Abstract
Recessive dystrophic epidermolysis bullosa (RDEB) is characterized by a functional deficit of type VII collagen protein due to gene defects in the type VII collagen gene (COL7A1). Gene augmentation therapies are promising, but run the risk of insertional mutagenesis. To abrogate this risk, we explored the possibility of using engineered transcription activator-like effector nucleases (TALEN) for precise genome editing. We report the ability of TALEN to induce site-specific double-stranded DNA breaks (DSBs) leading to homology-directed repair (HDR) from an exogenous donor template. This process resulted in COL7A1 gene mutation correction in primary fibroblasts that were subsequently reprogrammed into inducible pluripotent stem cells and showed normal protein expression and deposition in a teratoma-based skin model in vivo. Deep sequencing-based genome-wide screening established a safety profile showing on-target activity and three off-target (OT) loci that, importantly, were at least 10 kb from a coding sequence. This study provides proof-of-concept for TALEN-mediated in situ correction of an endogenous patient-specific gene mutation and used an unbiased screen for comprehensive TALEN target mapping that will cooperatively facilitate translational application.
Introduction
Monogenic disorders, characterized by small alterations to the genetic code, are the direct cause of hundreds of disorders afflicting millions of patients. For some of these disorders, restoration of the missing gene product can be achieved by solid organ or hematopoietic cell transplantation that can provide a therapeutic effect. These therapies are hindered by the paucity of donors, as well as by pre- and post-transplant-related complications that can cause morbidity and mortality. In many instances, this class of disease is also amenable to gene therapy whereby a functional copy of the missing gene is provided to the cell. This strategy also makes possible the ability to functionally correct a patient's own cells and deliver them back to them, thus minimizing allogeneic transplant-associated morbidities. However, gene therapy vehicles capable of mediating sustained transgene expression can be associated with significant risks that are derived from the properties of the specific platform. Viral vectors possess broad tropism; however, integrating vectors tend to insert at or near areas of transcriptional activity that can result in adverse events due to insertional mutagenesis.1,2 Non-integrating vectors largely avoid insertional mutagenic concerns due to their episomal nature; however, they may be limited in their use to organs/cells that divide at low rates.3,4 Similar to integrating viral vectors, non-viral integrating (e.g., transposon based) platforms can perturb the genomic sequence by virtue of the semirandomness of their integration pattern.5 In each instance, the expression cassette includes exogenous regulatory elements that mediate sustained transgene expression that is not subject to normal cellular regulatory control. Taken together, these drawbacks make being able to precisely target genes in a patient's own cells a highly desirable strategy that maximizes safety and efficacy.
Such a strategy is incumbent on the use of the cellular homologous recombination machinery that, at endogenous levels, is sufficiently low to preclude robust gene targeting.6 To overcome this hurdle, engineered nucleases specific for genomic targets have been used to generate a double-stranded DNA break (DSB) that increases the rate and efficiency of homologous recombination.7 Major classes of reagents capable of accomplishing this are the meganucleases, zinc finger nucleases (ZFN), clustered regularly interspaced short palindromic repeats (CRISPR), and transcription activator-like effector nucleases (TALEN). Meganucleases bind relatively large sequences of DNA and can be targeted to specific sequences by a complex engineering process that may be limiting to unspecialized laboratories.8,9 ZFNs are hybrid proteins comprised of a DNA-binding element that confers target specificity and a nuclease component that mediates DSBs. ZFNs can be easily assembled as modules; however, because the individual subunits influence the overall binding affinity of the reagent in a context-dependent manner, this iteration of ZFNs can be suboptimal in rates of gene targeting.10 ZFNs constructed in a context-dependent manner show higher rates of activity; however, the generation procedure requires the acquisition of specialized starting materials and experimental procedures.11,12 Furthermore, the targeting frequency of ZFNs is ~1 target site per 500 bp of DNA, which may make portions of the genome refractory to ZFN targeting.11,13 The recently described CRISPR/Cas9 system relies on a guide RNA to recruit the Cas9 helicase/nuclease to the target site and is available in a simplified generation protocol amenable for widespread use.14,15,16 However, the restricted targeting profile whereby the target site must contain a G(N)20GG motif may make aspects of the genome inaccessible to targeting.14,15 TALENs can be assembled rapidly from freely available modules, and the targeting capacity is such that a TALEN binding site exists, on average, every 35 bp of DNA.17,18 Because of their high rates of activity, expansive targeting repertoire, and ease of generation, we sought to determine the efficacy of TALENs using recessive dystrophic epidermolysis bullosa (RDEB) as a model disease.
RDEB is caused by mutations to the COL7A1 gene located on chromosome three. A lack of type VII collagen protein at the dermal-epidermal junction (DEJ) results in a loss of structural integrity of the skin. Patients with RDEB exhibit incurable, often fatal, skin blistering, and have an increased risk for aggressive squamous cell carcinoma.19 Restoration of deposition of the type VII collagen at the DEJ by allogeneic systemic hematopoietic cell or localized fibroblast transplantation can alleviate symptoms.20,21,22 However, suboptimal efficacy of allogeneic cell transplantation due to graft failure provides an impetus to develop new autologous-based therapies.20,21 Therefore, we used TALENs in a genome-editing strategy whereby we targeted a specific patient mutation in the COL7A1 gene for correction. Using primary, patient-derived fibroblasts, we were able to isolate gene-corrected clones of cells that could be expanded to clinically meaningful numbers. These data support the usefulness of TALENs for fibroblast correction that would be amenable for localized therapy. To expand the potential therapeutic impact of these cells, we differentiated the corrected fibroblasts into inducible pluripotent stem cells (iPSCs) and showed their ability to form skin-like structures in vivo that expressed the corrected type VII collagen protein at the DEJ. We also used an ultrasensitive deep sequencing/gene tagging methodology to, for the first time, establish an unbiased genome-wide screen to identify TALEN on- and off-target (OT) sites. In totality, these data show the potential of TALENs for personalized genomic-based medicine approaches.
Results
COL7A1 TALEN gene targeting, construction, and validation
Using the TAL Effector-Nucleotide Targeter software,23 we identified >50 potential TALEN sites (data not shown) for the human COL7A1 locus within 200 bp of a homozygous premature termination codon (PTC) causative of RDEB (Figure 1a). These in silico data and recent experimental data on a large series of human genes24 emphasize the high theoretical targeting capacity for TALENs, an important consideration for RDEB and other diseases that exhibit heterogeneity in the location and number of mutated sequences.25
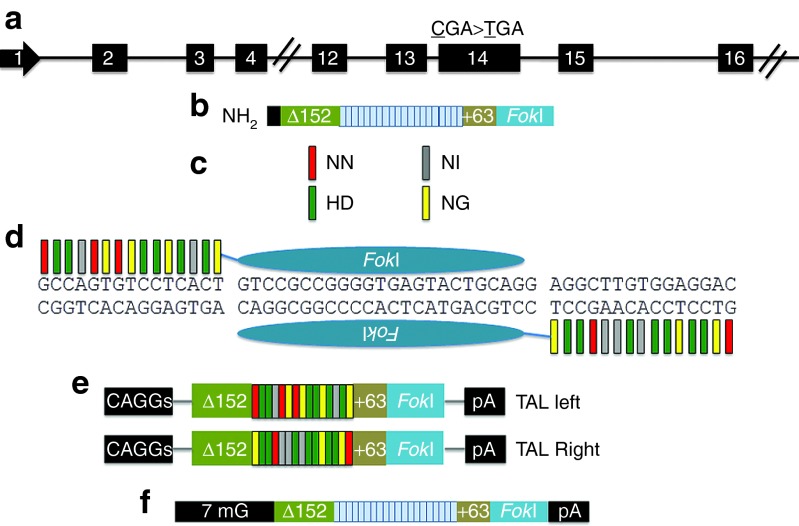
Figure 1.
TALEN targeting, nuclease architecture, and modification of COL7A1 gene. (a) COL7A1 TALEN target site in exon 14 of the COL7A1 gene on chromosome 3. The mutation at 1837 (CGA>TGA) is shown. (b) Core constituents of the nuclease complex: an N-terminal deletion of 152 residues of Xanthomonas TALEs, followed by the repeat domain, and a +63 C-terminal subregion fused to the catalytic domain of the FokI nuclease. (c) RVD base recognition. The RVDs NN, NI, HD, and NG bind guanine, adenine, cytosine, and thymine, respectively, and are color coded. (d) TALEN array binding to the COL7A1 gene target sequence. Colored bars represent the target-specific RVDs and correspond to the RVD:DNA code in c. TALEN expression platforms. (e) DNA plasmids with TALEN arrays in trans with gene expression controlled by the mini-CAGGs promoter and the bovine growth hormone polyadenylation signal. (f) mRNA species shown with TALEN architecture, 5′-7-methylguanosine cap (7 mG) and polyadenylation signal. RVD, repeat variable diresidue; TALEN, transcription activator-like effector nucleases.
A TALEN is composed of an engineered TALE repeat array fused to the FokI nuclease domain (Figure 1b). The repeat unit contains 33–35 amino acids that are constant save for two hypervariable residues (repeat variable diresidues) located at positions 12 and 13.18,26 These repeat variable diresidues dictate protein:DNA interactions such that the repeat variable diresidues NN, NI, HD, and NG recognize the DNA bases guanine, adenine, cytosine, and thymine, respectively (Figure 1c).26,27 The Golden Gate cloning methodology18 was used to generate a patient-specific nuclease proximal to the 1837 C>T PTC in exon 14 of the COL7A1 gene (Figure 1d), and the left and right individual TALEN arrays were cloned into the GoldyTALEN mini-CAGGs promoter-driven expression cassette (Figure 1e).28 Alternatively, the TALEN candidates were inserted into the RCIscript-GoldyTALEN vector to allow us to generate capped TALEN mRNA (Figure 1f).28
To test TALEN activity, we introduced the left and right TALEN arrays into primary fibroblasts obtained from a skin biopsy of an patient with RDEB and then analyzed the target locus for evidence of repair by the two major DNA repair pathways: error-prone non-homologous end-joining and homology-directed repair (HDR). Surveyor nuclease assay showed activity of TALENs when delivered as either DNA (Figure 2a) or mRNA (Figure 2b). Sanger sequencing confirmed the presence of insertions/deletions that are consistent with imperfect repair by the non-homologous end-joining pathway (Supplementary Figure S1).29 These data established that the nuclease is active at the target site and that delivery of the TALEN as a DNA species mediated higher rates of non-homologous end-joining. Accordingly, we next ascertained whether RDEB cells could undergo HDR following codelivery of TALEN DNA and an oligonucleotide donor (ODN) containing a unique primer sequence flanked by short donor arms bearing sequence homology to the COL7A1 locus (Figure 2c). RDEB fibroblasts transfected with TALEN plasmids and the ODN were then analyzed with a three-primer PCR approach that simultaneously detects the unmodified and modified alleles (Figure 2c,d). This assay showed that TALENs in RDEB cells can stimulate HDR to incorporate an exogenous sequence from the ODN donor (Figure 2d) and show the efficacy of TALEN use for the modification of the human COL7A1 gene in primary fibroblasts.
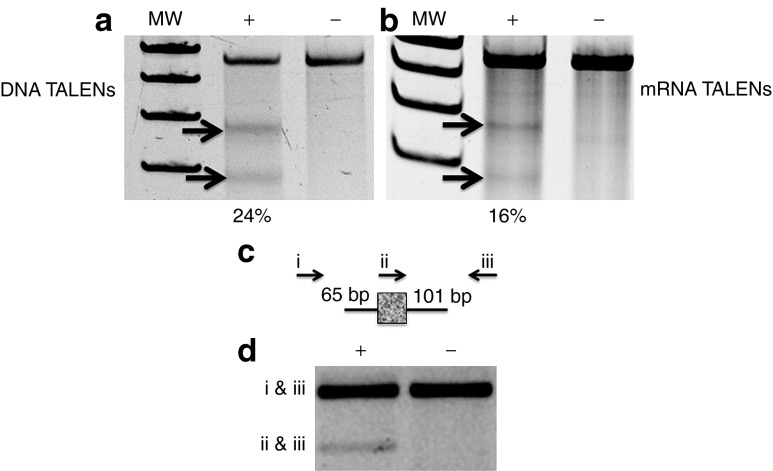
Figure 2.
TALEN activity in primary patient fibroblasts. Error-prone non-homologous end-joining assessment by Surveyor nuclease assay in cells that received (a) DNA TALENs or (b) mRNA TALENs. The COL7A1 locus in primary RDEB fibroblasts that received (“+”) or did not receive (“−”) TALENs was subjected to Surveyor nuclease treatment; arrows indicate the cleavage bands. The percentage of modification is indicated below the “+” lane. Molecular weight standards (MW) and was determined by densitometry. Homology-directed repair. (c) The single-stranded oligonucleotide donor (ssODN) contained 65 bp of COL7A1 gene homology on the left arm and 101 bp on the right, with a short foreign sequence that serves as a unique primer site and is indicated with the mottled, gray box. (d) Three-primer PCR results in amplification with endogenous primer pairs that are outside of the ODN donor sequence (indicated with arrows labeled i and iii). TALEN insertion of the ODN results in a second, smaller PCR product size generated by primer pairs ii and iii. Each treatment group received the ODN; the “+” or “−” refers to the presence or absence of the TALEN plasmid DNA. The data are representative gels of at least two experiments each. RDEB, recessive dystrophic epidermolysis bullosa; TALEN, transcription activator-like effector nucleases.
Corrective donor design and selection
To efficiently correct the gene mutation and be able to recover a homogenous population of corrected cells, we implemented a donor strategy that would allow for selective pressure to be applied to gene-modified cells. Our double-stranded plasmid DNA donor consisted of homology arms that spanned ~1 kb of the COL7A1 locus between exons 12 and 15 (Figure 3a). Within the donor was a floxed-PGK-puromycin cassette oriented such that it would be inserted into the intron between exons 12 and 13, thus allowing for cre-recombinase–mediated removal (Figure 3b). Within the right donor arm, we included the corrective base for the 1837 C>T PTC, as well as a downstream single base pair alteration (that we term a silent point mutation polymorphism) that removed an ApaI restriction enzyme site to allow for delineation of HDR-modified alleles versus unmodified ones (Figure 3b). We delivered the donor and TALENs as either plasmid DNA or mRNA to primary RDEB fibroblasts, and executed a selection strategy whereby cells were selected in bulk, sequestered into subpools, and finally isolated and expanded as clones Figure 3c–e.
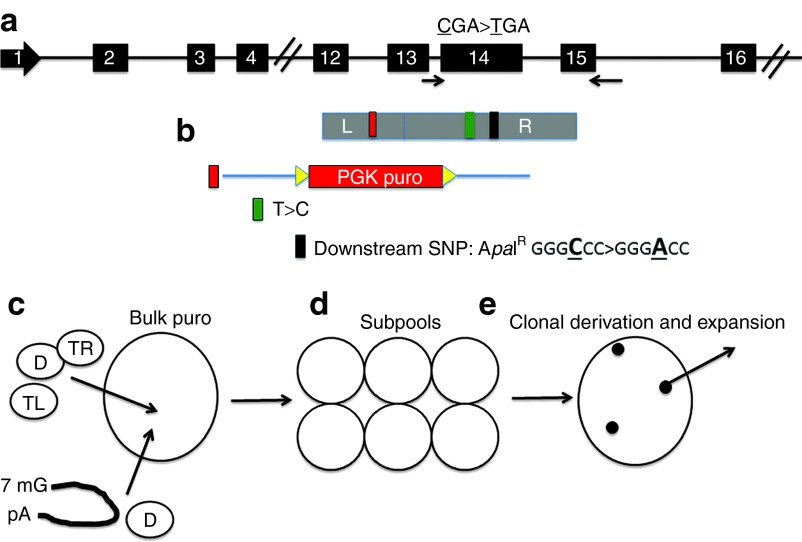
Figure 3.
TALEN COL7A1 donor design and schema for selection of gene-corrected cells. (a) The COL7A1 locus with exon 14 1837 CGA>TGA mutation indicated by asterisk. (b) Gene targeting/correction donor. The donor is shown in alignment to its relation with the endogenous locus. The plasmid donor is composed of COL7A1 genomic sequences comprising a left arm (L) that is 706 bp long. Following this was a PGK puromycin cassette (red box), flanked by loxp sites (yellow arrows), oriented so that it would be knocked into the intron between exons 12 and 13. The right arm (R) is 806 bp long and contained the normalized base for the 1837 mutation (green box) and a terminal SPMP that removed an ApaI restriction site (black box). (c) Selection schema. TALEN DNA (“TL”, TALEN Left, and “TR”, TALEN Right) or mRNA and donor plasmids (“D”) were delivered to cells followed by bulk puromycin selection. Cells were (d) split into subpools and (e) then plated at low density for clonal derivation and subsequent re-expansion. SPMP, silent point mutation polymorphism; TALEN, transcription activator-like effector nucleases.
Talen-edited cell screening and characterization
The cells were screened at the subpool stage for the presence of donor-mediated HDR. For each delivery platform (i.e., mRNA or DNA), five out of six subpool cell groups were positive for a PCR product that amplifies through the donor arm into the adjacent COL7A1 gene sequence (Figure 4a,b). In the case of the cells where the TALEN was delivered as mRNA, a second, lower molecular weight band was observed (Figure 4a, bottom arrow) that was analyzed by sequencing. These data revealed the insertion of the donor with an associated deletion of COL7A1 gene sequence (Supplementary Figure S2).
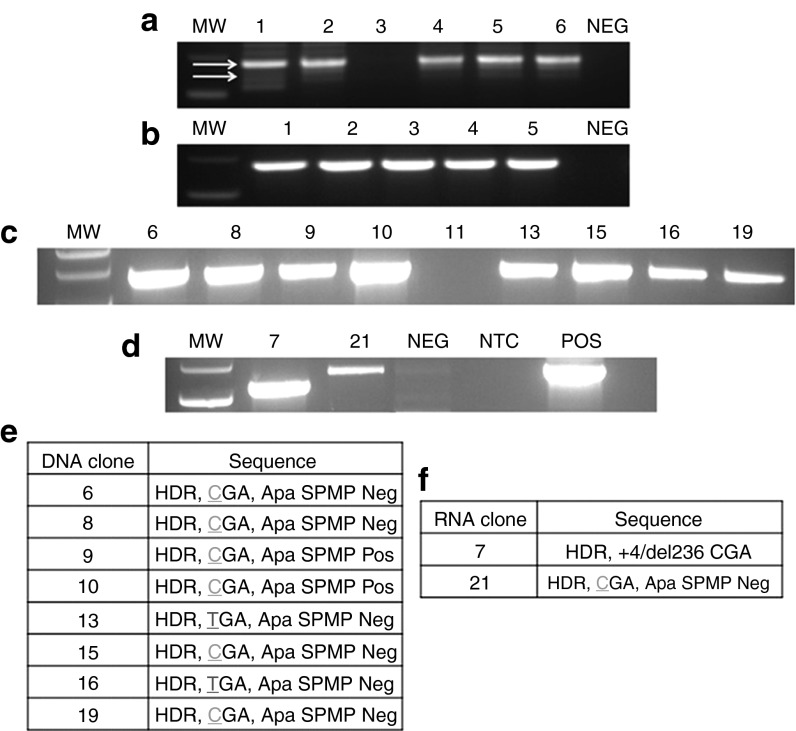
Figure 4.
Homology-directed repair, detection, and sequencing of genomic DNA. PCR analysis of post-TALEN-treated, puromycin-selected subpools in bulk for TALENs delivered as (a) mRNA or (b) DNA. Using the primers indicated with arrows in Figure 3a, one of which is within the donor and the second is outside the donor arm, individual clones were screened. (c) DNA TALEN and (d) mRNA TALEN clone screening. Numbers indicate clone identification designation. Clone 11 was a representative negative sample. Sanger sequence analysis summarization of PCR screen for (e) DNA TALENs or (f) mRNA TALENs. At left are the fibroblast (Fb) clone identification designators. At right are the sequences recovered and their status as to the occurrence of HDR (established by a positive PCR reaction), the status of the 1837 mutation location (green “C” indicates gene correction and red “T” shows maintenance of the mutation), and the presence or absence of the ApaI silent mutation polymorphism, as well as any associated insertions/deletions (“+4” = insertion of 4 bp and “del” refers to the deletion of 236 bp in mRNA clone 7). The clones derived are a representative data set of three individually performed experiments. MW, molecular weight standards; NTC, no template control; POS, positive control; TALEN, transcription activator-like effector nucleases.
The subpools were seeded at low density to allow for clonal segregation and homogeneous expansion, and were then screened for on-target HDR events. When the TALENs were delivered as plasmid DNA, the subsequent clones that were positive for HDR were 8/18 (Figure 4c). In contrast, when mRNA was the delivery platform, only 2/16 clones showed evidence of HDR (Figure 4d). One of these (clone 7) showed a truncation that corresponded to the deletion observed in Supplementary Figure S2. The remaining HDR-specific PCR products were sequenced (results summarized in Figure 4e,f) and revealed a mixture of on-target HDR events. These events are characterized thusly: (i) Early crossover/gene mutation maintenance event. This is defined as the presence of donor-derived sequences; however the COL7A1 1837 C>T mutation persists (Figure 4e,f and Supplementary Figure S3a–e). (ii) Early crossover/gene correction, defined as the presence of donor-derived sequence including the correction of the 1837 C>T mutation; however, the downstream silent point mutation polymorphism is absent (Figure 4e,f and Supplementary Figure S3f–h). (iii) Full-HDR, defined as the presence of the up and downstream silent point mutation polymorphisms, as well as correction of the C>T mutation (Figure 4e,f and Supplementary Figure S3i,k). These data show the ability of TALENs to mediate HDR in primary fibroblasts and show a higher frequency of correction when using DNA as the delivery vehicle.
To further classify the gene-targeting event, we removed the puromycin cassette with cre-recombinase and screened the clones by PCR, which revealed a modified and (Supplementary Figure S4a) unmodified allele (Supplementary Figure S4b), showing a heterozygous gene-correction event.
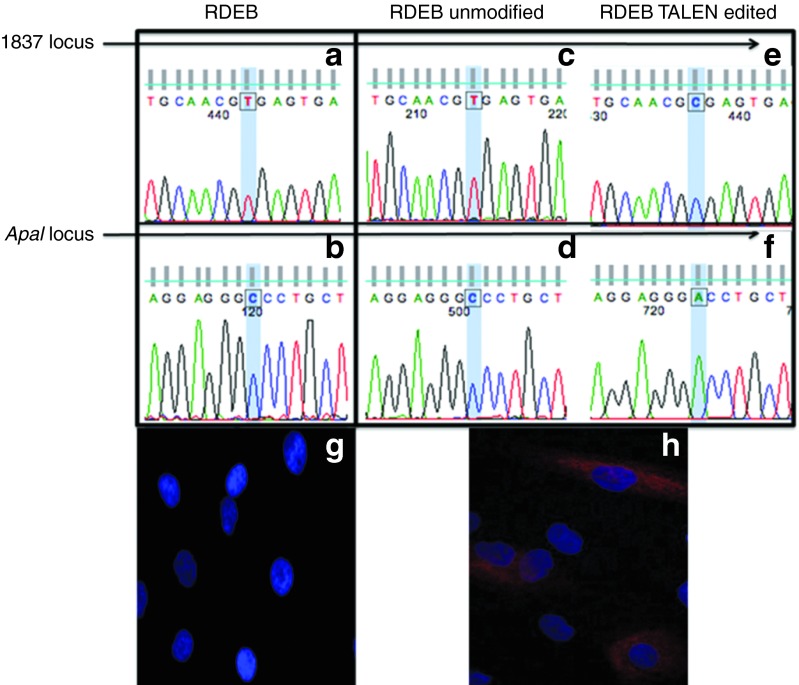
Genotypic and phenotypic correction
We then used a PCR strategy that allowed us to interrogate the state of the mutation locus as well as the ApaI locus in the same transcript. All of the transcripts in non-TALEN control cells showed the mutated base at the 1837 site and the unmodified cytosine at the ApaI locus (Figure 5a,b). In TALEN-treated cells, there was an equal mixture of gene-corrected and uncorrected transcripts that is consistent with heterozygous gene correction. The uncorrected allele resulted in the production of transcripts bearing thymine at position 1837 and cytosine at the ApaI locus (Figure 5c,d). The TALEN-corrected allele restored normal transcript status as evidenced by the presence of both donor-derived bases: the normal cytosine at the 1837 locus (Figure 5e) and the adenine that confers ApaI resistance (Figure 5f). To determine if protein expression was restored, we compared TALEN-treated with untreated RDEB mutant cells. Immunofluorescence-based analysis of type VII collagen showed no detectable type VII collagen protein in RDEB cells (Figure 5g), which is consistent with the homozygous PTC nature of the mutation. When the gene-edited RDEB cells were stained with anti-type VII collagen antibody, we observed the rescue of protein production (Figure 5h). These results show the ability of TALENs to mediate a genotypic correction that results in normalized transcripts that restore type VII collagen protein expression.
Figure 5.
TALEN-mediated gene editing resulting in normalized transcripts in primary fibroblasts. Uncorrected RDEB cell analysis showing the mutated base at (a) the 1837 locus and (b) the ApaI sensitive base. Non-targeted allele in TALEN-treated cells showing (c) presence of the mutation and (d) the absence of the ApaI-resistant base. Gene-edited allele in TALEN-treated cells showing (e) correction of the 1837 mutation and (f) the presence of the ApaI sensitive base. Immunofluorescence staining for type VII collagen protein. (g) Untreated RDEB fibroblasts showed a complete absence of detectable type VII collagen protein and (h) TALEN-corrected cells showed expression of type VII collagen. Antibody staining for each cell type was performed at the same time, and representative images were generated with identical microscopy settings. Blue, DAPI nuclear stain; red, type VII collagen. RDEB, recessive dystrophic epidermolysis bullosa; TALEN, transcription activator-like effector nucleases.
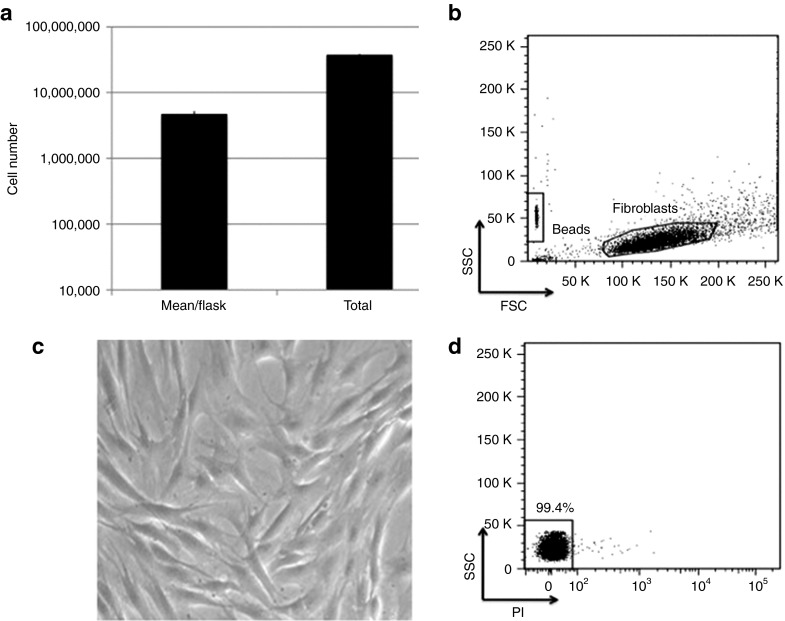
Cellular expansion
To date, one study has shown the potential of using fibroblasts as a delivery vehicle for type VII collagen in patients with RDEB.21 This strategy relied on the delivery of ~5 × 106 cells. To surpass this benchmark, we expanded a gene-corrected clone to ~58 × 106 cells (Figure 6a,b) that showed a normal morphology with high viability (Figure 6c,d). These data show our ability to derive, maintain, archive, and expand gene-edited cells to clinically meaningful numbers.
Figure 6.
Gene-corrected primary fibroblast expansion. Parallel samples in duplicate were sequentially expanded until they reached >85% confluency; absolute viable cell count was determined using bead-enhanced flow cytometry. (a) Total expanded cell numbers. Total cell numbers and data are represented as average cell count/T150 cm2 flask; the total number was from the sum of all flasks. (b) FACS analysis of forward (FSC) and side scatter (SSC) of a representative data plot of expanded fibroblasts. Shown at the lower left decade are the PKH26 reference microbeads used to calculate numbers of cells. (c) Cell morphology. Cells were visualized at 20× magnification using light microscopy and exhibited a normal elongated morphology. (d) Cell viability. Propidium iodide (PI) staining of gated cells in b.
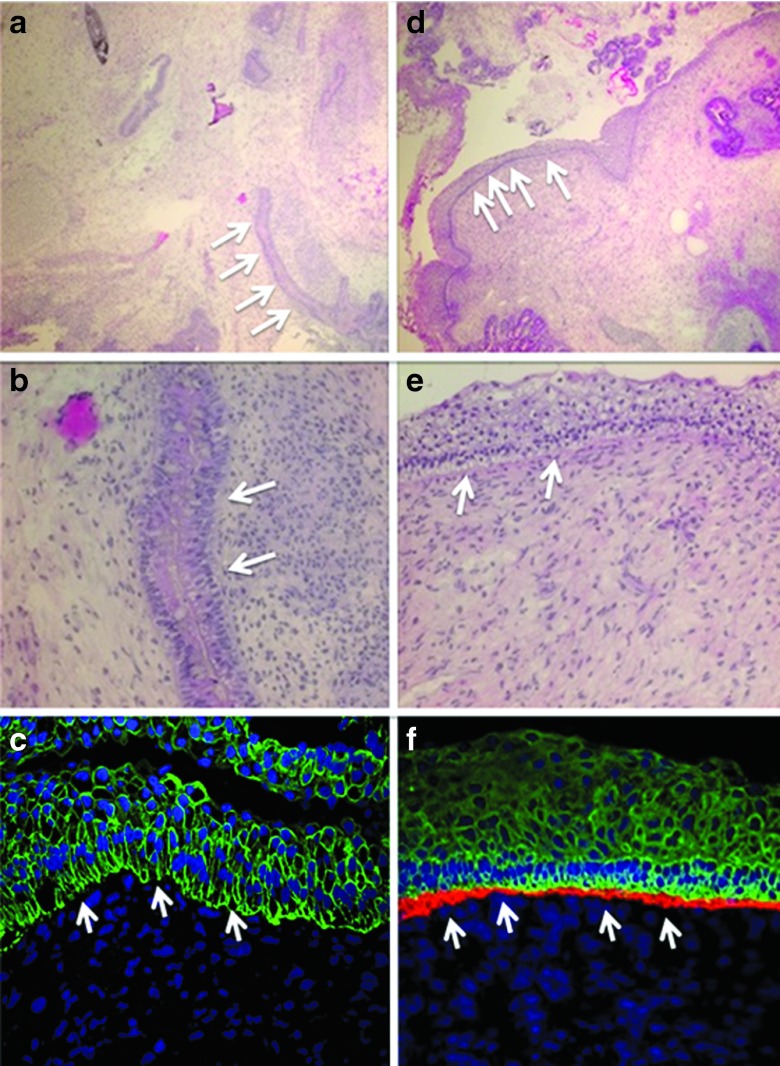
iPSC generation
In addition to their potential for direct cutaneous benefit, fibroblasts are also an ideal cell type for iPSC generation. Due to their broad differentiation potential, iPSCs hold great promise; therefore, we reprogrammed gene-edited fibroblasts and performed in vivo teratoma assays in murine recipients. Patient-derived, uncorrected fibroblasts used to generate iPSCs showed the formation of skin-like structures, but did not reveal any type VII collagen protein at the DEJ following staining with an anti-type VII collagen antibody (Figure 7a–c). When we generated iPSCs from gene-corrected cells, we also observed skin-like structures (Figure 7d,e) that when further analyzed showed the presence of type VII collagen protein at the DEJ, which confirmed the ability of TALENs to correct the primary genetic defect and restore functional protein production at the proper ultrastructural location in vivo (Figure 7f).
Figure 7.
iPSC teratoma and rescued type VII collagen contribution to the DEJ using gene-edited cells. iPSC-derived teratoma analysis. Homozygous 1837 C>T RDEB and TALEN gene-corrected fibroblasts were reprogrammed into iPSCs for teratoma differentiation assay. RDEB mutant cells: (a,b) iPSC light microscopy and (c) immunofluorescence. RDEB TALEN-corrected cells: (d,e) iPSC light microscopy and (f) immunofluorescence. (a,b,d,e) Hematoxylin and esosin stained light microscopy images are at 10× and 20× magnification, respectively and showed epidermal skin-like structures (white arrows). (c,f) Teratoma immunofluorescence shows the DEJ indicated with white arrows. Immunofluorescence markers were as follows: Blue, DAPI nuclear stain; green, cytokeratin 5; red, type VII collagen. Images are representative images from at least three animals. Antibody staining was performed from a single master mix on the same day using identical microscopy settings.
OT analysis
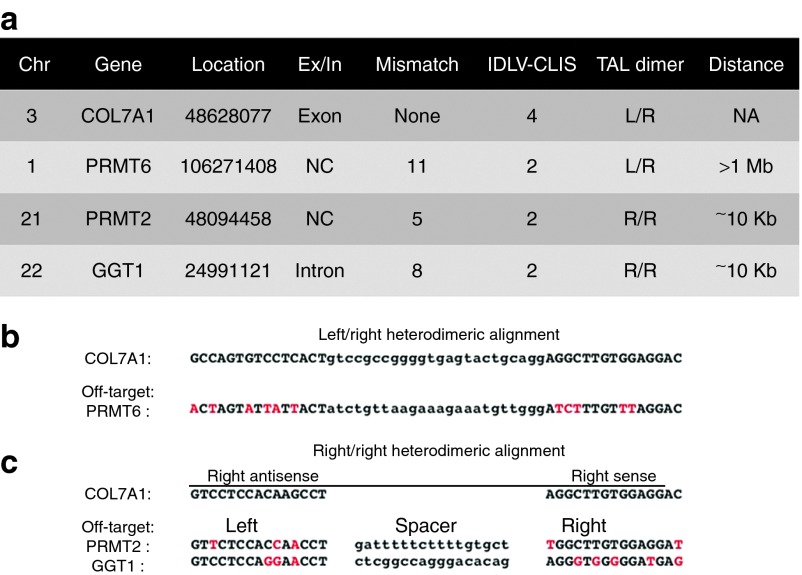
A critical consideration for the clinical use of genome-editing reagents is potential for OT effects. It has been previously shown that DSBs generated by ZFNs can result in the “trapping” of DNA at the site that permanently marks it, allowing for the subsequent detection and mapping of these on-target and OT sites.30 To determine if TALENs would function similarly, we used 293 cells and introduced an integrase-defective lentivirus (IDLV) expressing a green fluorescent protein (GFP) marker gene in the presence or absence of TALENs. We then performed PCR using an LTR-specific primer and a COL7A1 primer. This PCR showed that only when TALENs were cointroduced with the IDLV did the GFP IDLV insert into the spacer region of the COL7A1 target site (Supplementary Figure S5a–c).
These data were extended by monitoring GFP gene expression and showed that when the IDLV alone was delivered, a rapid loss of GFP occurred that showed the episomal nature of this species of DNA (Supplementary Figure S6a). In contrast, the cointroduction of IDLV and TALENs resulted in a stable population of GFP cells that were used for mapping the integration sites (IS) with classical and nonrestrictive linear amplification–mediated PCR (LAM and (nr)LAM-PCR) and deep sequencing30 (Supplementary Figure S6b,c). Vector:genome junctions were observed at the TALEN target site as well as three OT sites that share some homology to the COL7A1 TALEN and are located at chromosomes 1, 21, and 22 (Figure 8a). Analysis of the OT sites showed that the PRMT6 locus was the result of the left and right arrays of the COL7A1 TALEN binding (Figure 8b). The other two OT sites were the result of two right arrays of the COL7A1 TALEN dimerizing at the OT locus (Figure 8c). Importantly, none of the OT sites are within ~10 kb of an exon (Figure 8a), indicating that this TALEN reagent possesses a safety profile that is not predicted to negatively impact gene expression.
Figure 8.
TALEN target site LAM-PCR deep sequence analysis. (a) Summary of IDLV:genomic junction fragments and chromosomal location (“Chr”), gene name, gene location (UCSC BLAT Database numerical nomenclature), location of recovered site in regards to an exon (“Ex”) or intron (“In”), number of mismatches between the recovered site and the target site for which the TALEN was designed, number of clustered integration sites (IDLV-CLIS) recovered, manner in which the TALEN is predicted to bind to the candidate sequence (TAL dimer: L/R, left right array TALEN binding, R/R, two right TALEN arrays homodimerizing), and distance of recovered site from an exon. (b,c) TALEN on- and off-target sequences. For each, the gene name is at left; capital letters indicate the array binding sites and lower-case letters are the spacer region. Red letters indicate the mismatches between the PRMT2 and GGT1 off-target sites and the COL7A1 target. Sequence alignment of (b) Left/Right heterodimers and (c) Right/Right homodimers. A dimerization of two right arrays is predicted to be the manner in which two of the off-target effects will manifest. Because nuclease arrays bind to opposite strands (e.g., sense and antisense), both orientations of the COL7A1 right target site are shown. Alignments to these conformations (antisense at left and sense at right) for the off-target sites are shown with red letters indicating mismatches between the true (i.e., COL7A1) and off-target binding sites. LAM-PCR data were generated from duplicate samples. CLIS, clustered integration site; IDLV, integrase-defective lentivirus; NA, not applicable; TALEN, transcription activator-like effector nucleases.
Discussion
We present the following key findings: the generation of a TALEN generated for a specific mutation and the correction of a COL7AI gene mutation in patient-derived, primary fibroblasts with subsequent generation of iPSCs. Second, we have performed an unbiased deep sequencing-based OT analysis that shows three possible OT sites (Figure 8). These latter findings are neither unprecedented nor unexpected, as even nucleases used in clinical trials show OT effects.30,31
The platforms to map the OT sites of gene-editing nucleases include: (i) performing in vitro Systematic Evolution of Ligands by Exponential Enrichment with monomeric DNA-binding proteins of each nuclease in a pair and then using these data to predict potential OT sites,32 (ii) performing an in vitro cleavage site analysis,33 and (iii) using the propensity of an IDLV to integrate into nuclease-induced DSBs and then identifying points of insertion by LAM-PCR.30 Although methods (ii) and (iii) appear to be better at identifying nuclease OT sites than method (i), methods (ii) and (iii) failed to identify OT sites predicted by the other, suggesting that neither method is comprehensive in its detection of OT events.34 We used method (iii) with an IDLV with GFP gene that can be trapped into a nuclease-generated DSB (Supplementary Figures S5 and S6). The choice of cell types for such a strategy is an important consideration, and the use of transformed cells that rapidly proliferate is an attractive option as has been documented for ZFN OT analysis.30 This is based on the rapid dilution of the non-integrating IDLV that minimizes prolonged presence of the episome that, in slower dividing primary cells, may result in increased non-nuclease–dependent IS that occur from normal cellular physiology at fragile genomic loci.30,35 To differentiate between random IS and true nuclease-mediated IDLV trapping, this study showed the usefulness of clustered integration site analysis,30 and we used this methodology in TALEN-treated 293 cells. We identified 76 total IS with 66 of these fitting the category of a non-nuclease–mediated IS and the remaining 10 fitting the criteria of clustered integration site and showing on-target TALEN activity at the COL7A1 locus as well as three OT sites (Figure 8). The great distance between the OT sites and any coding sequence (≥10 kb) is significant in that nuclease repair by non-homologous end-joining favors small insertions/deletions, which has been shown previously as well as in this study where the maximum deletion observed was 51 bp (Supplementary Figure S1).30 Taken together, the ability of deep sequencing to differentiate between IS that are inherent to the methodology and clustered integration site that represent true nuclease-induced DSB:gene capture junctions makes it a powerful platform for establishing safety profiles.
We used this TALEN pair to mediate heterozygous correction of the 1837 C>T PTC COL7A1 gene mutation. Because this disorder is recessive and carriers exhibit no overt symptoms, a heterozygous gene-correction event is sufficient for phenotypic rescue, and we did not pursue a second round of gene targeting to achieve homozygous correction. These gene-correction events were characterized by variability in where HDR occurred on the donor plasmid used as the repair template. Previous reports have suggested that optimal rates of homologous recombination/HDR can be achieved when targeting nucleases within 200 bp of the target site.36,37,38 Our results support this notion as evidenced by the occurrence of HDR in two of our clones from a point on the donor plasmid upstream of the mutation site, which resulted in persistence of the mutation (Figure 4e and Supplementary Figure S3). Importantly, the majority of HDR events resulted in correction of the mutation and whether the downstream donor-derived silent point mutation polymorphism was present was variable, which further highlights the necessity of mutation proximal (<200 bp) targeting (Figure 4e,f and Supplementary Figure S3).
Our data also show a differential ability of TALENs to mediate HDR depending on whether they are delivered as DNA or mRNA (Figures 1, 2, and 4). Delivery of TALENs as mRNA is desirable as it is expressed for a short duration and there is no risk of random integration. Other studies have shown the ability of TALEN mRNA to effectively mediate HDR from an ODN donor.28 Our study shows a much higher frequency of clones bearing the corrected sequence when the TALENs were delivered as DNA (Figure 4c,e) than when delivered as mRNA (Figure 4d,f). This might suggest that the expression kinetics of the TALENs may impact the frequency of HDR alone or in conjunction with the format of the donor molecule (i.e., double versus single stranded). Future studies are warranted to further define the most efficient manner in which TALENs are used for HDR.
We chose dermal fibroblasts obtained from a minimally invasive punch biopsy of an individual with RDEB as the cell type of choice for genome modification. Clinically, these cells have been used as part of an allogeneic therapy due to the superior ability of fibroblasts to contribute to a functional DEJ than keratinocytes.21,39,40 We were able to document gene correction at the genomic locus resulting in normal mRNA transcript expression with concomitant type VII collagen protein expression in patient-derived cells that we were able to expand nearly tenfold above the dose used clinically (Figures 4, 5, and 6). As such, this allogeneic approach may allow for a prolonged presence of therapeutic cells in an immune-competent host. To extend this finding, we generated iPSCs from the gene-corrected cells and showed in an in vivo model the ability of TALEN-edited cells to deposit type VII collagen at the DEJ (Figure 7). These findings are key in that the plasticity of iPSCs represents a platform from which the non-mucocutaneous manifestations of RDEB can be addressed as part of autologous systemic therapies.
We provide proof-of-concept for the use of TALENs in individualized therapies. These data are especially relevant in regards to targeting genes such as COL7AI that generally have no mutational hotspots and where patients often possess unique mutations. As such, the platform chosen for gene correction will be dictated by the genomic locus being targeted. TALENs appear to be the most expansive and flexible targeting platform compared with ZFNs and the CRISPR/Cas9 system.11,14,15,16,24,25 In summary, we merged the technologies of TALENs, iPSCs, and LAM-PCR deep sequencing to show the ability of gene-edited cells to contribute functional type VII collagen to the DEJ with minimal OT effects. These data further establish TALENs as a viable tool for future ex vivo therapies.
Materials and Methods
Research subject and cell line derivation. Declaration of Helsinki protocols were followed, and parents gave their written, informed consent. After obtaining informed parental consent, we obtained a punch biopsy from the skin of a male patient with RDEB with a homozygous c.1837 C>T PTC mutation. Approval for research on human subjects was obtained from the University of Minnesota Institutional Review Board. A primary fibroblast cell line was derived by mincing the skin tissue, placing it under a glass microscope slide, and submerging in complete Dulbecco's modified Eagle media with 20% fetal bovine serum, and expanded and maintained in low oxygen concentration conditions.
TALEN and donor construction. The TALEN candidates were generated via the Golden Gate Assembly method and inserted into a homodimeric form of a CAGGs promoter-driven FokI endonuclease (GoldyTALEN mini-CAGGs).28 Alternatively, the TALEN candidates were inserted into the RCIscript-GoldyTALEN vector for mRNA production.18,28 The left donor arm was amplified with the LAF and LAR primers shown in Supplementary Table S1. The right arm was synthesized in two fragments (inner and outer) by means of an overlapping oligonucleotide assembly strategy41,42 using the primers shown in Supplementary Table S1. The left and right arms were then cloned into a floxed-PGK-puromycin cassette.
In vitro transcription. TALEN plasmids were linearized with Sac-I (New England Biolabs, Ipswich, MA) and then treated with RNAsecure (Ambion, Austin, TX) before purification over a QIAQuick Column (Qiagen, Valencia, CA). Linearized DNA of 1 μg was then used in an in vitro transcription reaction using the mMessagemMachine T3 Kit (Ambion) and purified before use with the RNeasy MinElute Cleanup Kit (Qiagen).
Gene transfer. All TALEN treatments consisted of delivery of 2.5 μg of each TALEN DNA or mRNA and 10 μg amount of donor via the Neon Transfection System (Invitrogen, Carlsbad, CA) with the following instrument settings: 1,500 V, 20 ms pulse width, and a single pulse. For 48 hours after gene transfer, the cells were incubated at 31 °C.43
Cell culture. Cells were maintained in growth media comprised Dulbecco's modified Eagle media supplemented with 20% fetal bovine serum, 100 U/ml nonessential amino acids, and 0.1 mg/ml each of penicillin and streptomycin (Invitrogen), and cultured at 2% O2, 5% CO2, and 37 °C.
Surveyor nuclease. Genomic DNA was isolated 48 hours after TALEN gene transfer and amplified for 30 cycles with Surveyor F and Surveyor R primers, and subjected to Surveyor nuclease (Transgenomic, Omaha, NE) treatment as described.29 Products were resolved on a 10% TBE PAGE gel (Invitrogen). Densitometry was performed using Image J as described.29
HDR analysis. For quantification of HDR, TALENs and 5 μl of a 40 μmol/l single-stranded ODN were transfected into cells under the Neon transfection conditions detailed above and screened by PCR at 48 hours using three primers: Surveyor F, Surveyor R, and linker forward primers. Densitometry was performed as described.29
Selection. Cells were selected in bulk in 0.2 μg/ml puromycin, segregated into subpools, screened for HDR, and then plated at low density (250–750 total cells) in a 10 cm2 dish. A cloning disk with silicone grease (all from Corning, Corning, NY) was placed over single cells in the presence of base media supplemented with 10 ng/ml epidermal growth factor and 0.5 ng/ml fibroblast growth factor. Cells were expanded to sequentially larger vessels in a manner such that they were never allowed to fall below a confluency percentage of 70%. This normally consisted of passage from the 10 cm dish to a 24-well dish to a six-well dish to a 25 cm flask and finally to T75 and T150 culture vessels, all while maintained at low oxygen culture conditions.
Cell correction molecular screening. Primer pairs were used to amplify a junction from the donor into the endogenous locus that was outside of the right donor arm. PCR products were either direct sequenced or cloned into the pCR 4 TOPO vector (Invitrogen) before sequencing. Messenger RNA from clonal isolates was converted to cDNA and screened with RT1 and RT2 and then digested with ApaI. ApaI-resistant amplicons were cloned and Sanger sequenced.
PGK puromycin removal. An adenoviral cre-recombinase (with GFP in trans) (Vector BioLabs, Philadelphia, PA) was added at an initial multiplicity of infection of 20 and then treated serially at multiplicity of infections up to 1,000.
Cell expansion analysis. Gene-corrected fibroblasts were expanded in T150 flasks and trypsinized to obtain single-cell suspensions. Cells were then resuspended in 100 µl phosphate-buffered saline + 0.5% bovine serum albumin + propidium iodide (eBiosciences, San Diego, CA), followed by addition of an equal volume of PKH26 reference microbeads (SIGMA, St Louis, MO). Five thousand bead events were collected, and absolute viable cell number was calculated per the manufacturer's protocol (SIGMA).
iPSC generation and teratoma assay. Gene-corrected fibroblasts (or uncorrected cells as a control) were reprogrammed to iPSCs as described44,45 and then placed in the flank of SCID mice (n = 3–5) until a visible mass formed. The mass was excised for embedding and staining by hematoxylin and eosin staining or immunofluorescence microscopy.
Immunofluorescence. Gene-corrected cells were plated on a chamber slide and were fixed 24 hours later with 4% paraformaldehyde, permeabilized with 0.2% Triton X, blocked with 1% bovine serum albumin and stained with a polyclonal antitype VII collagen antibody (1:1,500; generously provided by Drs David Woodley and Mei Chen). Secondary antibody staining was performed with donkey antirabbit IgG Cy3 (1:500; Jackson Immunoresearch, West Grove, PA). For teratomas, the tissue was embedded in Tissue-Tek O.C.T. Compound (AJ Alphen aan den Rijn, The Netherlands), sectioned at a thickness of six microns and fixed in acetone. An antitype VII monoclonal antibody (1:250; mouse antihuman; BD Biosciences, San Jose, CA) against amino acids 652–1,162 and rabbit antihuman keratin 5 (1:600; Covance, Emeryville, CA) were then added followed by secondary antibodies: goat antirabbit Alexa Fluor 488 (1:800; Invitrogen and antimouse Cy3 (1:500; Jackson Immunoresearch). For all staining procedures, isotype control staining was done using whole molecule rabbit IgG (Jackson Immunoresearch). Nuclei were stained with 4′, 6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA). Images were taken using a PMT voltage of 745 on an Olympus BX61 FV500 confocal microscope (Olympus Optical, Center Valley, PA) and analyzed using the Fluoview software version 4.3. Light microscopy was performed on a Leica microscope (Leica Microsystems, Buffalo Grove, IL).
IDLV and LAM-PCR/(nr)LAM-PCR. IDLV particles were produced in 293T cells via lipid-based cotransfection (Lipofectamine 2000; Invitrogen) of the CMV-GFP transfer vector, the pCMV-ΔR8.2 packaging plasmid harboring the D64V integrase mutation,30,46 and the pMD2.VSV-G envelope-encoding plasmid. Gene tagging was performed by nucleofection of HEK 293 cells with the TALENs, followed 24 hours later by a transduction of GFP IDLV at an multiplicity of infection of 7. Genomic DNA of 100 ng was analyzed in duplicate by LAM-PCR47 using enzymes MseI and Tsp509I and (nr)LAM-PCR31 to ensure genome-wide recovery of IDLV IS. (nr)LAM-PCR amplicons were sequenced by the Roche/454 pyrosequencing platform (Roche, Branford, CT) and integration site data were analyzed using the HISAP pipeline.48,49 Genomic position harboring >1 IS in close distance were scanned for potential TALEN OT binding sites using the pattern matcher scan-for-matches.48
SUPPLEMENTARY MATERIAL Figure S1. TALEN targeting of COL7A1 gene. Figure S2. mRNA TALEN associated deletion. Figure S3. Sequence analysis of HDR events and HDR mechanism. Figure S4. Selective transgene removal. Figure S5. IDLV capture at the TALEN target site. Figure S6. Integrase-deficient lentivirus. Table S1. Oligonucleotide sequences.
Acknowledgments
This study was supported in part by grants from Epidermolysis Bullosa Research Fund, Jackson Gabriel Silver Foundation, DebRA International, University of Minnesota Academic Health Center, Pioneering Unique Cures for Kids foundation, Children's Cancer Research Fund, Minneapolis, Minnesota, and the United States of America Department of Defense. D.F.V. was supported by National Institutes of Health (R01 GM098861). J.K.J. was supported by a National Institutes of Health Director's Pioneer Award DP1 OD006862, and the Jim and Ann Orr MGH Research Scholar Award. M.L.M was supported by a National Science Foundation Graduate Research Fellowship. D.F.V. is a listed inventor on a patent application titled “TAL effector–mediated DNA modification,” which is co-owned by Iowa State Univ. and the Univ. of Minnesota, and has been licensed to Cellectis, a European biotechnology company. J.K.J. has a financial interest in Transposagen Biopharmaceuticals. J.K.J.'s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. The other authors declare no conflict of interest.
Supplementary Materials
References
- Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Daya S, Berns KI. Gene therapy using adeno-associated virus vectors. Clin Microbiol Rev. 2008;21:583–593. doi: 10.1128/CMR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CE, Ehrhardt A, Kay MA. Progress and problems with the use of viral vectors for gene therapy. Nat Rev Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetschman T, Gregg RG, Maeda N, Hooper ML, Melton DW, Thompson S, et al. Targetted correction of a mutant HPRT gene in mouse embryonic stem cells. Nature. 1987;330:576–578. doi: 10.1038/330576a0. [DOI] [PubMed] [Google Scholar]
- Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- Stoddard BL. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure. 2011;19:7–15. doi: 10.1016/j.str.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F, Duchateau P. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr Gene Ther. 2007;7:49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- Ramirez CL, Foley JE, Wright DA, Müller-Lerch F, Rahman SH, Cornu TI, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Sander JD, Voytas DF, Joung JK. Oligomerized pool engineering (OPEN): an ‘open-source' protocol for making customized zinc-finger arrays. Nat Protoc. 2009;4:1471–1501. doi: 10.1038/nprot.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruett-Miller SM, Connelly JP, Maeder ML, Joung JK, Porteus MH. Comparison of zinc finger nucleases for use in gene targeting in mammalian cells. Mol Ther. 2008;16:707–717. doi: 10.1038/mt.2008.20. [DOI] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine JD, Mellerio JE. Extracutaneous manifestations and complications of inherited epidermolysis bullosa: part II. Other organs. J Am Acad Dermatol. 2009;61:387–402; quiz 403. doi: 10.1016/j.jaad.2009.03.053. [DOI] [PubMed] [Google Scholar]
- Wagner JE, Ishida-Yamamoto A, McGrath JA, Hordinsky M, Keene DR, Woodley DT, et al. Bone marrow transplantation for recessive dystrophic epidermolysis bullosa. N Engl J Med. 2010;363:629–639. doi: 10.1056/NEJMoa0910501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong T, Gammon L, Liu L, Mellerio JE, Dopping-Hepenstal PJ, Pacy J, et al. Potential of fibroblast cell therapy for recessive dystrophic epidermolysis bullosa. J Invest Dermatol. 2008;128:2179–2189. doi: 10.1038/jid.2008.78. [DOI] [PubMed] [Google Scholar]
- Tolar J, Ishida-Yamamoto A, Riddle M, McElmurry RT, Osborn M, Xia L, et al. Amelioration of epidermolysis bullosa by transfer of wild-type bone marrow cells. Blood. 2009;113:1167–1174. doi: 10.1182/blood-2008-06-161299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, Vandyk JK, et al. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: tools for TAL effector design and target prediction. Nucleic Acids Res. 2012;40 Web Server issue:W117–W122. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, et al. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci USA. 2012;109:17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- Paruzynski A, Arens A, Gabriel R, Bartholomae CC, Scholz S, Wang W, et al. Genome-wide high-throughput integrome analyses by nrLAM-PCR and next-generation sequencing. Nat Protoc. 2010;5:1379–1395. doi: 10.1038/nprot.2010.87. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Blazar B, High K, Porteus M. Zinc fingers hit off target. Nat Med. 2011;17:1192–1193. doi: 10.1038/nm1011-1192. [DOI] [PubMed] [Google Scholar]
- Mátrai J, Cantore A, Bartholomae CC, Annoni A, Wang W, Acosta-Sanchez A, et al. Hepatocyte-targeted expression by integrase-defective lentiviral vectors induces antigen-specific tolerance in mice with low genotoxic risk. Hepatology. 2011;53:1696–1707. doi: 10.1002/hep.24230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Elliott B, Jasin M. Repair of double-strand breaks by homologous recombination in mismatch repair-defective mammalian cells. Mol Cell Biol. 2001;21:2671–2682. doi: 10.1128/MCB.21.8.2671-2682.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott B, Richardson C, Winderbaum J, Nickoloff JA, Jasin M. Gene conversion tracts from double-strand break repair in mammalian cells. Mol Cell Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M, Sawamura D, Ito K, Abe M, Nishie W, Sakai K, et al. Fibroblasts show more potential as target cells than keratinocytes in COL7A1 gene therapy of dystrophic epidermolysis bullosa. J Invest Dermatol. 2006;126:766–772. doi: 10.1038/sj.jid.5700117. [DOI] [PubMed] [Google Scholar]
- Yan WF, Murrell DF. Fibroblast-based cell therapy strategy for recessive dystrophic epidermolysis bullosa. Dermatol Clin. 2010;28:367–70, xii. doi: 10.1016/j.det.2010.01.015. [DOI] [PubMed] [Google Scholar]
- Osborn MJ, DeFeo AP, Blazar BR, Tolar J. Synthetic zinc finger nuclease design and rapid assembly. Hum Gene Ther. 2011;22:1155–1165. doi: 10.1089/hum.2011.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Choi VM, Xia DF, Vo TD, Gregory PD, Holmes MC. Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat Methods. 2010;7:459–460. doi: 10.1038/nmeth.1456. [DOI] [PubMed] [Google Scholar]
- Tolar J, Xia L, Lees CJ, Riddle M, McElroy A, Keene DR, et al. Keratinocytes from induced pluripotent stem cells in junctional epidermolysis bullosa. J Invest Dermatol. 2013;133:562–565. doi: 10.1038/jid.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolar J, Xia L, Riddle MJ, Lees CJ, Eide CR, McElmurry RT, et al. Induced pluripotent stem cells from individuals with recessive dystrophic epidermolysis bullosa. J Invest Dermatol. 2011;131:848–856. doi: 10.1038/jid.2010.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas J, Jr, Gusella GL, Najfeld V, Klotman ME, Cara A. Novel integrase-defective lentiviral episomal vectors for gene transfer. Hum Gene Ther. 2004;15:361–372. doi: 10.1089/104303404322959515. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Schwarzwaelder K, Bartholomae C, Zaoui K, Ball C, Pilz I, et al. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR). Nat Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- Dsouza M, Larsen N, Overbeek R. Searching for patterns in genomic data. Trends Genet. 1997;13:497–498. doi: 10.1016/s0168-9525(97)01347-4. [DOI] [PubMed] [Google Scholar]
- Arens A, Appelt JU, Bartholomae CC, Gabriel R, Paruzynski A, Gustafson D, et al. Bioinformatic clonality analysis of next-generation sequencing-derived viral vector integration sites. Hum Gene Ther Methods. 2012;23:111–118. doi: 10.1089/hgtb.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.