Abstract
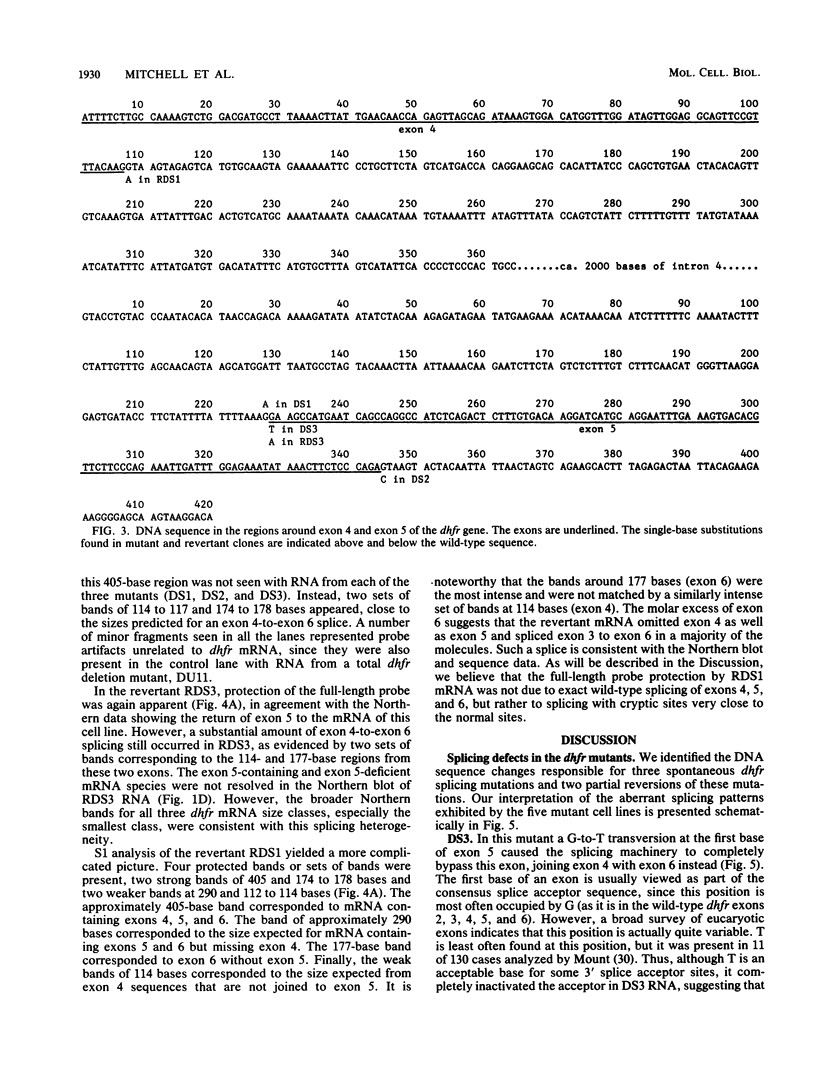
We isolated and characterized three spontaneous mutants of Chinese hamster ovary cells that were deficient in dihydrofolate reductase activity. All three mutants contained no detectable enzyme activity and produced dihydrofolate reductase mRNA species that were shorter than those of the wild type by about 120 bases. Six exons are normally represented in this mRNA; exon 5 was missing in all three mutant mRNAs. Nuclease S1 analysis of the three mutants indicated that during the processing of the mutant RNA, exon 4 was spliced to exon 6. The three mutant genes were cloned, and the regions around exons 4 and 5 were sequenced. In one mutant, the GT dinucleotide at the 5' end of intron 5 had changed to CT. In a second mutant, the first base in exon 5 had changed from G to T. In a revertant of this mutant, this base was further mutated to A, a return to a purine. Approximately 25% of the mRNA molecules in the revertant were spliced correctly to produce an enzyme with one presumed amino acid change. In the third mutant, the AG at the 3' end of intron 4 had changed to AA. A mutation that partially reversed the mutant phenotype had changed the dinucleotide at the 5' end of intron 4 from GT to AT. The splicing pattern in this revertant was consistent with the use of cryptic donor and acceptor splice sites close to the original sites to produce an mRNA with three base changes and a protein with two amino acid changes. These mutations argue against a scanning model for the selection of splice site pairs and suggest that only a single splice site need be inactivated to bring about efficient exon skipping (a regulatory mechanism for some genes). The fact that all three mutants analyzed exhibited exon 5 splicing mutations indicates that these splice sites are hot spots for spontaneous mutation.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adrian G. S., Wiginton D. A., Hutton J. J. Structure of adenosine deaminase mRNAs from normal and adenosine deaminase-deficient human cell lines. Mol Cell Biol. 1984 Sep;4(9):1712–1717. doi: 10.1128/mcb.4.9.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton W. D., Davis R. W. Screening lambdagt recombinant clones by hybridization to single plaques in situ. Science. 1977 Apr 8;196(4286):180–182. doi: 10.1126/science.322279. [DOI] [PubMed] [Google Scholar]
- Black D. L., Chabot B., Steitz J. A. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985 Oct;42(3):737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- Brandt C. R., Morrison S. L., Birshtein B. K., Milcarek C. Loss of a consensus splice signal in a mutant immunoglobulin gene eliminates the CH1 domain exon from the mRNA. Mol Cell Biol. 1984 Jul;4(7):1270–1277. doi: 10.1128/mcb.4.7.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Benoist C., O'Hare K., Gannon F., Chambon P. Ovalbumin gene: evidence for a leader sequence in mRNA and DNA sequences at the exon-intron boundaries. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4853–4857. doi: 10.1073/pnas.75.10.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart R. E., Nguyen H. T., Medford R. M., Destree A. T., Mahdavi V., Nadal-Ginard B. Intricate combinatorial patterns of exon splicing generate multiple regulated troponin T isoforms from a single gene. Cell. 1985 May;41(1):67–82. doi: 10.1016/0092-8674(85)90062-5. [DOI] [PubMed] [Google Scholar]
- Brody E., Abelson J. The "spliceosome": yeast pre-messenger RNA associates with a 40S complex in a splicing-dependent reaction. Science. 1985 May 24;228(4702):963–967. doi: 10.1126/science.3890181. [DOI] [PubMed] [Google Scholar]
- Capizzi R. L., Jameson J. W. A table for the estimation of the spontaneous mutation rate of cells in culture. Mutat Res. 1973 Jan;17(1):147–148. doi: 10.1016/0027-5107(73)90265-0. [DOI] [PubMed] [Google Scholar]
- Carothers A. M., Urlaub G., Ellis N., Chasin L. A. Structure of the dihydrofolate reductase gene in Chinese hamster ovary cells. Nucleic Acids Res. 1983 Apr 11;11(7):1997–2012. doi: 10.1093/nar/11.7.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi E., Kuehl M., Wall R. RNA splicing generates a variant light chain from an aberrantly rearranged kappa gene. Nature. 1980 Aug 21;286(5775):776–779. doi: 10.1038/286776a0. [DOI] [PubMed] [Google Scholar]
- Chu G., Sharp P. A. A gene chimaera of SV40 and mouse beta-globin is transcribed and properly spliced. Nature. 1981 Jan 29;289(5796):378–382. doi: 10.1038/289378a0. [DOI] [PubMed] [Google Scholar]
- Collins F. S., Weissman S. M. The molecular genetics of human hemoglobin. Prog Nucleic Acid Res Mol Biol. 1984;31:315–462. doi: 10.1016/s0079-6603(08)60382-7. [DOI] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H., Farabaugh P. J., Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978 Aug 24;274(5673):775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Crouse G. F., Simonsen C. C., McEwan R. N., Schimke R. T. Structure of amplified normal and variant dihydrofolate reductase genes in mouse sarcoma S180 cells. J Biol Chem. 1982 Jul 10;257(13):7887–7897. [PubMed] [Google Scholar]
- Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., Hood L. Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell. 1980 Jun;20(2):313–319. doi: 10.1016/0092-8674(80)90617-0. [DOI] [PubMed] [Google Scholar]
- Fradin A., Jove R., Hemenway C., Keiser H. D., Manley J. L., Prives C. Splicing pathways of SV40 mRNAs in X. laevis oocytes differ in their requirements for snRNPs. Cell. 1984 Jul;37(3):927–936. doi: 10.1016/0092-8674(84)90427-6. [DOI] [PubMed] [Google Scholar]
- Frendewey D., Keller W. Stepwise assembly of a pre-mRNA splicing complex requires U-snRNPs and specific intron sequences. Cell. 1985 Aug;42(1):355–367. doi: 10.1016/s0092-8674(85)80131-8. [DOI] [PubMed] [Google Scholar]
- Grabowski P. J., Seiler S. R., Sharp P. A. A multicomponent complex is involved in the splicing of messenger RNA precursors. Cell. 1985 Aug;42(1):345–353. doi: 10.1016/s0092-8674(85)80130-6. [DOI] [PubMed] [Google Scholar]
- Krangel M. S. Unusual RNA splicing generates a secreted form of HLA-A2 in a mutagenized B lymphoblastoid cell line. EMBO J. 1985 May;4(5):1205–1210. doi: 10.1002/j.1460-2075.1985.tb03761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A., Keller W., Appel B., Lührmann R. The 5' terminus of the RNA moiety of U1 small nuclear ribonucleoprotein particles is required for the splicing of messenger RNA precursors. Cell. 1984 Aug;38(1):299–307. doi: 10.1016/0092-8674(84)90551-8. [DOI] [PubMed] [Google Scholar]
- Kühne T., Wieringa B., Reiser J., Weissmann C. Evidence against a scanning model of RNA splicing. EMBO J. 1983;2(5):727–733. doi: 10.1002/j.1460-2075.1983.tb01492.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang K. M., Spritz R. A. RNA splice site selection: evidence for a 5' leads to 3' scanning model. Science. 1983 Jun 24;220(4604):1351–1355. doi: 10.1126/science.6304877. [DOI] [PubMed] [Google Scholar]
- Lazar E., Jacob M., Krol A., Branlant C. Accessibility of U1 RNA to base pairing with a single-stranded DNA fragment mimicking the intron extremities at the splice junction. Nucleic Acids Res. 1982 Feb 25;10(4):1193–1201. doi: 10.1093/nar/10.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melera P. W., Davide J. P., Hession C. A., Scotto K. W. Phenotypic expression in Escherichia coli and nucleotide sequence of two Chinese hamster lung cell cDNAs encoding different dihydrofolate reductases. Mol Cell Biol. 1984 Jan;4(1):38–48. doi: 10.1128/mcb.4.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melera P. W., Hession C. A., Davide J. P., Scotto K. W., Biedler J. L., Meyérs M. B., Shanske S. Antifolate-resistant Chinese Hamster Cells. mRNA directed overproduction of multiple dihydrofolate reductases from a series of independently derived sublines containing amplified dihydrofolate reductase genes. J Biol Chem. 1982 Nov 10;257(21):12939–12949. [PubMed] [Google Scholar]
- Milbrandt J. D., Azizkhan J. C., Greisen K. S., Hamlin J. L. Organization of a Chinese hamster ovary dihydrofolate reductase gene identified by phenotypic rescue. Mol Cell Biol. 1983 Jul;3(7):1266–1273. doi: 10.1128/mcb.3.7.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Carothers A. M., Han J. H., Harding J. D., Kas E., Venolia L., Chasin L. A. Multiple transcription start sites, DNase I-hypersensitive sites, and an opposite-strand exon in the 5' region of the CHO dhfr gene. Mol Cell Biol. 1986 Feb;6(2):425–440. doi: 10.1128/mcb.6.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M. A catalogue of splice junction sequences. Nucleic Acids Res. 1982 Jan 22;10(2):459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima Y., Fujii-Kuriyama Y., Muramatsu M., Ogata K. Alternative transcription and two modes of splicing results in two myosin light chains from one gene. Nature. 1984 Mar 22;308(5957):333–338. doi: 10.1038/308333a0. [DOI] [PubMed] [Google Scholar]
- Orkin S. H., Goff S. C., Kelley W. N., Daddona P. E. Transient expression of human adenosine deaminase cDNAs: identification of a nonfunctional clone resulting from a single amino acid substitution. Mol Cell Biol. 1985 Apr;5(4):762–767. doi: 10.1128/mcb.5.4.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin S. H., Kazazian H. H., Jr The mutation and polymorphism of the human beta-globin gene and its surrounding DNA. Annu Rev Genet. 1984;18:131–171. doi: 10.1146/annurev.ge.18.120184.001023. [DOI] [PubMed] [Google Scholar]
- Orlofsky A., Chasin L. A. A domain of methylation change at the albumin locus in rat hepatoma cell variants. Mol Cell Biol. 1985 Jan;5(1):214–225. doi: 10.1128/mcb.5.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett R. A., Konarska M. M., Grabowski P. J., Hardy S. F., Sharp P. A. Lariat RNA's as intermediates and products in the splicing of messenger RNA precursors. Science. 1984 Aug 31;225(4665):898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- Padgett R. A., Mount S. M., Steitz J. A., Sharp P. A. Splicing of messenger RNA precursors is inhibited by antisera to small nuclear ribonucleoprotein. Cell. 1983 Nov;35(1):101–107. doi: 10.1016/0092-8674(83)90212-x. [DOI] [PubMed] [Google Scholar]
- Pikielny C. W., Rosbash M. mRNA splicing efficiency in yeast and the contribution of nonconserved sequences. Cell. 1985 May;41(1):119–126. doi: 10.1016/0092-8674(85)90066-2. [DOI] [PubMed] [Google Scholar]
- Rautmann G., Matthes H. W., Gait M. J., Breathnach R. Synthetic donor and acceptor splice sites function in an RNA polymerase B (II) transcription unit. EMBO J. 1984 Sep;3(9):2021–2028. doi: 10.1002/j.1460-2075.1984.tb02085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R., Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985 May;41(1):95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- Rosenfeld M. G., Amara S. G., Evans R. M. Alternative RNA processing: determining neuronal phenotype. Science. 1984 Sep 21;225(4668):1315–1320. doi: 10.1126/science.6089345. [DOI] [PubMed] [Google Scholar]
- Ruskin B., Krainer A. R., Maniatis T., Green M. R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984 Aug;38(1):317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman J. G., Leder P. A mutant immunoglobulin light chain is formed by aberrant DNA- and RNA-splicing events. Nature. 1980 Aug 21;286(5775):779–783. doi: 10.1038/286779a0. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Su T. S., Beaudet A. L., O'Brien W. E. Abnormal mRNA for argininosuccinate synthetase in citrullinaemia. Nature. 1983 Feb 10;301(5900):533–534. doi: 10.1038/301533a0. [DOI] [PubMed] [Google Scholar]
- Urlaub G., Carothers A. M., Chasin L. A. Efficient cloning of single-copy genes using specialized cosmid vectors: isolation of mutant dihydrofolate reductase genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1189–1193. doi: 10.1073/pnas.82.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub G., Chasin L. A. Isolation of Chinese hamster cell mutants deficient in dihydrofolate reductase activity. Proc Natl Acad Sci U S A. 1980 Jul;77(7):4216–4220. doi: 10.1073/pnas.77.7.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urlaub G., Käs E., Carothers A. M., Chasin L. A. Deletion of the diploid dihydrofolate reductase locus from cultured mammalian cells. Cell. 1983 Jun;33(2):405–412. doi: 10.1016/0092-8674(83)90422-1. [DOI] [PubMed] [Google Scholar]
- Urlaub G., Landzberg M., Chasin L. A. Selective killing of methotrexate-resistant cells carrying amplified dihydrofolate reductase genes. Cancer Res. 1981 May;41(5):1594–1601. [PubMed] [Google Scholar]
- Wieringa B., Hofer E., Weissmann C. A minimal intron length but no specific internal sequence is required for splicing the large rabbit beta-globin intron. Cell. 1984 Jul;37(3):915–925. doi: 10.1016/0092-8674(84)90426-4. [DOI] [PubMed] [Google Scholar]
- Wieringa B., Meyer F., Reiser J., Weissmann C. Unusual splice sites revealed by mutagenic inactivation of an authentic splice site of the rabbit beta-globin gene. Nature. 1983 Jan 6;301(5895):38–43. doi: 10.1038/301038a0. [DOI] [PubMed] [Google Scholar]
- Yang J. K., Masters J. N., Attardi G. Human dihydrofolate reductase gene organization. Extensive conservation of the G + C-rich 5' non-coding sequence and strong intron size divergence from homologous mammalian genes. J Mol Biol. 1984 Jun 25;176(2):169–187. doi: 10.1016/0022-2836(84)90419-4. [DOI] [PubMed] [Google Scholar]
- Zeitlin S., Efstratiadis A. In vivo splicing products of the rabbit beta-globin pre-mRNA. Cell. 1984 Dec;39(3 Pt 2):589–602. doi: 10.1016/0092-8674(84)90466-5. [DOI] [PubMed] [Google Scholar]