The WD domain protein Rrp9 associates with the U3 snoRNA, which is required for 18S rRNA processing and small subunit ribosome formation. In this article, the authors analyze the structure and functional site of Rrp9. They locate an RNA-binding site on Rrp9 essential for specific recognition of U3, nucleolar localization, and yeast growth. They also find a nonessential bipartite nuclear localization signal.
Keywords: crystal structure, WD domain, RNA-binding protein, nuclear localization signal, eukaryotic ribosome biogenesis
Abstract
The U3 snoRNA is required for 18S rRNA processing and small subunit ribosome formation in eukaryotes. Different from other box C/D snoRNAs, U3 contains an extra 5′ domain that pairs with pre-rRNA and a unique B/C motif essential for recruitment of the U3-specific Rrp9 protein. Here, we analyze the structure and function of Rrp9 with crystallographic, biochemical, and cellular approaches. Rrp9 is composed of a WD repeat domain and an N-terminal region. The crystal structures of the WD domain of yeast Rrp9 and its human ortholog U3-55K were determined, revealing a typical seven-bladed propeller fold. Several conserved surface patches on the WD domain were identified, and their function in RNP assembly and yeast growth were analyzed by mutagenesis. Prior association of Snu13 with the B/C motif was found to enhance the specific binding of the WD domain. We show that a conserved 7bc loop is crucial for specific recognition of U3, nucleolar localization of Rrp9, and yeast growth. In addition, we show that the N-terminal region of Rrp9 contains a bipartite nuclear localization signal that is dispensable for nucleolar localization. Our results provide insight into the functional sites of Rrp9.
INTRODUCTION
Eukaryotic ribosomes, which are composed of four rRNAs and approximately 80 ribosomal proteins, are synthesized through a highly complicated multistep process (for recent reviews, see Henras et al. 2008; Kressler et al. 2010; Phipps et al. 2011). This process, which is thought to be generally conserved in eukaryotes, is best understood in the yeast Saccharomyces cerevisiae. Ribosome synthesis starts in the nucleolus with the transcription of a 35S precursor rRNA (pre-rRNA) that contains 18S, 5.8S, and 25S rRNAs and four spacer sequences. These spacer sequences are excised through a series of endo- and exonucleolytic cleavages (Venema and Tollervey 1999). The nascent pre-rRNA is extensively 2′-O-methylated and pseudouridylated by box C/D small nucleolar (sno) RNPs and box H/ACA snoRNPs, respectively. Each of these RNPs uses a distinct H/ACA or C/D snoRNA to select the modification target (Kiss 2001). In addition, a handful of snoRNAs, including the universally conserved U3, U14, and snR30/U17, are required for pre-rRNA processing.
In yeast, approximately 200 nonribosomal proteins have been identified to transiently associate with pre-rRNA processing intermediates. The earliest detectable 35S pre-rRNA is cotranscriptionally assembled into the small subunit (SSU) processome or 90S preribosomal particle (Dragon et al. 2002; Grandi et al. 2002; Osheim et al. 2004). Following cleavage at sites A0 and A1 in the 5′-external transcribed spacer (5′-ETS) and at site A2 in the internal transcribed spacer 1 (ITS1), the pre-rRNA is separated into the 20S and 27SA2 pre-rRNAs. The 20S pre-rRNA is assembled into pre-40S particles, which are exported to the cytoplasm and maturated into small subunit ribosomes. The 27SA2 pre-rRNA, which is the precursor of the 5.8S and 25S rRNAs, is assembled into pre-60S particles, which undergo a series of maturation steps to yield large subunit ribosomes.
The U3 box C/D snoRNA is essential for the early cleavages of 18S rRNA (sites A0, A1, and A2 in yeast) in yeast and vertebrates (Kass et al. 1990; Savino and Gerbi 1990; Hughes and Ares 1991). The U3 snoRNA functions in the context of the large SSU processome that contains nearly 50 protein assembly factors (Dragon et al. 2002; Grandi et al. 2002; Phipps et al. 2011). The 5′ domain of U3 base-pairs with four sites in the 5′-ETS and 18S regions of pre-rRNA (Beltrame and Tollervey 1995; Hughes 1996; Sharma and Tollervey 1999; Borovjagin and Gerbi 2000; Dutca et al. 2011; Marmier-Gourrier et al. 2011). These pairing interactions of U3 are thought to direct the processing and folding of pre-rRNA, yet the exact role of U3 is still speculative. In addition to the large SSU processome, the mature U3 snoRNP also exists as a 12S monoparticle, which contains four common C/D RNP proteins, namely Nop1 (fibrillarin), Nop56, Nop58, and Snu13 (15.5K in humans and L7Ae in Archaea), and the U3-specific Rrp9 (U3-55K in humans) (Watkins et al. 2000). U3-55K is required for the assembly of human U3 into the SSU processome (Granneman et al. 2004).
The 3′-domain of U3 contains four conserved sequence elements: boxes C′, B, C, and D. Boxes C′ and D juxtapose into a kink-turn (K-turn) structure at the base of the 3′ domain, and boxes B and C form another K-turn in the internal region (Watkins et al. 2000). The K-turn is a widespread RNA structural motif that is composed of one stem with canonical Watson-Crick pairs (stem I), another stem with tandem sheared G-A pairs (stem II), and a 3-nt bulge (Vidovic et al. 2000; Klein et al. 2001). Many K-turns are specifically recognized by L7Ae family proteins. The C′/D motif, which corresponds to the box C/D motif in other C/D snoRNAs, associates with Snu13, nucleating the assembly of Nop58, Nop56, and Nop1 (Watkins et al. 2000, 2002). The B/C motif associates with another copy of Snu13/15.5K, recruiting Rrp9/U3-55K (Lubben et al. 1993; Granneman et al. 2002). The C′/D motif, not the B/C motif, is essential for U3 stability in yeast (Samarsky and Fournier 1998), but both the C′/D and B/C motifs are required for U3 stability and RNP assembly in humans (Knox et al. 2011). Cross-linking analyses have shown that the paralogous Nop58 and Nop56 proteins bind at the C′/D and B/C motifs, respectively (Cahill et al. 2002; Granneman et al. 2009; van Nues et al. 2011). Nop58 and Nop56 are expected to dimerize via their coiled-coil domains in a similar manner to their single archaeal homolog, Nop5 (Aittaleb et al. 2003). This prediction was recently supported by protein-protein cross-linking results (Yang et al. 2012).
The Rrp9/U3-55K protein is composed of a WD repeat domain that is preceded by an N-terminal region (Pluk et al. 1998). The WD domain is responsible for binding the B/C motif (Lubben et al. 1993; Lukowiak et al. 2000; Venema et al. 2000; Granneman et al. 2002). The association of U3-55K with human U3 depends on prior binding of 15.5K with the B/C motif (Granneman et al. 2002). Mutational analysis of human and yeast U3 snoRNAs has shown that certain conserved sequences within the B/C motif and stem I are important for Rrp9/U3-55K binding (Granneman et al. 2002; Clery et al. 2007). However, little is known about structural elements of Rrp9/U3-55K that mediate U3 binding and function. The N-terminal region of U3-55K was predicted to contain a nuclear localization signal (NLS) (Pluk et al. 1998), but the composition and functional role of the NLS have not been characterized.
In this study, we performed a structure-function analysis of Rrp9. We determined the crystal structure of the WD domain of yeast Rrp9 and human U3-55K, revealing a typical β-propeller fold. We analyzed conserved residues on the surface of the WD domain for their function in U3 RNA binding and yeast growth by mutagenesis and identified a prominent loop that is essential for specific recognition of U3 and Rrp9 function. In addition, we identified a novel bipartite NLS in the N-terminal region of Rrp9 using a GFP reporter. Rrp9 still localizes in the nucleolus in the absence of NLS, which suggests the presence of an alternative nuclear import route.
RESULTS
Structural determination and overall structure of Rrp9 and U3-55K WD domains
As WD domains commonly mediate protein-protein interactions, it is of interest to ask whether the WD domain of Rrp9/U3-55K possesses any special structural element for RNA binding. To this end, we determined the crystal structures of the Rrp9 WD domain (residues 127–573) and the U3-55K WD domain (residues 137–475). The proteins of full-length Rrp9 and U3-55K and their WD domains were expressed in insect cells using baculovirus vectors because these proteins were insoluble when expressed in Escherichia coli. The N-terminal region of Rrp9/U3-55K lacks significant secondary structure by prediction and was not included in crystallization.
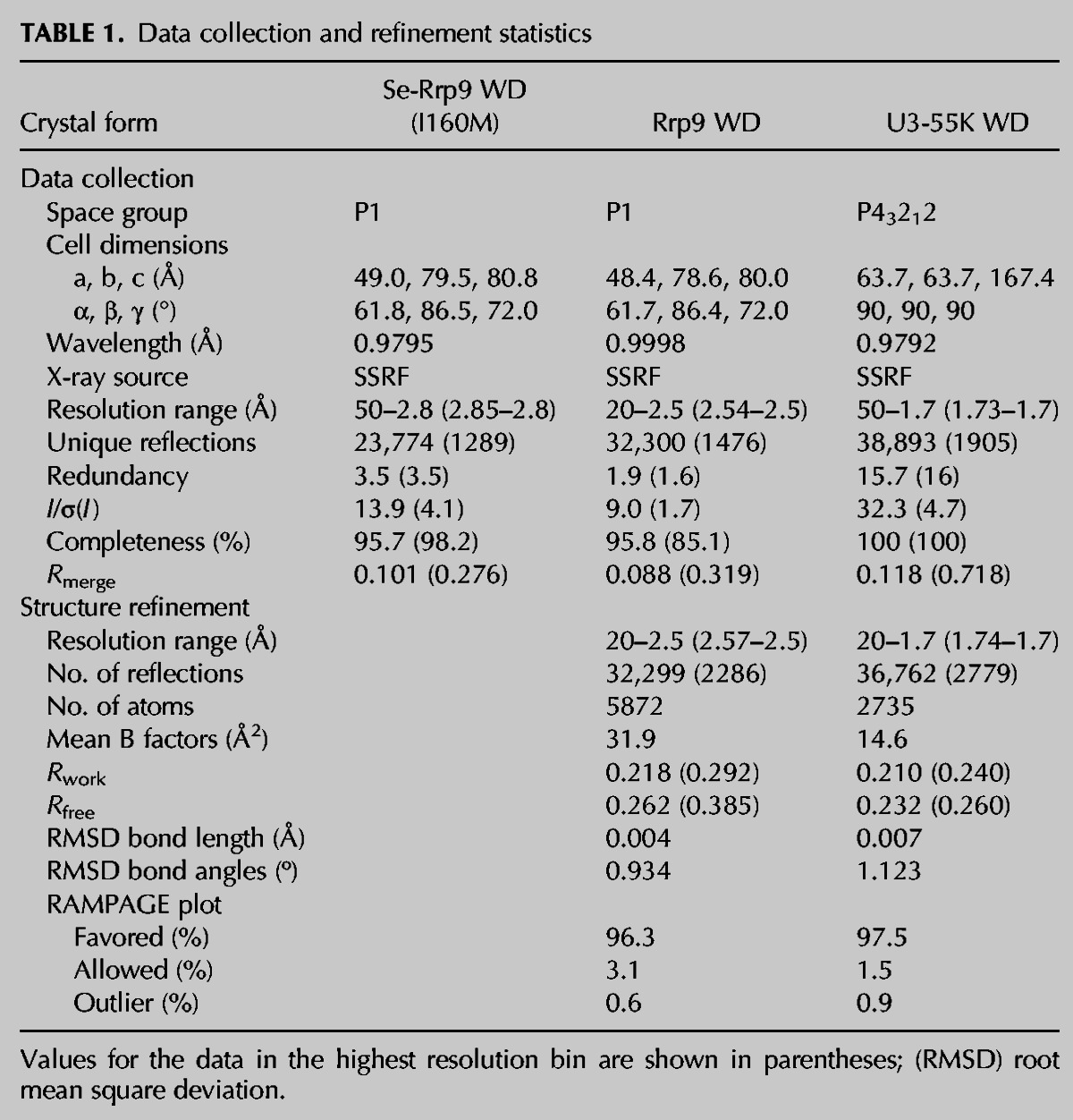
The Rrp9 WD structure was determined by a combination of molecular replacement (MR) using the yeast RACK1 structure as a search model and single-wavelength anomalous dispersion (SAD) phasing of seleno-methionine derivative crystals. The structure of the Rrp9 WD domain was refined to an Rwork/Rfree of 0.218/0.262 at 2.5 Å resolution. The structure of the U3-55K WD domain was determined by molecular replacement using the Rrp9 WD structure as a search model and refined to an Rwork/Rfree of 0.210/0.232 at 1.7 Å resolution. Table 1 lists the statistics of the diffraction data and refinement for the two structures.
TABLE 1.
Data collection and refinement statistics
The WD domain of Rrp9 adopts a seven-bladed propeller structure with a channel at the center (Fig. 2A, see below), typical of other WD domain structures. Each blade is composed of four antiparallel β-strands. Characteristic of WD domains, the first N-terminal strand of a WD repeat constitutes the outmost strand of one blade and the three C-terminal strands form the innermost strands of the succeeding blade. In particular, the first strand (7d) of repeat 1 aligns with the last strand (7c) of repeat 7 to close the propeller structure.
FIGURE 2.
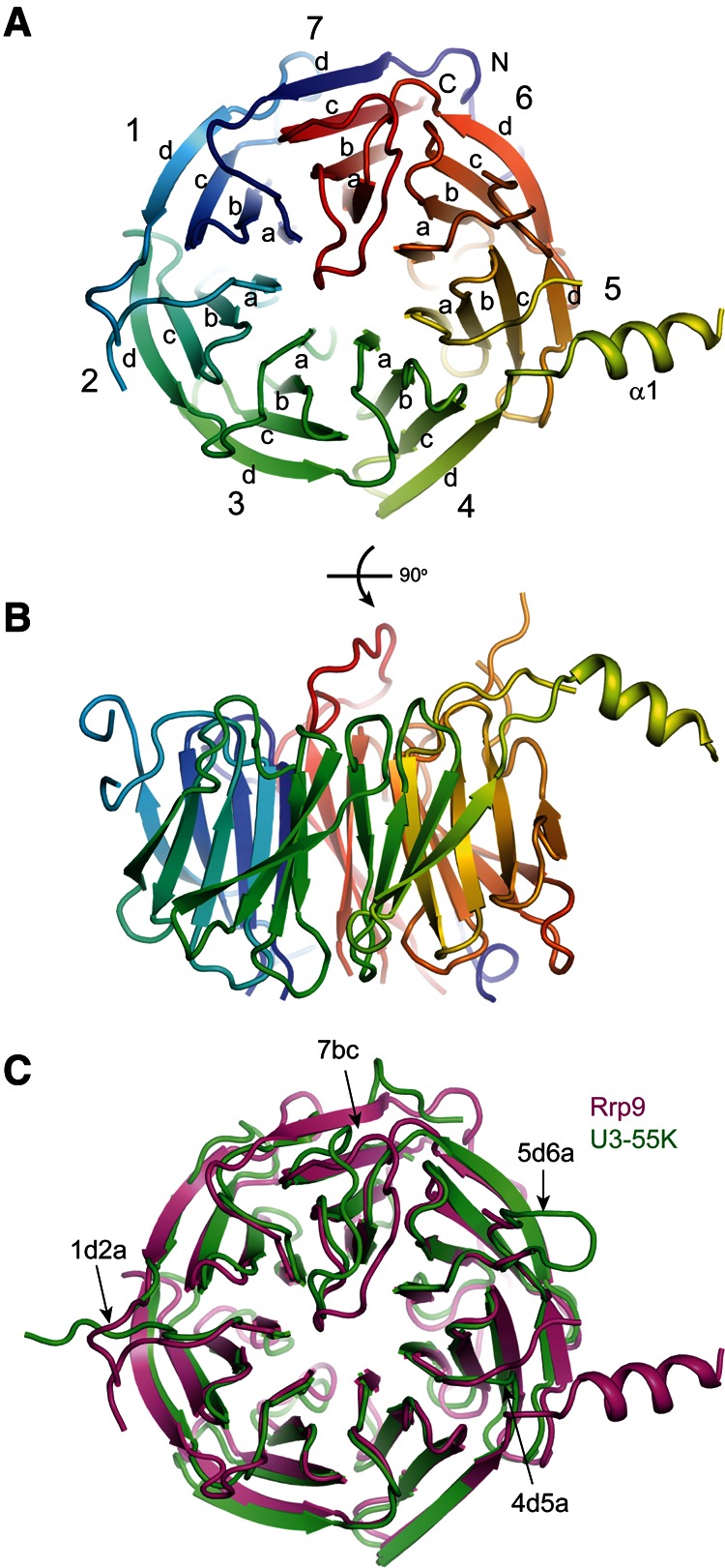
Crystal structures of the yeast Rrp9 and human U3-55K WD domains. (A,B) Ribbon representation of the yeast Rrp9 WD structure viewed from the top (A) and side (B). The polypeptide chain is colored blue to red from the N- to the C terminus. The seven β-sheet blades are numbered 1 to 7, and the four β-strands in each blade are labeled a to d in the outward direction. The N- and C termini are labeled. Each loop is named after the secondary structural elements it connects. (C) Structural alignment of the Rrp9 and U3-55K WD domains. Rrp9 is magenta and U3-55K is green. The long loops on the top are labeled.
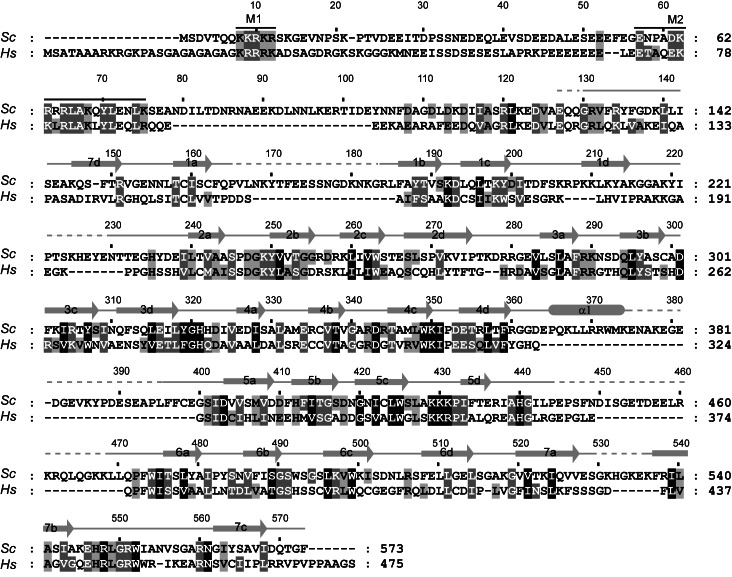
The structures of the Rrp9 and U3-55K WD domains are highly similar to each other and can be superimposed with a root mean square deviation (RMSD) of 1.105 Å over 270 Cα pairs. When compared to the U3-55K structure, Rrp9 contains an extra α helix that is inserted between strands 4d and 5a. Rrp9 is approximately 100 residues larger than U3-55K, which is mainly due to the presence of several elongated loops in the WD domain (1ab, 1d2a, 4d5a, 5d6a, and 7ab) (Figs. 1, 2C). These loops are not conserved and partially disordered in the Rrp9 structure.
FIGURE 1.
Sequence alignment of Rrp9 orthologs. A total of 131 sequences were aligned, and only the sequences of Saccharomyces cerevisiae (Sc) Rrp9 and Homo sapiens (Hs) U3-55K are shown. The residues that are conserved in 97%, 80%, and 60% of all aligned sequences are shaded in black, gray, and light gray, respectively. The secondary structures observed in the crystal structure of the Rrp9 WD domain are displayed on the top. The dashed lines denote disordered regions. Similarity groups are defined as follows: S and T, D and E, K and R, L, I, V, and M, and F, Y, and W. The two conserved nuclear localization signals, M1 and M2, are marked with lines.
The specific association of Rrp9 with the U3 B/C motif is enhanced by Snu13
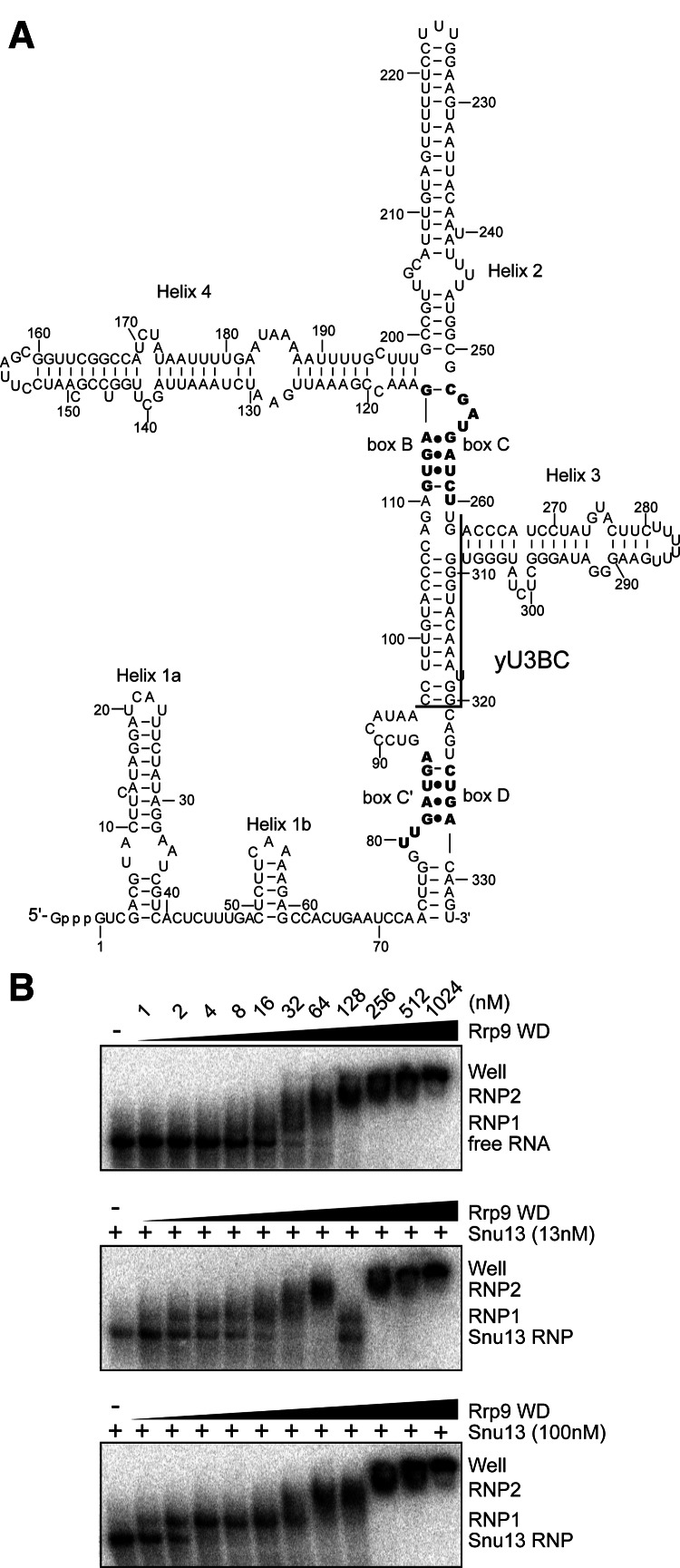
U3-55K is dependent upon 15.5K for binding to the B/C motif (Granneman et al. 2002). It is unclear whether the yeast Rrp9 protein has a similar requirement of Snu13 for U3 association. Using recombinant proteins, we analyzed binding of the Rrp9 WD domain with a fragment of yeast U3 RNA that spanned the B/C motif (yU3BC) (Fig. 3A) using electrophoretic mobility shift assays (EMSA).
FIGURE 3.
Binding of Rrp9 to the U3 B/C motif is enhanced by Snu13. (A) Secondary structural model of yeast U3 RNA. Boxes C′, B, C, and D are shown in bold. The boundaries of the yU3BC construct, which contains nucleotides 96–262 and 309–320, are marked. (B) EMSA of yU3BC RNA with Rrp9 WD domain. The 5′-32P-labeled RNA was incubated with 0, 13, or 100 nM of Snu13 and 0, 1, 2, 4, 8, 16, 32, 64, 128, 256, 512, and 1024 nM of the Rrp9 WD domain. The RNA species were resolved in native gels and visualized with phosphorimaging. The binding reactions contained 0.1 mg/mL yeast tRNA as a nonspecific competitor. Free RNA and various RNP species are labeled. RNP2 could include multiple poorly resolved species.
The WD domain of Rrp9 formed multiple RNP species with yU3BC in a protein concentration-dependent manner (Fig. 3B). A fast-migrating RNP1 species was transitively formed in the presence of 8–32 nM of Rrp9. Incubation with higher concentrations of Rrp9 led to the formation of slow-migrating RNPs and very large complexes stuck in the well. The binding of Rrp9 was also assayed in the presence of Snu13, which recognizes the K-turn structure formed by the B/C motif. The association of Snu13 alone caused little mobility shift of yU3BC. Preassembly of Snu13 with yU3BC significantly enhanced the formation of RNP1 but did not change the distribution patterns of other larger RNPs (Fig. 3B). This suggests that RNP1 represents the physiologically relevant complex in which Rrp9 recognizes the B/C motif. These larger RNPs appear to result from association of additional Rrp9 molecules at high protein concentrations and may not be functionally relevant. The dissociation constant (Kd) of RNP1 with respect to Rrp9 could be roughly estimated as the concentration of Rrp9 at which the fraction of free or Snu13-bound RNA equals that of RNP1. In this way, the Kd of RNP1 was estimated at 30 nM in the absence of Snu13, 4 nM in the presence of 13 nM Snu13, and 2 nM in the presence of 100 nM Snu13. The assembly of Rrp9 into RNP1 was also more cooperative when Snu13 had a higher concentration. These data indicate that binding of Rrp9 to the B/C motif was enhanced ∼10-fold by Snu13.
Structural features of Rrp9/U3-55K WD domain
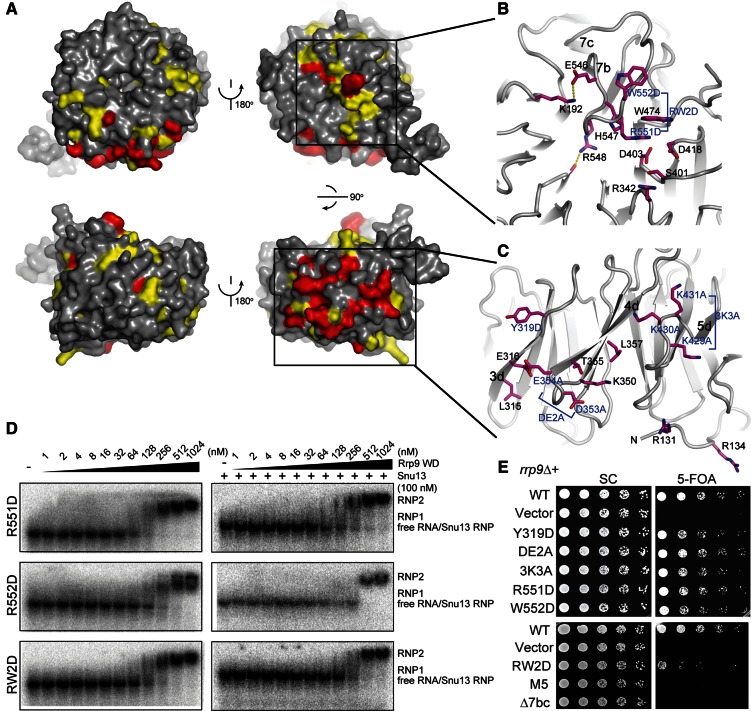
On the top face of the Rrp9/U3-55K WD domain, the 7bc loop that connects β-strands 7b and 7c is remarkably long and covers one end of the central channel (Fig. 4A,B). The highly conserved Arg548 residue in loop 7bc inserts into the central channel and forms a hydrogen bond with a carbonyl group lining the wall of the channel. Interestingly, the central channel of the WD protein WDR5 was shown previously to recognize an arginine of the histone H3 tail in a similar manner (Han et al. 2006). The central opening on the top face is a frequent protein-binding site of WD domains, but the 7bc loop seems to block Rrp9/U3-55K from making such an intermolecular protein interaction.
FIGURE 4.
Functional sites in the Rrp9 WD domain. (A) The conserved surface of the Rrp9 WD domain shown in four orientations. The residues with 97% and 80% conservation as defined in Figure 1 are colored red and yellow, respectively. (B,C) Close-up views of the top surface (B) and the side surface that spans blades 3 to 5 (C). The residues with at least 80% conservation are shown as sticks. Those residues analyzed by mutagenesis are indicated with blue labels. (D) EMSA of the yU3BC RNA with the mutant Rrp9 WD domains. The 5′-32P-labeled yU3BC RNA was incubated with the Rrp9 WD domain of indicated concentrations in the absence or presence of 100 nM Snu13. (E) Yeast growth assay of rrp9 mutants. The rrp9Δ strain complemented by wild-type (WT) RRP9 on a URA3 plasmid was transformed with a LEU2 vector encoding WT or mutant Rrp9. Fivefold serial dilutions of yeast cells were grown at 30°C on synthetic complete (SC) medium with or without 5-FOA, which counter-selects the URA3 plasmid carrying WT RRP9. DE2A is the D353A and E354A double mutant. 3K3A is the K429A, K430A, and K431A triple mutant. RW2D is the R515D and W552 double mutant. M5 is a combination of Y319D, ED2A, 3K3A, and RW2D.
The 7bc loop and nearby residues on the top face of the propeller are highly conserved, suggesting that this region is of functional importance. The C-terminal part of the 7bc loop, which is downstream from Arg548, adopts different conformations in the Rrp9 and U3-55K structures (Fig. 2C) and may undergo a conformational change when assuming different functional states. In addition to the top surface, analysis of the conserved residues reveals that the side of the WD propeller that spans blades 3, 4, and 5 is highly conserved (Fig. 4A,C).
The 7bc loop mediates U3 RNP assembly and is essential for yeast growth
The conserved surface residues of Rrp9 may participate in U3 RNP assembly. To test this possibility, we made mutations in four spatially separated surface patches on the WD domain and analyzed the effect on U3 RNA binding by EMSA (Fig. 4B,C). Specifically, on the side of blades 3–5, the Tyr319 residue in the 3d strand was mutated to Asp (Y319D), two acidic residues, i.e., Asp353 and Glu354, located in the 4cd loop were both mutated to Ala (DE2A), and the three consecutive lysine residues at positions 429–431 in the 5cd loop were all mutated to Ala (3K3A). On the top face, residues Arg551 and Trp552 in the 7bc loop were replaced individually by Asp (R551D, W552D) or in combination (RW2D). These mutations were incorporated into the Rrp9 WD domain. All of the mutant proteins displayed the same elution profile as wild-type proteins in size exclusion chromatography, suggesting that these mutations did not grossly disrupt the structure.
The mutant WD domains were assessed for yU3BC RNA binding in the absence and presence of Snu13 by EMSA. The Y319D, ED2A, and 3K3A mutations showed a minor effect on binding of either RNA alone or the yU3BC-Snu13 complex (data not shown). In contrast, when the 7bc loop contains mutations R551D, W552D, or their combination, only large RNPs were observed both in the absence and presence of Snu13 (Fig. 4D). These mutations appear to specifically impair the formation of RNP1 such that multiple Rrp9 molecules bind the RNA simultaneously at high protein concentrations. The results indicate that the 7bc loop is important for Rrp9 to specifically recognize the B/C motif.
To analyze the in vivo effect of Rrp9 mutations, we complemented the rrp9 knockout haploid with plasmid-encoded rrp9 mutants and assessed yeast growth (Fig. 4E). The rrp9Δ yeast was lethal as expected for RRP9, being an essential gene (Venema et al. 2000). Yeasts expressing Rrp9 mutants Y319D, DE2A, 3K3A, R551D, and W552D grew only slightly worse at 30°C than the yeast containing wild-type Rrp9. The growth phenotype of these mutants was similar at 20°C and 37°C (data not shown). The R551D/W552D double mutant resulted in a clear growth defect at 30°C (Fig. 4E) and was lethal at 37°C (data not shown). Moreover, cell growth was not supported upon removal of residues 547–552 in the 7bc loop (Δ7bc). These data indicate that the 7bc loop is essential for Rrp9 function, underscoring the importance of its role in U3 RNP assembly. Although mutations of Y319D, D353A/E354A, and K429A/K430A/K431A alone caused only a weak growth defect, a combination of them with R551D and W552D led to lethality (Fig. 4E, M5). Hence, at least one of these mutated residues is functionally important.
The N-terminal region of Rrp9 contains a bipartite nuclear localization signal
As a core component of U3 snoRNP, U3-55K/Rrp9 is localized in the nucleolus (Pluk et al. 1998; Huh et al. 2003). The WD domain of U3-55K is required for nucleolar localization of the protein (Pluk et al. 1998; Lukowiak et al. 2000). The U3-55K mutants with a disrupted WD domain failed to bind U3 but were still localized in the nucleus (Pluk et al. 1998; Lukowiak et al. 2000), suggesting that U3-55K has a NLS. The N-terminal region of U3-55K was predicted to contain a classic NLS, but the deletion of residues 1–44, which includes the predicted NLS, did not prevent the nucleolar localization of U3-55K (Pluk et al. 1998). One possible reason for this observation is that U3-55K contains multiple NLSs and was still imported in the absence of one NLS.
To clarify this problem, we analyzed the composition and function of NLS in Rrp9. Rrp9 and its variants were fused to the N terminus of green fluorescence protein (GFP) and expressed from the centromere plasmid pRS415 under the control of the native promoter in strain BY4741. The localization of GFP-fusions was examined by fluorescence imaging (Fig. 5A). The GFP protein alone, which contains no NLS, was distributed throughout the cell. The GFP fusion of wild-type Rrp9 was concentrated in crescent-shaped nucleoli and weakly distributed in the surrounding ellipse-shaped nuclei, as was expected (Pluk et al. 1998; Huh et al. 2003).
FIGURE 5.
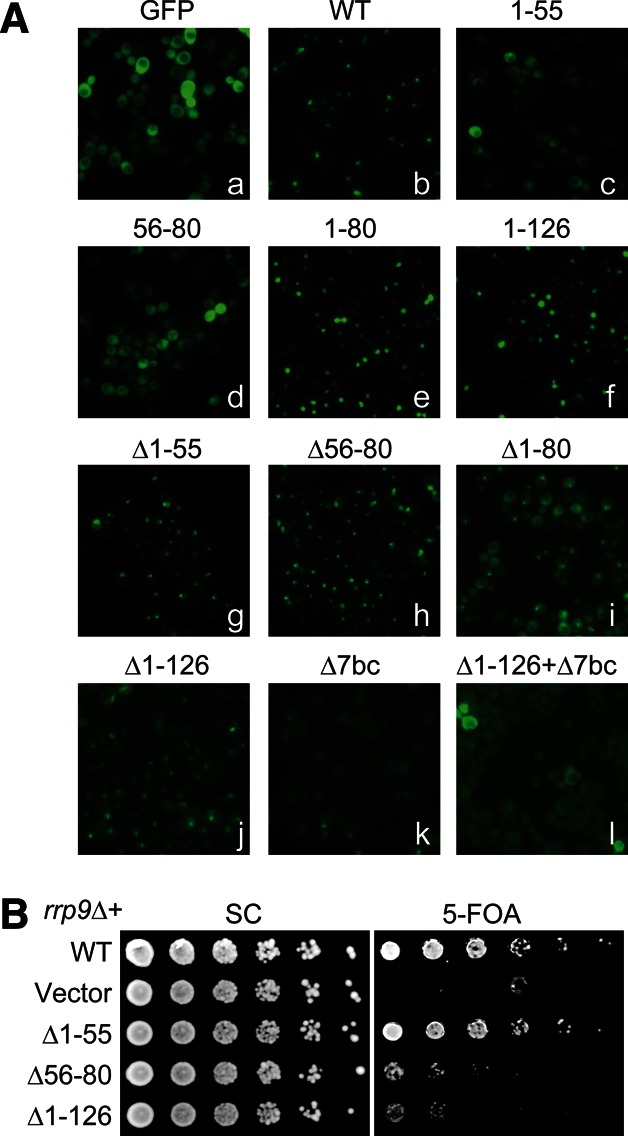
A bipartite nuclear localization signal at the N-terminal region of Rrp9. (A) Localization of Rrp9-GFP fusions. Fluorescence images were taken for GFP (a); GFP-fusion with wild-type Rrp9 (b); GFP-fusions with Rrp9 residues 1–55 (c), 56–80 (d), 1–80 (e), or 1–126 (f); or GFP-fusions with mutant Rrp9 that contains deletion (Δ) of residues 1–55 (g), 56–80 (h), 1–80 (i), 1–126 (j), the 7bc loop (residues 547–552) (k), or both residues 1–126 and the 7bc loop (l). These GFP fusion proteins were expressed under the endogenous promoter of Rrp9 from plasmid pRS415 in strain BY4741. (B) Yeast growth assay of rrp9 mutants. The rrp9Δ strain, complemented by wild-type (WT) RRP9 on a URA3 plasmid, was transformed with LEU2 vectors encoding WT RRP9 or the indicated mutants. Fivefold serial dilutions of yeast cells were grown at 30°C on synthetic complete (SC) medium with or without 5-FOA, which counter-selects the URA3 plasmid carrying WT RRP9.
The N-terminal 80 or 126 residues of Rrp9 were sufficient to drive GFP into the nucleus, indicating that this region contains a functional NLS. Multiple sequence alignment showed that the N-terminal region contains two conserved motifs, hereafter called M1 and M2 (Fig. 1). The M1 motif (Rrp9 residues 8–12), which corresponds to the previously predicted NLS (Pluk et al. 1998), is composed of a track of five basic residues and conforms to the classic monopartite NLS (Chook and Suel 2011). The M2 motif (Rrp9 residues 57–75) has a consensus sequence of E-T-A-X-[E/D]-[K/R]-[R/K]-L-R-L-A-K-X-Y-L-X-X-Ψ-[K/R], where X is any amino acid and Ψ is hydrophobic. The M2 motif is rich in basic residues but does not match a known NLS motif.
To dissect the composition of the NLS, residues 1–55 and 56–80 were fused separately to GFP and examined for localization in yeast. Fragment 1–55, which contains the M1 motif, was much less enriched in the nucleus compared with fragment 1–80. Fragment 56–80, which harbors the M2 motif, was evenly distributed in the cell, indicating that M2 is not an independent NLS. Our data indicate that the M1 motif is a weak NLS by itself and needs to function together with the M2 motif to constitute an efficient bipartite NLS.
The deletion of either the M1 or M2 motif (Δ1–55 and Δ56–80) caused a slight increase in the cytoplasmic localization of Rrp9-GFP but did not affect the nucleolar distribution much. When the entire NLS was removed (Δ1–126 and Δ1–80), the Rrp9 WD domain showed a clear distribution in the cytoplasm, indicating a defect in nuclear import. Nevertheless, a prominent fluorescence signal persisted in the nucleolus. This persistence indicates that the N-terminal bipartite NLS is not essential for nucleolar localization of Rrp9-GFP.
Deletion mutations in the U3-55K WD domain were previously shown to abolish the nucleolar localization of the protein (Pluk et al. 1998; Lukowiak et al. 2000). These deletion mutations most likely disrupted the WD domain structure, which raised the question as to whether U3 assembly or another function of the WD domain was required for nucleolar localization. To clarify this issue, we made the 7bc deletion mutation (Δ7bc) that would specifically disrupt the U3 assembly while minimally affecting the structure of the WD domain. The Δ7bc mutant of the full-length Rrp9-GFP fusion lost nucleolar enrichment, indicating that association of U3 was a prerequisite for Rrp9 nucleolar localization. The GFP fusion of Rrp9-Δ7bc still localized to the nucleus, due to the presence of the NLS; the deletion of both the NLS and the 7bc loop (Δ1–126 + Δ7bc) led to an even distribution in the cell.
We investigated the functional importance of the NSL by complementing the rrp9Δ strain with the N-terminal deletion mutants of Rrp9 (Fig. 5B). The Δ1–55 mutant, like wild-type RRP9, fully restored the growth of the rrp9Δ strain at 30°C, indicating that the M1 motif is dispensable. The growth of the Δ1–55 mutant yeast was not affected either at 20°C or 37°C (data not shown). In contrast, the Δ56–80 mutant only partially restored the growth of the rrp9Δ strain. The deletion of both the M1 and M2 motifs (Δ1–126) showed a similar growth defect as the Δ56–80 mutant. These data indicate that the M2 motif is more important in function than the M1 motif. Notably, the functional importance of the M2 motif is not correlated with its role in nuclear import. The isolated M1 motif, but not the M2 motif, shows intrinsic nuclear import activity. Deletion of the M1 and M2 motifs caused a similar minor effect on the nucleolar localization of Rrp9. One possible reason is that the M2 motif may have an additional function other than nuclear import that contributes to the observed phenotype.
DISCUSSION
We have determined the structure of the Rrp9 and U3-55K WD domains and analyzed functional sites in Rrp9. We show that the conserved 7bc loop on the top surface of the propeller is important for Rrp9 to recognize the B/C motif. In the absence of the 7bc loop, it is likely that Rrp9 cannot assemble into U3 snoRNP, causing the loss of nucleolar localization and lethality.
Our data show that binding of Rrp9 to the B/C motif was enhanced by prior binding of Snu13, which is consistent with the previous result that U3-55K depends on 15.5K to bind human U3 RNA (Granneman et al. 2002). Rrp9 may simultaneously recognize the B/C motif and Snu13 in the context of U3 snoRNP. Such a binding mode would be similar to that adopted by Nop5, which binds both L7Ae and the K-turn in the archaeal box C/D RNP (Ye et al. 2009; Xue et al. 2010; Lin et al. 2011). In support of this model, a weak direct interaction was found between U3-55K and 15.5K (Schultz et al. 2006). No interaction between Rrp9 and Snu13 was detected in our study (data not shown), but their interaction could occur in the assembled RNP, as is the case for Nop5 and L7Ae. Alternatively, binding of Snu13 may induce the RNA into a conformation that is specifically recognized by Rrp9. The exact structural mechanism by which Snu13 enhances Rrp9 binding of U3 requires further study.
In the core U3 snoRNP, the B/C motif associates with at least three proteins: Snu13, Rrp9, and Nop56. While the interaction of Snu13 with K-turn RNA has been well characterized structurally, it is not known how Rrp9 and Nop56 bind the B/C motif. Several previous experiments suggest that Rrp9/U3-55K is placed at the stem I side of the B/C motif. First, the association of U3 with U3-55K was affected by structural alternations of stem I (Granneman et al. 2002). Second, U3-55K binding was reduced by mutations in helix 4 of 15.5K (Schultz et al. 2006); this helix faces the stem I side in the K-turn complex structure of 15.5K (Vidovic et al. 2000). Last, Rrp9 is cross-linked to helices in the stem I region (Granneman et al. 2009). Nop56 would bind at the stem II side of B/C motif, if Nop56 resembles the homologous archaeal Nop5 in RNP assembly (Ye et al. 2009; Xue et al. 2010; Lin et al. 2011). Such an organization is compatible with Rrp9 bound at the other side. Nevertheless, a better understanding of U3 snoRNP organization would need further reconstitution and structural studies.
The 5′ domain of U3 has a well-established role in pairing with the pre-rRNA and orchestrating the assembly of the SSU processome, whereas the function of the U3 3′ domain is less clear. A study of human U3 snoRNA showed that the disruption of box C, hence the U3-55K binding, would abolish association of U3 with the SSU processome (Granneman et al. 2004). More recently, a region in the U3 3′ domain, which corresponds to the guide sequence in other box C/D snoRNAs, was shown to cross-link to 18S rRNA (Kudla et al. 2011). These data suggest that the 3′ domain of U3 has close interaction with the SSU processome. Consequently, some regions of Rrp9 could function in the context of the SSU processome, such as the conserved surface around blades 3–5 at the side of the Rrp9 WD domain, which has no detectable role in U3 RNP assembly, and the M2 motif in the N-terminal region, whose role in nuclear localization seems to not account for the observed deletion phenotype.
Using a GFP reporter, we have shown that the N-terminal region of Rrp9 contains a bipartite NLS that includes the previously predicted NLS, i.e., the M1 motif, and a novel M2 motif. Although lacking an intrinsic nuclear localization activity, the M2 motif enhances the otherwise weak localization activity of the M1 motif. The M2 motif may increase the binding affinity of the M1 NLS to importin.
We show that Rrp9 is able to localize to the nucleolus without the entire bipartite NLS. There must be alternative, although less efficient, routes to import Rrp9 into the nucleus. One possibility is that a small fraction of Rrp9 can passively diffuse into the nucleus through the nuclear pore complex. Once in the nucleoplasm, Rrp9 is assembled into U3 snoRNP (Watkins et al. 2004) and brought into the nucleolus, which, in turn, lowers the protein concentration in the nucleoplasm and drives further diffusion. Another possibility is that Rrp9 is imported via association with other proteins that have their own NLS. The U3 snoRNP is primarily assembled in the nucleoplasm, but whether it is ever exported to the cytoplasm is unclear (Watkins and Bohnsack 2011). The nuclear export factors CRM1 and PAXH have been shown to be associated with the U3 precursor snoRNP (Boulon et al. 2004; Watkins et al. 2004). A small fraction of the U3 precursor snoRNA was found in the cytoplasmic extract of HeLa cells (Watkins et al. 2007). If U3 has a cytoplasmic phase, there is a chance for Rrp9 to associate with U3 in the cytoplasm and be imported as part of pre-snoRNP. In addition, U3-55K was recently shown to interact with Nop58 and Nop56 (Knox et al. 2011). Both Nop56 and Nop58 contain nuclear and nucleolar localization signals at their C-terminal tail (Pradet-Balade et al. 2011) and may carry the associated U3-55K into the nucleus.
MATERIALS AND METHODS
Protein expression and purification
The genes of full-length Rrp9 and its WD domain (127–573) were PCR-amplified from yeast genomic DNA and cloned using the In-Fusion method (TaKaRa) into a modified pFastBac1 plasmid with an N-terminal six-His tag and PreScission cleavage site. The U3-55K gene was amplified from a human cDNA library and cloned in a similar way as Rrp9. The baculoviruses were prepared using the Bac-to-Bac system (Invitrogen), and the proteins were expressed in High Five cells following the manufacturer’s instructions.
The cells were harvested 48 h post-infection and lysed by sonication in buffer A (10 mM Tris-HCl, pH 8.0, 200 mM NaCl, and 5% glycerol). After centrifugation, the supernatant was applied to a HisTrap column (GE Healthcare). The column was washed with 30 mM imidazole in buffer A, and the bound protein was eluted with 500 mM imidazole in buffer A. The pooled sample was loaded onto a Q column, and the flowthrough was collected. After overnight digestion with PreScission at 4°C to remove the His-tag, the sample was loaded onto a heparin column and eluted with a 0–1 M NaCl gradient in 10 mM Tris-HCl, pH 8.0. The protein was further purified with a Superdex 200 column that was equilibrated in 10 mM Tris, pH 8.0 and 200 mM NaCl and was concentrated to 6 mg/mL using Millipore Ultrafree spin concentrators.
Seleno-methionine (SeMet) was incorporated into a mutant Rrp9 WD domain with Ile160 substituted by Met. The cells were initially grown in SF-900 II SFM medium. After virus infection for 8 h, the cells were spun down and resuspended in an equal volume of SF-900 II SFM methionine-free and cysteine-free medium with 200 mg/L L-cysteine. After 8 h of growth, SeMet was added to 250 mg/L, and the cells were harvested 37 h later. The SeMet-labeled protein was purified in the same way as the unlabeled protein.
Crystallization and structure determination
The native and SeMet-labeled I160M Rrp9 WD domains were crystallized at 4°C using the hanging drop vapor diffusion method by mixing 1 μL of recombinant protein (6 mg/mL in 10 mM Tris-HCl, pH 8.0 and 200 mM NaCl) with 1 μL of reservoir solution (100 mM HEPES-K, pH 7.5, 25% (v/v) ethylene glycol). The crystals grew as needle clusters within 3 d, and single crystals were obtained through gentle separation of the clusters. The crystals were transferred to a cryoprotectant solution containing four parts of the reservoir solution and one part of 100% glycerol and flash-frozen in liquid nitrogen. The crystals of the U3-55K WD domain were grown and cryoprotected in the same conditions as the Rrp9 crystals.
The diffraction data were collected at the Shanghai Synchrotron Radiation Facility at beamline BL17U and processed using HKL2000 (Otwinowski and Minor 1997). The Rrp9 crystal belongs to the P1 space group and contains two protomers in the asymmetric unit. The structure was first solved by molecular replacement in Phaser (McCoy et al. 2007), with the RACK1 structure (PDB ID 3FRX) as a search model (Coyle et al. 2009). The obtained RFZ score was 6.3 and 5.6 for the first and second copies of the model. However, the electron density map calculated from the MR phases was poor and precluded model building. The MR phases were used to locate selenium sites in a SeMet derivative crystal. The SAD phases were calculated and combined with the MR phases in SHARP (Vonrhein et al. 2007), yielding an interpretable electron density map. The model was built in Coot based on the MR model (Emsley and Cowtan 2004) and refined using Phenix and Refmac (Murshudov et al. 1999; Adams et al. 2010). The current model was refined to 2.5 Å with a maximum-likelihood-based coordinate error of 0.32 Å and contains two copies of the Rrp9 WD domain with residues 129 or 130 to 165, 184–222, 230–375, 396–443, 469–529, and 536–570 and 34 water molecules.
The U3-55K WD domain crystal belongs to the P43212 space group with one protomer in the asymmetric unit. The structure was solved by molecular replacement using the yeast Rrp9 WD structure as the search model in Phaser. The model was refined to 1.7 Å with a maximum-likelihood-based coordinate error of 0.069 Å and contained U3-55K residues 137–187 and 193–464, four residues from the vector (GPEA, as residues 133–136), and 216 water molecules.
RNA preparation and EMSA
The U3 snR17A gene was cloned from yeast genomic DNA into plasmid pBC-SK downstream from the T7 promoter, and the intron of U3 was removed by QuickChange. The yU3BC fragment containing residues 96–262 and 309–333 was generated. RNA was transcribed in vitro by T7 RNA polymerase using PCR products as a template and purified by denaturing polyacrylamide gel and electroelution. After dephosphorylation by calf intestinal phosphatase, 20 pmol of RNA was 5′-labeled with [γ-32P] ATP using T4 kinase. The 5′-labeled RNA (∼10 fmol) was incubated with 0, 13, or 100 nM of Snu13 and various concentrations of the Rrp9 WD domain in 20 μL of 25 mM HEPES-K, pH 7.3, 100 mM KCl, 2 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 0.1 mg/mL yeast tRNA, and 0.025% NP40. The binding reactions were incubated at 4°C for 10 min and separated with 5% native polyacrylamide gels in Tris-glycine (pH 8.3) buffer. The 32P-labeled RNA was visualized by phosphorimaging.
Yeast genetic experiments
The DNA sequences spanning the RRP9 gene, its upstream 340 nt, and its downstream 500 nt were cloned into the LEU2 plasmid pRS415 and the URA3 pRS416 plasmid (Brachmann et al. 1998). Point mutations and deletions were introduced into pRS415-RRP9 with the QuikChange method. A heterozygous RRP9/rrp9::kanMX4 diploid strain was purchased from Euroscarf and was subjected to tetrad dissection to construct a haploid rrp9Δ shuffle strain containing a pRS416-RRP9 plasmid as previously described (Li et al. 2011). The rrp9Δ shuffle strain was transformed with pRS416 plasmids encoding RRP9 or mutants and assayed for growth as previously described (Li et al. 2011). Yeast growth, transformations, and genetic manipulations were performed according to standard procedures. The generated plasmids, primers, and yeast strains are listed in Supplemental Tables 1–3.
GFP fluorescence imaging
An enhanced GFP gene was inserted downstream from the C terminus of RRP9 in pRS415-RRP9 by overlap PCR. The RRP9 gene in the resulting plasmid was mutated using QuikChange. Strain BY4741 was transformed with the Rrp9-GFP reporter construct and cultured to an OD of 0.8 in Leu-deficient synthetic defined complete medium. GFP-tagged fusion proteins were detected in vivo in a DeltaVision personal DV microscope. Images were acquired with a 100×, 1.4 NA objective, a GFP filter set (Chroma 89006 set), and a Photometrics CoolSNAP HQ2 camera. Four Z-sections at 0.5-μm intervals were merged into one image using the maximum intensity projection method with the softWoRx software.
DATA DEPOSITION
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession numbers 4J0X for the Rrp9 WD domain and 4J0W for the U3-55K WD domain.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank the staff at the Beijing Synchrotron Radiation Facility beamline BL17U1 for assistance in data collection. We thank Drs. Li-Lin Du and Jinqiu Zhou for help in yeast experiments. This research was supported by the National Basic Research Program of China (973 Program, 2010CB835402) and the Beijing Municipal Government.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aittaleb M, Rashid R, Chen Q, Palmer JR, Daniels CJ, Li H 2003. Structure and function of archaeal box C/D sRNP core proteins. Nat Struct Biol 10: 256–263 [DOI] [PubMed] [Google Scholar]
- Beltrame M, Tollervey D 1995. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J 14: 4350–4356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovjagin AV, Gerbi SA 2000. The spacing between functional Cis-elements of U3 snoRNA is critical for rRNA processing. J Mol Biol 300: 57–74 [DOI] [PubMed] [Google Scholar]
- Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, Ospina JK, Kiss T, Matera AG, Bordonne R, et al. 2004. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell 16: 777–787 [DOI] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 [DOI] [PubMed] [Google Scholar]
- Cahill NM, Friend K, Speckmann W, Li ZH, Terns RM, Terns MP, Steitz JA 2002. Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO J 21: 3816–3828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook YM, Suel KE 2011. Nuclear import by karyopherin-βs: Recognition and inhibition. Biochim Biophys Acta 1813: 1593–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clery A, Senty-Segault V, Leclerc F, Raue HA, Branlant C 2007. Analysis of sequence and structural features that identify the B/C motif of U3 small nucleolar RNA as the recognition site for the Snu13p-Rrp9p protein pair. Mol Cell Biol 27: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle SM, Gilbert WV, Doudna JA 2009. Direct link between RACK1 function and localization at the ribosome in vivo. Mol Cell Biol 29: 1626–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon F, Gallagher JE, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417: 967–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutca LM, Gallagher JE, Baserga SJ 2011. The initial U3 snoRNA:pre-rRNA base pairing interaction required for pre-18S rRNA folding revealed by in vivo chemical probing. Nucleic Acids Res 39: 5164–5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schafer T, Kuster B, Tschochner H, Tollervey D, et al. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol Cell 10: 105–115 [DOI] [PubMed] [Google Scholar]
- Granneman S, Pruijn GJ, Horstman W, van Venrooij WJ, Luhrmann R, Watkins NJ 2002. The hU3-55K protein requires 15.5K binding to the box B/C motif as well as flanking RNA elements for its association with the U3 small nucleolar RNA in vitro. J Biol Chem 277: 48490–48500 [DOI] [PubMed] [Google Scholar]
- Granneman S, Vogelzangs J, Luhrmann R, van Venrooij WJ, Pruijn GJ, Watkins NJ 2004. Role of pre-rRNA base pairing and 80S complex formation in subnucleolar localization of the U3 snoRNP. Mol Cell Biol 24: 8600–8610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Kudla G, Petfalski E, Tollervey D 2009. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc Natl Acad Sci 106: 9613–9618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Guo L, Wang H, Shen Y, Deng XW, Chai J 2006. Structural basis for the specific recognition of methylated histone H3 lysine 4 by the WD-40 protein WDR5. Mol Cell 22: 137–144 [DOI] [PubMed] [Google Scholar]
- Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y 2008. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65: 2334–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JM 1996. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J Mol Biol 259: 645–654 [DOI] [PubMed] [Google Scholar]
- Hughes JM, Ares M Jr 1991. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J 10: 4231–4239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Kass S, Tyc K, Steitz JA, Sollner-Webb B 1990. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell 60: 897–908 [DOI] [PubMed] [Google Scholar]
- Kiss T 2001. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J 20: 3617–3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Schmeing TM, Moore PB, Steitz TA 2001. The kink-turn: A new RNA secondary structure motif. EMBO J 20: 4214–4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox AA, McKeegan KS, Debieux CM, Traynor A, Richardson H, Watkins NJ 2011. A weak C′ box renders U3 snoRNA levels dependent on hU3-55K binding. Mol Cell Biol 31: 2404–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D, Hurt E, Bassler J 2010. Driving ribosome assembly. Biochim Biophys Acta 1803: 673–683 [DOI] [PubMed] [Google Scholar]
- Kudla G, Granneman S, Hahn D, Beggs JD, Tollervey D 2011. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc Natl Acad Sci 108: 10010–10015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Duan J, Li D, Ma S, Ye K 2011. Structure of the Shq1-Cbf5-Nop10-Gar1 complex and implications for H/ACA RNP biogenesis and dyskeratosis congenita. EMBO J 30: 5010–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Lai S, Jia R, Xu A, Zhang L, Lu J, Ye K 2011. Structural basis for site-specific ribose methylation by box C/D RNA protein complexes. Nature 469: 559–563 [DOI] [PubMed] [Google Scholar]
- Lubben B, Marshallsay C, Rottmann N, Luhrmann R 1993. Isolation of U3 snoRNP from CHO cells: A novel 55 kDa protein binds to the central part of U3 snoRNA. Nucleic Acids Res 21: 5377–5385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowiak AA, Granneman S, Mattox SA, Speckmann WA, Jones K, Pluk H, Venrooij WJ, Terns RM, Terns MP 2000. Interaction of the U3-55k protein with U3 snoRNA is mediated by the box B/C motif of U3 and the WD repeats of U3-55k. Nucleic Acids Res 28: 3462–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmier-Gourrier N, Clery A, Schlotter F, Senty-Segault V, Branlant C 2011. A second base pair interaction between U3 small nucleolar RNA and the 5′-ETS region is required for early cleavage of the yeast pre-ribosomal RNA. Nucleic Acids Res 39: 9731–9745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ 1999. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr 55: 247–255 [DOI] [PubMed] [Google Scholar]
- Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL 2004. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell 16: 943–954 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Phipps KR, Charette JM, Baserga SJ 2011. The SSU processome in ribosome biogenesis—progress and prospects. WIREs RNA 2: 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluk H, Soffner J, Luhrmann R, van Venrooij WJ 1998. cDNA cloning and characterization of the human U3 small nucleolar ribonucleoprotein complex-associated 55-kilodalton protein. Mol Cell Biol 18: 488–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradet-Balade B, Girard C, Boulon S, Paul C, Azzag K, Bordonne R, Bertrand E, Verheggen C 2011. CRM1 controls the composition of nucleoplasmic pre-snoRNA complexes to licence them for nucleolar transport. EMBO J 30: 2205–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarsky DA, Fournier MJ 1998. Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol Cell Biol 18: 3431–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savino R, Gerbi SA 1990. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J 9: 2299–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz A, Nottrott S, Watkins NJ, Luhrmann R 2006. Protein-protein and protein-RNA contacts both contribute to the 15.5K-mediated assembly of the U4/U6 snRNP and the box C/D snoRNPs. Mol Cell Biol 26: 5146–5154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Tollervey D 1999. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol Cell Biol 19: 6012–6019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nues RW, Granneman S, Kudla G, Sloan KE, Chicken M, Tollervey D, Watkins NJ 2011. Box C/D snoRNP catalysed methylation is aided by additional pre-rRNA base-pairing. EMBO J 30: 2420–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema J, Tollervey D 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet 33: 261–311 [DOI] [PubMed] [Google Scholar]
- Venema J, Vos HR, Faber AW, van Venrooij WJ, Raue HA 2000. Yeast Rrp9p is an evolutionarily conserved U3 snoRNP protein essential for early pre-rRNA processing cleavages and requires box C for its association. RNA 6: 1660–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidovic I, Nottrott S, Hartmuth K, Luhrmann R, Ficner R 2000. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol Cell 6: 1331–1342 [DOI] [PubMed] [Google Scholar]
- Vonrhein C, Blanc E, Roversi P, Bricogne G 2007. Automated structure solution with autoSHARP. Methods Mol Biol 364: 215–230 [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Bohnsack MT 2011. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 3: 397–414 [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Segault V, Charpentier B, Nottrott S, Fabrizio P, Bachi A, Wilm M, Rosbash M, Branlant C, Luhrmann R 2000. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103: 457–466 [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Dickmanns A, Luhrmann R 2002. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol Cell Biol 22: 8342–8352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, Luhrmann R 2004. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell 16: 789–798 [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Lemm I, Luhrmann R 2007. Involvement of nuclear import and export factors in U8 box C/D snoRNP biogenesis. Mol Cell Biol 27: 7018–7027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue S, Wang R, Yang F, Terns RM, Terns MP, Zhang X, Maxwell ES, Li H 2010. Structural basis for substrate placement by an archaeal box C/D ribonucleoprotein particle. Mol Cell 39: 939–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Wu YJ, Zhu M, Fan SB, Lin J, Zhang K, Li S, Chi H, Li YX, Chen HF, et al. 2012. Identification of cross-linked peptides from complex samples. Nat Methods 9: 904–906 [DOI] [PubMed] [Google Scholar]
- Ye K, Jia R, Lin J, Ju M, Peng J, Xu A, Zhang L 2009. Structural organization of box C/D RNA-guided RNA methyltransferase. Proc Natl Acad Sci 106: 13808–13813 [DOI] [PMC free article] [PubMed] [Google Scholar]