HORNA2′p ligation by T1001A (k = 0.090 ± 0.006 sec−1) was virtually identical to that of wild-type AtRNL. Because the kinase step is required for and precedes end sealing, the observed rate of HORNA2′p ligation (0.089 sec−1) establishes a lower bound for the rate of 5′ phosphorylation of the HORNA2′p strand (Fig. 1C), which is approximately threefold greater than the rate of phosphorylation of the HORNA>p substrate by the T1001A enzyme (0.026 sec−1). Indeed, one can detect the transient accumulation and disappearance of a pRNA2′p intermediate (migrating just faster than the input substrate, but slower than the ligated circle) during the reactions of wild-type AtRNL and T1001A with the HORNA2′p substrate (Fig. 4A).
FIGURE 4.
Ligation of RNAs with pre-healed 2′-PO4 ends. (A) Kinetics of HORNA2′p ligation and bypass of the T1001A CPD mutation. Reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 10 mM MgCl2, 0.1 mM ATP, 2 mM DTT, 20 nM HORNA2′p substrate (as shown), and 1 µM wild-type AtRNL or T1001A mutant as specified were incubated at 22°C. The reactions were quenched at the times indicated and the products analyzed by urea-PAGE (top panel). The extents of ligation by wild-type (WT) and T1001A AtRNL are plotted as a function of reaction time (bottom panel). Each datum is the average of four separate experiments ±SEM. Nonlinear regression curve fits of the data to a single exponential are shown. (B) Kinetics of pRNA2′p ligation. Reaction mixtures contained 20 nM pRNA2′p substrate (as shown) and 1 µM wild-type AtRNL. Each datum in the graph is the average of four separate experiments ±SEM. (C) Kinetics of AppRNA2′p ligation. Reaction mixtures contained 20 nM AppRNA2′p or AppRNA>p substrates (as shown) and 1 µM wild-type AtRNL. Each datum in the graph is the average of either four (AppRNA2′p) or five (AppRNA>p) separate time-course experiments ±SEM.
Kinetics of pRNA2′p ligation
To focus our analysis on the end-sealing phase of the plant ligase pathway, we prepared a 32P-labeled 20-mer pRNA2′p substrate with “pre-healed” 5′-PO4 and 3′-OH, 2′-PO4 ends. Reaction of 20 nM pRNA2′p with 1 µM AtRNL in the presence of Mg2+ and ATP generated circular and multimeric ligation products in a time-dependent fashion (Fig. 4B) with an apparent rate constant of 0.056 ± 0.004 sec−1. Because the rate of sealing a pre-healed pRNA2′p substrate was quite similar to the composite healing and sealing pathway for HORNA>p and to the ligation of HORNA2′p (Fig. 1C), we surmise that the sealing steps catalyzed by the ligase domain are either rate limiting or colimiting (e.g., with the kinase reaction) for the overall splicing pathway.
A 5′-PO4 bypasses inactivating mutations of the kinase domain
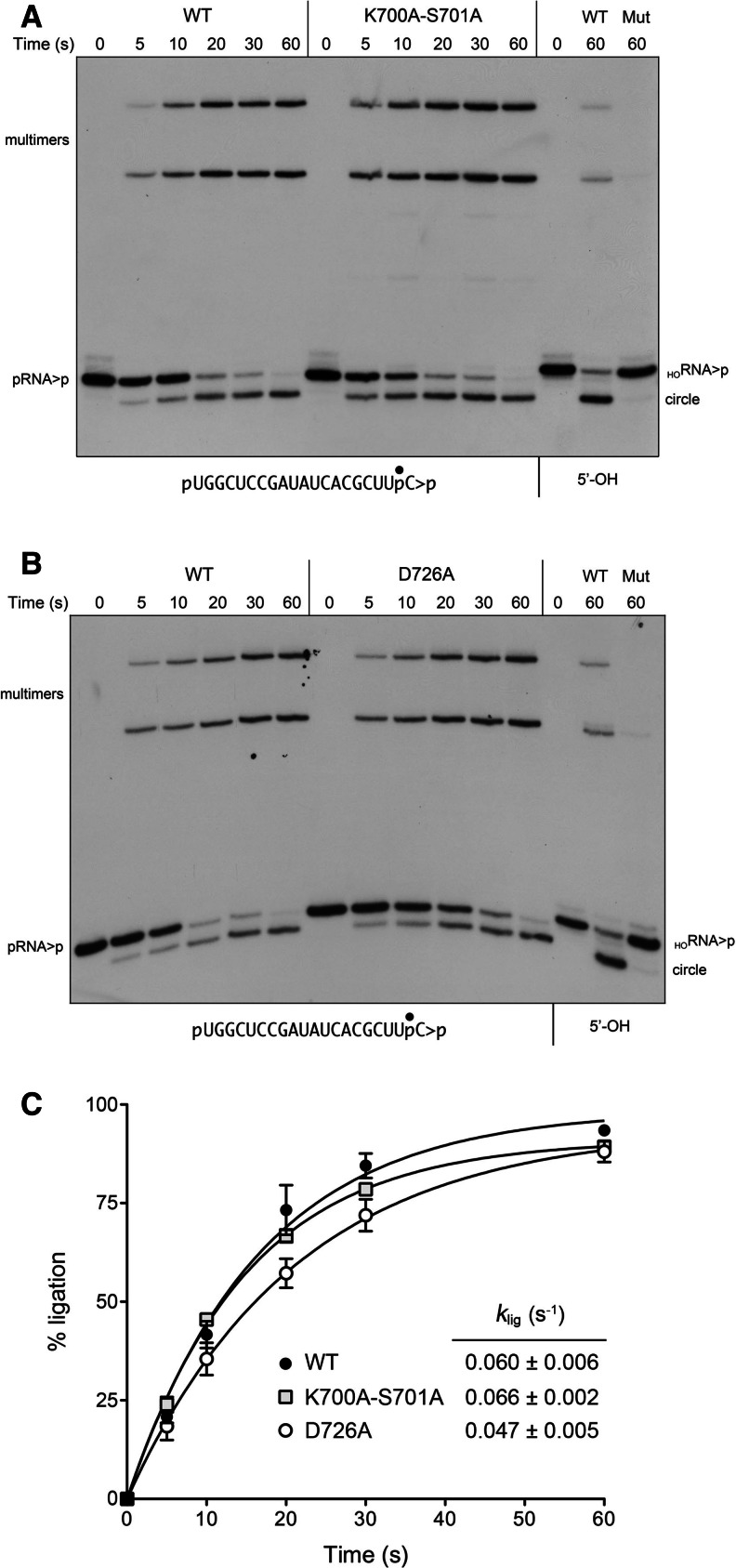
The kinase domain of plant tRNA ligase is homologous to the kinase domains of the RNA repair enzymes T4 polynucleotide kinase-phosphatase (T4Pnkp) and Clostridium thermocellum polynucleotide kinase-phosphatase (CthPnkp) that have been characterized structurally and biochemically (Galburt et al. 2002; Wang et al. 2002, 2012). Phosphoryl transfer during the kinase reaction entails an in-line attack of the polynucleotide 5′-OH on the ATP γ-phosphorus, aided by an aspartate general base (corresponding to Asp726 in AtRNL) that activates the 5′-OH. The P-loop lysine (Lys700 in AtRNL) and the catalytic Mg2+ bridge the ATP β- and γ-phosphates and stabilize the transition state. The P-loop serine (Ser701 in AtRNL) is a constituent of the Mg2+ coordination complex. AtRNL mutations S701A and D726A were shown to abolish tRNA splicing activity in vivo (Wang et al. 2006). Here we tested the effects of a double-alanine P-loop mutation K700A-S701A and a single D726A mutation on AtRNL splicing activity in vitro. Whereas wild-type AtRNL efficiently healed and sealed the HORNA>p substrate, the mutant enzymes failed to do so (Fig. 5A,B, cf. lanes WT and Mut). However, when the kinase-defective K700A-S701A and D726A mutants were presented with a substrate with a pre-formed 5′-PO4 (pRNA>p), they recovered ligation activity. The kinetics of pRNA>p ligation by wild-type and kinase-defective enzymes are shown in Figure 5, A and B, and quantified in Figure 5C. The rate constant of 0.060 sec−1 for pRNA>p ligation by wild-type AtRNL echoed the value of 0.056 sec−1 for pRNA2′p ligation (see Fig. 1C), again attesting to the fact that the CPD step is not rate limiting. The rate constants for pRNA>p ligation by K700A-S701A (0.066 sec−1) and D726A (0.047 sec−1) were similar to wild-type AtRNL. The apparently complete gain of RNA ligation activity of the CPD-defective and kinase-defective mutants with RNAs containing pre-healed 3′ and 5′ ends weighs against the healing steps being kinetically or physically coupled to the downstream sealing steps.
FIGURE 5.
A 5′-PO4 bypasses inactivating mutations of the kinase domain. (A,B) Reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 10 mM MgCl2, 0.1 mM ATP, 2 mM DTT, 20 nM pRNA>p substrate (as shown), and 1 µM wild-type AtRNL (A,B) or mutants K700A-S701A (A) or D726A (B) as specified were incubated at 22°C. The reactions were quenched at the times indicated, and the products were analyzed by urea-PAGE. Control reactions containing 20 nM HORNA>p substrate (5′-OH) and 1 µM wild-type (WT) or mutant AtRNLs were incubated for 60 sec and analyzed in parallel in the lanes at right. (C) The extents of pRNA>p ligation by WT, K700A-S701A, and D726A AtRNL are plotted as a function of reaction time. Each datum is the average of three (WT and K700A-S701A) or four (D726A) separate experiments ±SEM. Nonlinear regression curve fits of the data and the derived rate constants are shown.
ATP-independent sealing of a pre-adenylylated RNA
ATP is consumed chemically at two discrete stages during the plant tRNA ligase pathway: it is converted to ADP during the kinase reaction and to AMP during the sealing reaction (Pick et al. 1986). Use of an RNA substrate with a pre-formed 5′-PO4 end circumvented the need for a kinase activity but did not eliminate the ATP requirement for sealing, as evinced by the pRNA>p reactions depicted in Figure 6A. There, we see that 1 µM wild-type AtRNL efficiently converted 20 nM pRNA>p to circular and multimeric ligation products during a 2-min reaction in the presence of Mg2+ and ATP (lane +), but was unreactive when ATP was omitted (lane −). The role of ATP during the sealing reaction is to sequentially adenylate the ligase active site (step 1 in Fig. 1C) and then transfer AMP to the RNA 5′-PO4 (step 2), to form an AppN terminus that is attacked by an RNA 3′-OH to form the splice junction (step 3). Because the AppN-terminated species is not detected during single-turnover reaction of AtRNL with pRNA>p (Fig. 5A,B), we infer that step 3 is fast compared with step 2 when sealing initiates at a 5′-PO4 end.
FIGURE 6.
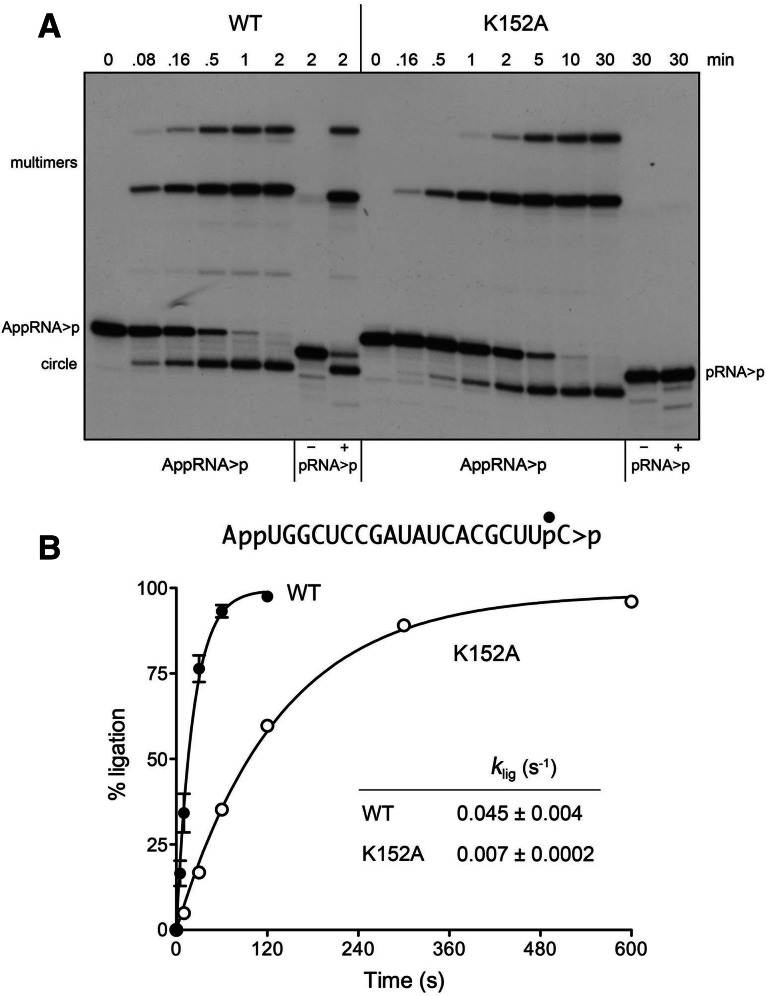
A pre-adenylylated RNA bypasses the need for ATP and the active-site lysine. (A) Reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 10 mM MgCl2, 0.1 mM ATP, 2 mM DTT, 20 nM AppRNA>p substrate (as shown), 1 µM WT or K152A AtRNL, and no added ATP were incubated at 22°C. The reactions were quenched at the times specified, and the products were analyzed by urea-PAGE. Control reactions contained 20 nM pRNA>p substrate, 1 µM WT, or K152A AtRNL, and either no added ATP (lanes −) or 0.1 mM ATP (lanes +). (B) The extents of AppRNA>p ligation by WT and K152A AtRNL are plotted as a function of reaction time. Nonlinear regression curve fits of the data (±SEM) and the derived rate constants are shown.
To study the isolated step of phosphodiester synthesis, we prepared a pre-adenylylated 20-mer substrate, AppRNA>p. The 32P-labeled AppRNA>p strand migrated slower than pRNA>p species during denaturing PAGE (Fig. 6A). Reaction of 20 nM AppRNA>p with 1 µM wild-type AtRNL yielded circular and multimeric ligation products in a time-dependent fashion (Fig. 6A) with an apparent rate constant of 0.045 sec−1 (Fig. 6B). This experiment establishes two salient points: (1) that the ATP requirement for ligation is bypassed by pre-adenylylation of the 5′ end; and (2) that the reaction of wild-type AtRNL at an AppN terminus is unidirectional, with no detectable reversal of the step 2 reaction to regenerate a 5′-PO4 end. (The experiment also disclosed the nature of the ligated multimers, as follows. Whereas the monomer RNA circles migrated identically whether the substrate was AppRNA>p or pRNA>p, the lower multimer species formed with the AppRNA>p substrate migrated slower than the corresponding species generated with the pRNA>p substrate [Fig. 6A]. This identifies the lower multimers as linear dimers with either 5′-AppN or 5′-PO4 termini, respectively. Because the upper multimer migrated identically whether originating from AppRNA>p or pRNA>p [Fig. 6A] and its appearance was delayed compared with the linear dimer, we suspect that the upper multimer is a dimer circle.)
RNA pre-adenylylation bypasses the Lys152 active-site nucleophile
The first of three steps in the polynucleotide ligase reaction is the nucleophilic attack of an active-site lysine (Lys152 in AtRNL) on the ATP α-phosphorus to form a covalent ligase-(lysyl-Nζ)-AMP intermediate and release pyrophosphate (Fig. 1C). Changing Lys152 to alanine abolishes AtRNL activity in vivo (Wang et al. 2006). Here we purified AtRNL-K152A and queried its sealing activity (Fig. 6A). A 30-min reaction of 1 µM K152A with 20 nM pRNA>p substrate in the presence or absence of ATP yielded no circular or multimeric ligation products, as expected from the nature of 5′ RNA end activation via a lysyl-AMP adduct. However, incubation of K152A with pre-adenylylated AppRNA>p in the absence of ATP readily generated circular and multimeric ligation products in a time-dependent fashion, that attained an endpoint in ∼10 min (Fig. 6A), with an apparent rate constant of 0.007 sec−1 (Fig. 6B). Side-by-side comparison of the wild-type and K152A proteins in AppRNA>p sealing revealed that whereas Lys152 is not essential for catalysis of phosphodiester bond formation, it contributes an approximately sevenfold rate enhancement of the step 3 reaction, presumably via its predicted noncovalent interactions with the phosphate oxygens of the AMP leaving group of RNA-adenylate (Nandakumar et al. 2006).
Ligation of AppRNA2′p with a pre-healed 2′-PO4 end
To study the step 3 phosphodiester synthesis step in an “on-pathway” fashion (assuming that a 2′,3′>p will normally be hydrolyzed by the CPD domain well before a kinased 5′-PO4 end becomes accessible to the ligase domain), we prepared a 5′-adenylylated RNA substrate with a pre-healed 3′-OH, 2′-PO4 end. The observed rate constant for ATP-independent ligation of AppRNA2′p was 0.13 sec−1, a value twice that of the rate of AppRNA>p sealing (0.056 sec−1) assayed in parallel (Fig. 4C). The step 3 rate of 0.13 sec−1 at a preformed AppN terminus can be regarded as a lower bound for the step 3 rate when the RNA adenylate intermediate is formed in situ (via step 2) in the ligase active site (Fig. 1C).
The 2′-PO4 requirement for sealing is enforced at the RNA adenylylation step
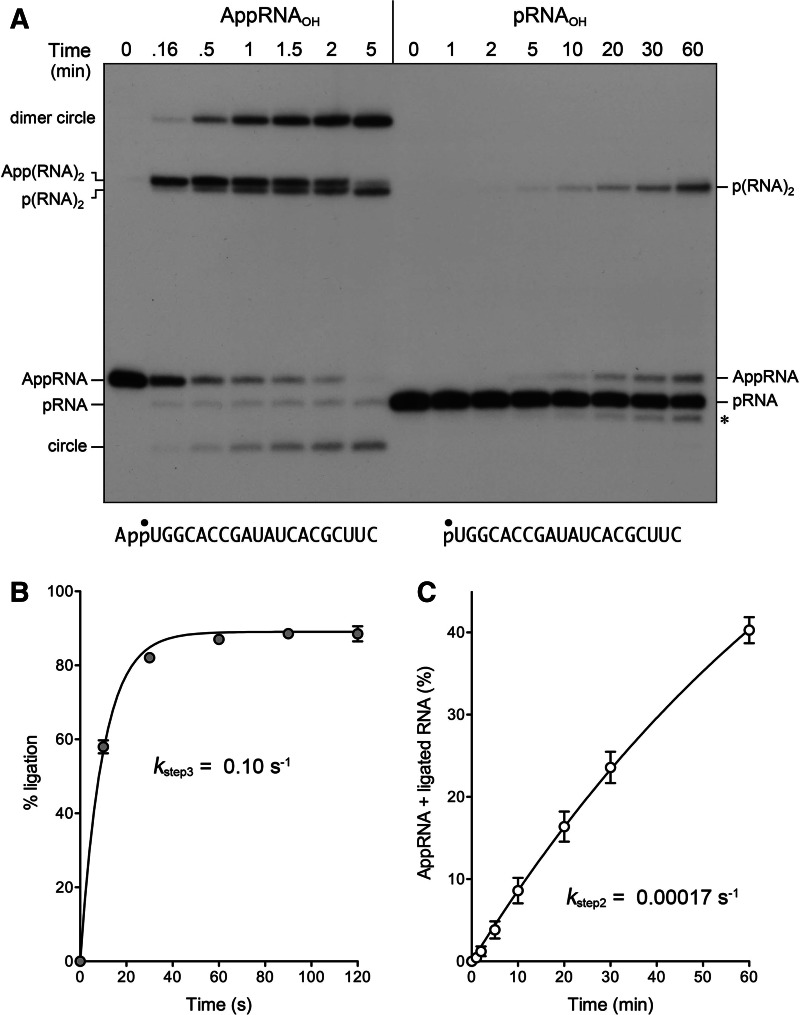
The defining feature of plant and yeast tRNA ligases vis à vis other ATP-dependent polynucleotide ligases is their requirement for a 2′-PO4 to synthesize a 3′–5′ phosphodiester bond. The biochemical and structural basis for this requirement is uncharted. It is conceivable that 2′-PO4 specificity is enforced uniquely during either step 2 or step 3 of the RNA sealing pathway; or 2′-PO4 specificity might be applied during both steps. To investigate this issue, we prepared a 5′ 32P-labeled AppRNAOH substrate and tested its reactivity with AtRNL. The AppRNAOH strand was efficiently ligated in the absence of ATP to yield circular and multimeric products (Fig. 7A). Ninety-three percent of the input substrate was sealed in 5 min; the apparent step 3 rate constant was 0.10 ± 0.005 sec−1 (Fig. 7B). Comparison to the AppRNA2′p ligation rate of 0.13 sec−1 (Fig. 1C) highlights that the 2′-PO4 moiety exerts little effect on the rate of step 3 phosphodiester synthesis. A subtle but notable difference in the outcome of the AppRNAOH reaction, compared with AppRNA2′p ligation, was that 5% of the input AppRNAOH substrate was deadenylylated, by reversal of step 2, to yield a pRNA product (Fig. 7A). A fraction of the linear dimer product of intermolecular joining, App(RNA)2, also underwent deadenylylation in a time-dependent fashion to form a faster-migrating p(RNA)2 species; this reverse step 2 reaction occurred in parallel with a forward step 3 reaction in which App(RNA)2 was converted to dimer circle (Fig. 7A). As expected, the deadenylylated linear pRNA and p(RNA)2 species were not formed when the AppRNAOH substrate was reacted with the K152A mutant, which cannot perform the step 2 reaction in either forward or reverse direction; however, the K152A mutant was adept at ligating the AppRNAOH substrate to form a mixture of dimer circle, App(RNA)2, and monomer circle (data not shown).
FIGURE 7.
The 2′-PO4 requirement for sealing is enforced at the RNA 5′-adenylylation step. (A) AppRNAOH and pRNAOH ligation. Reaction mixtures containing 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 10 mM MgCl2, 2 mM DTT, 20 nM 20-mer AppRNAOH, or pRNAOH substrate (shown at bottom with the 32P label denoted by •), 1 µM AtRNL, and either no added ATP (AppRNAOH reaction) or 0.1 mM ATP (pRNAOH reaction) were incubated at 22°C. The reactions were quenched with formamide, EDTA at the times specified, and the products were analyzed by urea-PAGE. The positions and identities of the radiolabeled substrates and products are indicated at left and right. (The asterisk at right denotes an n − 1 decay product derived from pRNAOH during prolonged incubation with AtRNL; the formation of this species, comprising 8% of the total labeled RNA at 60 min, was unaffected by omission of ATP but was eliminated by omission of Mg2+ [data not shown].) (B) AppRNAOH ligation kinetics. The extent of ligation is plotted as a function of reaction time. A nonlinear regression curve fit of the data (average of four experiments ±SEM) and the derived step 3 rate constant are shown. (C) pRNAOH reaction kinetics. The levels of AppRNA plus ligated RNA (as percent of total labeled RNA) are plotted as a function of reaction time. A nonlinear regression curve fit of the data (average of three experiments ±SEM) and the derived step 2 rate constant are shown.
To interrogate the need for a 2′-PO4 during the RNA 5′-adenylylation step of ligation, we presented AtRNL with a 5′ 32P-labeled pRNAOH substrate. A 60-min reaction in the presence of Mg2+ and ATP resulted in conversion of 15% of the pRNAOH strand to AppRNAOH and 25% ligation (Fig. 7A). Ligated RNA and AppRNA were not produced when ATP was omitted (data not shown). The kinetic profile of total product formation (AppRNA plus ligated RNA) is shown in Figure 7C. Fitting the data to a single exponential yielded a step 2 rate constant of 0.00017 ± 0.00005 sec−1 for adenylylation of the pRNAOH substrate. This value is 330-fold slower than the rate of the pRNA2′p ligation reaction under the same conditions (Fig. 1C). These experiments show that the 2′-PO4 requirement for end sealing is stringently enforced at the level of RNA adenylylation (step 2) and that this requirement can be bypassed by pre-adenylylating the RNA 5′ end.
DISCUSSION
Plant and fungal tRNA ligases are essential agents of informational and stress response pathways involving the repair of programmed tRNA and mRNA breaks. The biochemical outcomes and end specificities of the healing and sealing reactions they catalyze are unique compared with the tRNA repair systems elaborated by viruses, bacteria, and metazoans (Popow et al. 2012). Indeed, mammalian proteomes have no discernable homologs of the sealing domain of plant/fungal tRNA ligase. These features highlight tRNA ligases as untapped targets for the discovery of new antifungals and herbicides.
The present study fills gaps in our understanding of the mechanism and specificity of tRNA ligases, exemplified by the Arabidopsis enzyme AtRNL. By exploiting defined RNA substrates with different 5′ and 3′ terminal structures and mutant enzymes with step-specific defects to analyze reaction outcomes under conditions of enzyme excess, we provide a kinetic framework for the chemical steps along the reaction pathway. The rate constants for the individual steps or subsets of steps linked by arrows are summarized in Figure 1C.
The CPD reaction is evidently much faster than the kinase and ligase reactions, making it most likely that the inciting 2′,3′>p // 5′-OH break is converted to a 2′-PO4,3′-OH // 5′-OH configuration prior to its conversion by the kinase to a 2′-PO4,3′-OH // 5′-PO4 that can be joined by the ligase module. The rates of sealing of the partially healed (pRNA>p and HORNA2′p) and fully healed (pRNA2′p) repair intermediates are similar to one another and to the rate of the complete HORNA>p repair pathway, implying that the kinase reaction (which occurs on the same time scale as the three-step reaction executed by the ligase module) and the ligase reaction are jointly rate limiting for the overall pathway. Earlier studies of the plant tRNA ligase isolated from wheat germ had reported conflicting conclusions regarding the role of the 5′ end in dictating the rate of sealing, whereby one group found that a pRNA>p substrate is sealed ∼100-fold more efficiently than HORNA>p (Schwartz et al. 1983), while other investigators found that HORNA>p was sealed fivefold faster than pRNA>p (Pick and Hurwitz 1986). These early studies were performed under what we surmise are conditions of RNA substrate excess, and they relied on short homo-oligomeric RNAs—either oligo(A) or oligo(C)—as substrates. The present study of recombinant AtRNL under conditions of enzyme excess makes clear that the rate of sealing by this plant tRNA ligase is not influenced significantly by the initial configuration of the 5′ terminus.
However, we did note a consistent difference in the ligation product distribution when AtRNL reacted with a 5′-OH substrate versus a 5′-PO4 RNA or a 5′ pre-adenylylated RNA. Whereas sealing of 5′-OH substrates yielded a monomer RNA circle as the majority product rather than the slower migrating multimers (Figs. 1D, 4A), the multimers comprised the majority products when AtRNL sealed a 5′-PO4 or 5′-adenylylated substrate (Figs. 5, 6). Quantifying the endpoint product distribution from multiple experiments revealed that 5′-OH substrates yielded a monomer circle to a multimer ratio of 2.9, while 5′-PO4 and AppN substrates yielded ratios of 0.37 and 0.33, respectively.
To account for this difference, we speculate that the ligase domain of AtRNL has independent and specific binding sites for the 2′-PO4 and 5′-PO4 RNA ends. When presented with a HORNA2′p substrate (added as such or generated rapidly by CPD action on HORNA>p), the 2′-PO4 terminus is quickly engaged by the ligase module but the 5′-OH end is not. Subsequent phosphorylation of the 5′-OH by the kinase while the 2′-PO4 is engaged by ligase will tend to favor intramolecular capture of the 5′-PO4 end and the formation of a monomer circle product. However, when AtRNL is presented with a pRNA2′p substrate, the binding sites on the ligase domain can capture (without delay) the 2′-PO4 and 5′-PO4 ends of two different RNA substrates, providing better opportunity for intermolecular sealing.
Invocation of a binding site for the 2′-PO4 in the AtRNL ligase domain is attractive in light of the specificity for sealing 3′-OH, 2′-PO4 RNA ends, but it doesn’t explain how this specificity is enforced, e.g., at the level of initial binding of one or both RNA ends, a post-binding conformational step, or one or more chemical steps of the multistep RNA sealing reaction. Here we have provided the first mechanistic insights to the 2′-PO4 specificity of tRNA ligase, by showing that AtRNL seals a pre-adenylylated AppRNAOH substrate with a 3′-OH, 2′-OH end. The insignificant rate differential between AppRNA2′p and AppRNAOH sealing indicates that the 2′-PO4 moiety makes little contribution to the rate of step 3 catalysis, although it does add value by inhibiting RNA deadenylylation. Our experiments show that AtRNL 2′-PO4 specificity is enforced at the level of RNA 5′ adenylylation, insofar as the conversion of pRNAOH to AppRNAOH and ligated RNA in the presence of ATP was several hundredfold slower than the sealing of pRNA2′p under the same reaction conditions. Because the 2′-PO4 moiety is not chemically transformed during the sealing reaction, we surmise that its essentiality for step 2 reflects a mechanism of substrate-assisted catalysis in which the 2′-PO4 either (1) promotes proper geometry of the lysyl152-AMP and 5′-PO4 reactants during step 2; (2) assists in binding the Mg2+ cofactor during step 2; or (3) aids expulsion of the Lys152 leaving group. (A definitive account of the 2′-PO4 requirement hinges on obtaining atomic structures of the AtRNL ligase bound to an RNA substrate at discrete functional steps along the sealing pathway.)
The choice of step 2 RNA 5′ adenylylation to discriminate 3′-OH, 2′-PO4 versus standard 3′-OH, 2′-OH RNA ends makes sense as a quality-control checkpoint to ensure that only purposeful RNA breaks are sealed and nonspecific 5′ modifications are suppressed. By coupling the conversion of 5′-PO4 RNA ends to AppRNA to the engagement of a 2′-PO4 end on the ligase, the plant RNA repair system avoids conferring a protective adenylate “cap” on the many cellular RNAs that have 5′-PO4 ends, some of which are stable products of RNA processing reactions, but many of which are destined for rapid decay by 5′-exoribonucleases that require a 5′-PO4 and are inhibited by inverted 5′–5′ nucleotide caps. In sum, the present study of the kinetic mechanism and chemical specificity of plant tRNA ligase highlights the logical design of a eukaryal RNA repair machine.
MATERIALS AND METHODS
tRNA ligase purification
Recombinant wild-type AtRNL and mutants K152A, H999A, T1001A, H1060A, T1062A, D726A, and K700A/S701A were produced in E. coli as His10Smt3 fusions, which were isolated from soluble bacterial extracts by Ni-agarose chromatography, followed by treatment with Smt3-specific protease Ulp1 to cleave the tag (Nandakumar et al. 2008). After depletion of the tag by adsorption to Ni-agarose, the AtRNL proteins were subjected to gel filtration through a Superdex-200 column equilibrated in 50 mM Tris-HCl (pH 7.5), 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.05% Triton X-100, and 10% glycerol. Peak AtRNL fractions were pooled and stored at −80°C. Protein concentrations were determined by using the Bio-Rad dye reagent with BSA as the standard.
Preparation of 3′ 32P-labeled RNA substrates
The 20-mer HORNA3′p oligonucleotide labeled with 32P at the penultimate phosphate was prepared by T4 Rnl1-mediated addition of [5′-32P]pCp to a 19-mer synthetic oligoribonucleotide 5′-HOUGGCUCCGAUAUCACGCUUOH. The RNA3′p was treated with E. coli RNA 3′-terminal phosphate cyclase (RtcA) and ATP to generate a 2′,3′-cyclic phosphate derivative, HORNA>p, which was gel-purified, eluted from an excised gel slice, and recovered by ethanol precipitation. A 2′-PO4 terminated derivative, HORNA2′p, was produced by reacting HORNA>p with purified AtRNL in the presence of EDTA and no added ATP, then recovered by phenol extraction and ethanol precipitation. The 32P-labeling and end-modification reactions were performed as described previously (Tanaka et al. 2011; Das and Shuman 2013). The 5′-PO4 derivatives of the same 20-mer oligonucleotides (pRNA3′p, pRNA>p, and pRNA2′p) were prepared in parallel starting from a 5′-monophosphate-terminated 19-mer RNA 5′-pUGGCUCCGAUAUCACGCUUOH (Dharmacon). The 19-mer pRNAOH strand was 5′ adenylylated by reaction with ATP and RtcA (Chakravarty and Shuman 2011) prior to the step of 3′ 32P-labeling with pCp to form AppRNA3′p. The AppRNA3′p strand was reacted again with RtcA to form AppRNA>p, then gel purified. AppRNA>p was converted to AppRNA2′p by reaction with AtRNL, and the AppRNA2′p strand was recovered by phenol extraction and ethanol precipitation.
Preparation of 5′ 32P-labeled RNA substrates
A 20-mer HORNAOH strand, 5′-HOUGGCACCGAUAUCACGCUUCOH, was 5′ 32P-labeled by using T4 polynucleotide kinase and [γ-32P]ATP. The radiolabeled pRNAOH derivative was converted to AppRNAOH by reaction with RtcA and cold ATP. The pRNAOH and AppRNAOH oligonucleotides were gel purified, eluted, and recovered by ethanol precipitation.
Assay of CPD activity
A Kintek RQF3 rapid chemical quench apparatus was used to assay the reaction of HORNA>p with a 50-fold molar excess of AtRNL at 22°C in the absence of added ATP, in the range of 0.1-sec to 5-sec reaction times. Where applicable, longer reaction times were assayed manually. The reaction mixtures (40 µL) contained 50 mM Tris-HCl (pH 8.0), 50 mM NaCl, 2 mM DTT, 10 mM EDTA, 20 nM HORNA>p substrate, and 1 µM AtRNL. The rapid kinetic measurements were initiated by mixing two buffer solutions (20 µL each). One solution contained 50 mM Tris-HCl (pH 8.0), 2 mM DTT, 20 mM EDTA, and 40 nM HORNA>p substrate. The other solution contained 50 mM Tris-HCl (pH 8.0), 2 mM DTT, 100 mM NaCl, and 2 µM AtRNL. The reactions were quenched by rapid mixing with 110 μL of 0.7% SDS. The products were extracted with phenol:chloroform; the aqueous phase was removed and digested with RNase T1 (1000 units; Fermentas) for 15 min at 37°C. The samples were mixed with an equal volume of 90% formamide, 30 mM EDTA and analyzed by electrophoresis through a 20% polyacrylamide gel containing 7 M urea in TBE. The 32P-labeled oligonucleotides HOCUUpC>p and HOCUUpC2′p were visualized by autoradiography and quantified by scanning the gel with a PhosphorImager.
ACKNOWLEDGMENTS
This work was supported by NIH grant GM42498. S.S. is an American Cancer Society Research Professor.
REFERENCES
- Apostol BL, Westaway SK, Abelson J, Greer CL 1991. Deletion analysis of a multifunctional yeast tRNA ligase polypeptide: Identification of essential and dispensable functional domains. J Biol Chem 266: 7445–7455 [PubMed] [Google Scholar]
- Chakravarty AK, Shuman S 2011. RNA 3′-phosphate cyclase (RtcA) catalyzes ligase-like adenylylation of DNA and RNA 5′-monophosphate ends. J Biol Chem 286: 4117–4122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das U, Shuman S 2013. Mechanism of RNA 2′,3′-cyclic phosphate end-healing by T4 polynucleotide kinase-phosphatase. Nucleic Acids Res 41: 355–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englert M, Beier H 2005. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res 33: 388–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galburt EA, Pelletier J, Wilson G, Stoddard BL 2002. Structure of a tRNA repair enzyme and molecular biology workhorse: T4 polynucleotide kinase. Structure 10: 1249–1260 [DOI] [PubMed] [Google Scholar]
- Mazumder R, Iyer L, Vasudevan S, Aravind L 2002. Detection of novel members, structure–function analysis and evolutionary classification of the 2H phosphoesterase family. Nucleic Acids Res 30: 5229–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Ogasawara C, Inada T, Englert M, Beier H, Takezawa M, Endo T, Yoshihisa T 2010. Dual functions of yeast tRNA ligase in the unfolded protein response: Unconventional cytoplasmic splicing of HAC1 pre-mRNA is not sufficient to release translational attenuation. Mol Biol Cell 21: 3722–3734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllykoski M, Raasakka A, Han H, Kursula P 2012. Myelin 2′,3′-cyclic nucleotide 3′-phosphodiesterase: Active site ligand binding and molecular conformation. PLoS ONE 7: e32336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar J, Shuman S, Lima CD 2006. RNA ligase structures reveal the basis for RNA specificity and conformational changes that drive ligation forward. Cell 127: 71–84 [DOI] [PubMed] [Google Scholar]
- Nandakumar J, Schwer B, Schaffrath R, Shuman S 2008. RNA repair: An antidote to cytotoxic eukaryal RNA damage. Mol Cell 31: 278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pick L, Hurwitz J 1986. Purification of wheat germ RNA ligase: Characterization of a ligase-associated 5′-hydroxyl polynucleotide kinase activity. J Biol Chem 261: 6684–6693 [PubMed] [Google Scholar]
- Pick L, Furneaux H, Hurwitz J 1986. Purification of wheat germ RNA ligase: Mechanism of action of wheat germ RNA ligase. J Biol Chem 261: 6694–6704 [PubMed] [Google Scholar]
- Popow J, Schleiffer A, Martinez J 2012. Diversity and roles of (t)RNA ligases. Cell Mol Life Sci 18: 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya R, Schwer B, Shuman S 2003. Genetic and biochemical analysis of the functional domains of yeast tRNA ligase. J Biol Chem 278: 43298–43398 [DOI] [PubMed] [Google Scholar]
- Schwartz RC, Greer CL, Gegenheimer P, Abelson J 1983. Enzymatic mechanism of an RNA ligase from wheat germ. J Biol Chem 258: 8374–8383 [PubMed] [Google Scholar]
- Schwer B, Sawaya R, Ho CK, Shuman S 2004. Portability and fidelity of RNA-repair systems. Proc Natl Acad Sci 101: 2788–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Aronova A, Ramirez A, Braun P, Shuman S 2008. Mammalian 2′,3′ cyclic nucleotide phosphodiesterase (CNP) can function as a tRNA splicing enzyme in vivo. RNA 14: 204–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman S, Lima CD 2004. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr Opin Struct Biol 14: 757–764 [DOI] [PubMed] [Google Scholar]
- Spinelli SL, Consaul SA, Phizicky EM 1997. A conditional lethal yeast phosphotransferase mutant accumulates tRNA with a 2′-phosphate and an unmodified base at the splice junction. RNA 3: 1388–1400 [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Chakravarty AK, Maughan B, Shuman S 2011. A novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-hydroxyl ligation reactions. J Biol Chem 286: 43134–43143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LK, Lima CD, Shuman S 2002. Structure and mechanism of T4 polynucleotide kinase—an RNA repair enzyme. EMBO J 21: 3873–3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LK, Schwer B, Englert M, Beier H, Shuman S 2006. Structure–function analysis of the kinase-CPD domain of yeast tRNA ligase (Trl1) and requirements for complementation of tRNA splicing by a plant Trl1 homolog. Nucleic Acids Res 34: 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LK, Das U, Smith P, Shuman S 2012. Structure and mechanism of the polynucleotide kinase component of the bacterial Pnkp-Hen1 RNA repair system. RNA 18: 2277–2286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westaway SK, Belford HG, Apostol BL, Abelson J, Greer CL 1993. Novel activity of a yeast ligase deletion polypeptide: Evidence for GTP-dependent tRNA splicing. J Biol Chem 268: 2435–2443 [PubMed] [Google Scholar]
- Xu Q, Teplow D, Lee TD, Abelson J 1990. Domain structure in yeast tRNA ligase. Biochemistry 29: 6132–6138 [DOI] [PubMed] [Google Scholar]