Abstract
Bone marrow transplantation (BMT) can give rise to donor-derived osteopoiesis in mice and humans; however, the source of this activity, whether a primitive osteoprogenitor or a transplantable marrow cell with dual hematopoietic and osteogenic potential, has eluded detection. To address this issue, we fractionated whole BM from mice according to cell surface immunophenotype and assayed the hematopoietic and osteopoietic potentials of the transplanted cells. Here, we show that a donor marrow cell capable of robust osteopoiesis possesses a surface phenotype of c-Kit+ Lin− Sca-1+ CD34−/lo, identical to that of the long-term repopulating hematopoietic stem cell (LTR-HSC). Secondary BMT studies demonstrated that a single marrow cell able to contribute to hematopoietic reconstitution in primary recipients also drives robust osteopoiesis and LT hematopoiesis in secondary recipients. These findings indicate that LTR-HSC can give rise to progeny that differentiate to osteoblasts after BMT, suggesting a mechanism for prompt restoration of the osteoblastic HSC niche following BM injury, such as that induced by clinical BMT preparative regimens. An understanding of the mechanisms that regulate this differentiation potential may lead to novel treatments for disorders of bone as well as methods for preserving the integrity of endosteal hematopoietic niches.
Introduction
Bone and bone marrow (BM) are anatomically contiguous and harbor cell types that are functionally interrelated.1 Conceivably, then, a stem cell could give rise to both hematopoietic and osteopoietic progeny under the control of a specific genetic program or particular environmental cues. Several investigators have independently demonstrated that BM transplantation (BMT) results in donor-derived osteopoiesis early after this procedure in mice,2,3,4,5,6 whereas others have identified donor osteoblasts after transplantation in humans.7,8,9 Molecular analysis of transplanted, gene-marked marrow cells in mice revealed a common retroviral integration site in hematopoietic and osteopoietic cells suggesting a dual differentiation capacity of primitive marrow progenitors.2 The functional capacity of the differentiated osteopoietic cells has been demonstrated by their ability to secure clinical improvement in children with osteogenesis imperfecta7,8,10 and, more recently, by amelioration of the osteogenesis imperfecta phenotype in a mouse model.5 These reports establish a link between transplanted marrow cells and osteopoiesis, but lack the necessary evidence to identify the source of this osteopoietic activity. Identifying a transplantable osteoprogenitor or perhaps a putative dual hematopoietic-osteopoietic progenitor could be key to our understanding of the biology of marrow transplantation and the hematopoietic stem cell (HSC) niche. Such insights could lead in turn to the development of novel cell therapies based on endogenous biologic differentiation potential.
Using secondary BMT assays, we show here that a single marrow cell able to contribute to hematopoietic reconstitution in primary recipients drives both osteopoiesis and long-term (LT) hematopoiesis in secondary recipients. These findings, together with evidence that this bipotential cell satisfies stringent criteria for stemness, suggest a novel mechanism for hematopoietic-osteopoietic maintenance that could be harnessed for medical interventions.
Results
Transplantable osteoprogenitor activity resides within the primitive hematopoietic progenitor population
Our previous studies indicated that marrow cells unable to adhere to plastic in vitro are more robust transplantable osteoprogenitors than are adherent mesenchymal stem/stromal cells (MSCs) after systemic transplantation.2 This finding, together with detection of the Sca-1 marker on primary osteoblasts derived from bone explants (Figure 1a) and MSCs (Figure 1b), suggested that the putative transplantable marrow osteoprogenitor resides within the nonadherent Sca-1+ population. To identify this osteoprogenitor population in the non(plastic)-adherent BM cells, we transplanted 2 × 105 Lin− (Gr1, CD11b, CD4, CD8, B220, Ter119) Sca-1+ cells from a green fluorescent protein (GFP) expressing transgenic mouse11 into lethally irradiated recipient mice (Figure 1c,d). Short-term and LT hematopoiesis were reconstituted as was a mean (± SD) osteopoietic engraftment of 15.4 ± 4.3% (Figure 1e). In contrast the Lin− Sca1− fraction of marrow, reconstituted short-term but not LT hematopoiesis and did not give rise to osteoblasts (Figure 1f). To exclude contamination of these grafts by a rare, unidentified highly proliferative osteoprogenitor among the adherent MSCs, we transplanted 1 × 106 MSCs from a transgenic GFP-expressing mouse and found a median of only 1.8% donor-derived osteopoiesis (range, 0–2.5%; n = 5) consistent with our prior results.2 These data indicate that the Lin− Sca-1+ fraction of nonadherent cells contains all, or is at least highly enriched for, the transplantable osteoprogenitor activity.
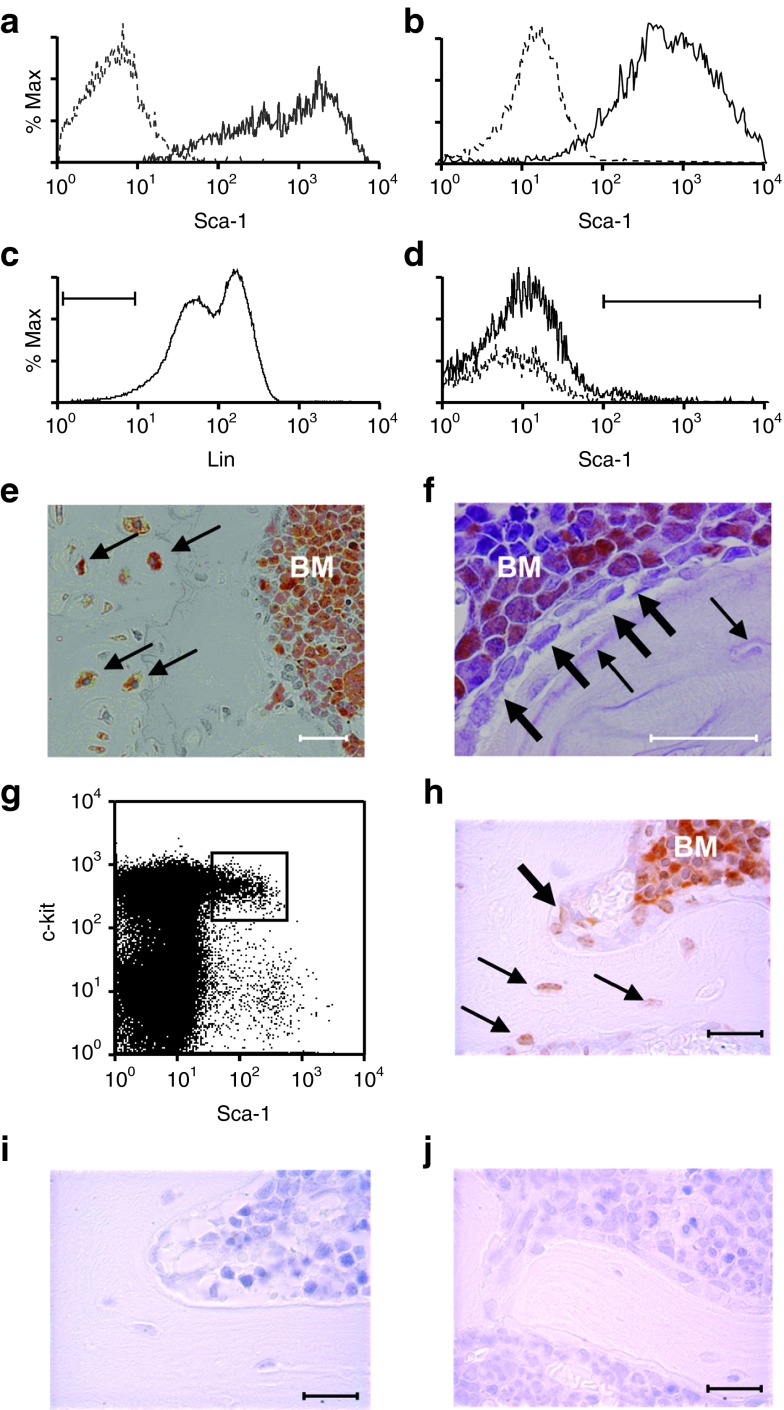
Figure 1.
Sca-1+ marrow cells engraft in bone and bone marrow (BM). Flow cytometric analysis demonstrating Sca-1 expression in primary cultures of (a) osteoblasts and (b) mesenchymal stem cells. Sca-1+ (—), isotype control (- - -). (c,d) Flow cytometric analysis of marrow cells showing the Lin− gate (~4.5%) and of Lin− cells showing the Sca-1+ gate (~1–2%), respectively. Representative immunohistochemical staining of green fluorescent protein (GFP) revealing donor-derived cells (orange) (e) in the bone and marrow cavity of mice (n = 5) transplanted with Lin− Sca-1+ cells (f) but only in the BM of mice (n = 5) transplanted with Sca-1− cells. Osteoblasts and osteocytes are indicated by thick and thin arrows, respectively. (g) Flow cytometric dot plot showing the gate of c-Kit+ Sca-1+ cells sorted from the Lin− cells analyzed in c. (h) Representative immunohistochemical staining of bone and BM from mice (n = 7) transplanted with GFP+ KLS cells (orange). Osteocytes and osteoblasts are designated as in f. (i) Negative control specimens from a mouse transplanted with KLS cells and stained with an isotype primary antibody or (j) from a wild-type mouse and stained with anti-GFP antiserum. Bar = 50 μm.
To further define the phenotype of the transplantable osteoprogenitor, we fluorescence-activated cell sorting (FACS)-sorted c-Kit+ Sca-1+ cells from lineage-depleted (Lin−) GFP transgenic marrow (Figure 1g) and transplanted 2,000 of these KLS cells (c-Kit+ Lin− Sca-1+) into each of five lethally irradiated mice. At 3 weeks post-transplantation, we observed >90% hematopoietic engraftment in each mouse and a mean (± SD) osteopoietic engraftment of 17.2 ± 5.6% (Figure 1h).
To investigate whether all of the osteopoietic activity of transplanted BM resides within the KLS cell fraction, we transplanted 1,000 FACS-sorted GFP+ KLS cells or 1,000 GFP+ BM cells depleted of KLS cells into lethally irradiated mice (n = 10 per group). At 3 weeks post-transplantation, the KLS grafts contributed to both hematopoiesis and osteopoiesis in all mice identical to the outcome shown in Figure 1h, whereas the marrow graft devoid of KLS cells gave rise to short-term hematopoiesis, but failed to engraft in bone, indistinguishable from the outcome after transplantation of Lin− Sca-1− cells (Figure 1f).
Transplantable osteoprogenitor activity exclusively resides among multilineage hematopoietic stem/progenitor cells
We FACS-sorted KLS cells into CD34+ and CD34− fractions (Figure 2a) and transplanted 200 of each cell population into lethally irradiated mice. In recipients of the CD34− KLS cells (n = 7), the mean (± SD) osteopoietic engraftment was 3.29 ± 1.34%, compared with 0.91 ± 0.38% in recipients (n = 4) of the CD34+ KLS cells (P = 0.007, Figure 2b). The low level of engraftment by the CD34+ cells was significantly higher (P = 0.02) than in the negative control (0.17 ± 0.06%, n = 3), indicating that the CD34+ KLS cells, although much less robust osteoprogenitors than their CD34− counterparts, still possessed osteopoietic activity.
Figure 2.
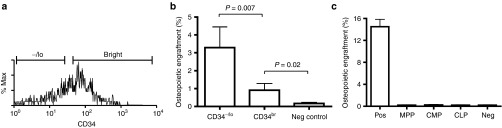
Engraftment of donor KLS cells. (a) Flow cytometric analysis of KLS cells showing the sort gates for the CD34−/lo and CD34bright subsets. (b) Comparison of osteopoietic engraftment rates for CD34bright versus CD34−/lo KLS cells. Mean (± SD) percentages of donor osteoblasts and osteocytes are shown. (c) Osteopoietic engraftment of multipotent progenitors (MPP), common myeloid progenitors (CMP), and common lymphoid progenitors (CLP). Positive controls (Pos) were mice transplanted with 1 × 106 unfractionated GFP+ marrow cells; negative controls (Neg) were mice transplanted with unlabeled marrow. GFP, green fluorescent protein.
To determine the extent of apparent hematopoietic cell differentiation while retaining osteoprogenitor potential, we transplanted multipotent progenitors (CD244+ CD150− CD48−),12 common lymphoid progenitors (c-Kit+ Lin− Sca-1+ CD127+)13 or common myeloid progenitors (c-Kit+ Lin− Sca-1−/lo CD127− CD34+FcγRlo)14 into lethally irradiated mice (n = 7 per group). None of these three cell populations contributed to osteopoiesis by 3 weeks post-transplantation (Figure 2c). These findings implicate the population of CD34− KLS cells, which are highly enriched for LT repopulating HSCs (LTR-HSCs),15 as the most robust transplantable osteoprogenitors within BM.
Hematopoietic stem/primitive progenitor cells specifically block transplantable osteoprogenitor activity
If the CD34− KLS marrow cells, indicative of the pool of primitive hematopoietic stem/progenitor cells, are the potent transplantable marrow osteoprogenitors as our data suggests, then donor-derived osteopoiesis after BMT, experimentally identified by GFP expression, should be competitively inhibited by cotransplantation of unlabeled CD34− KLS cells. To test this prediction, we transplanted 2 × 105 unfractionated, GFP-expressing marrow cells into lethally irradiated recipients either alone or with 2,000 unlabeled KLS cells, 20,000 KLS cells or 300 CD34− KLS cells (n = 5 mice per group) as competitor cells. At 3 weeks post-transplantation, the animals transplanted without competitor cells showed >95% GFP+ cells in whole BM as well as in the KLS marrow population (Figure 3a,b) and a mean (± SD) of 14.14 ± 2.11% donor osteopoiesis (Figure 3c). Both the lower and higher doses of unlabeled KLS competitors significantly reduced the percent contribution of the unfractionated GFP+ cells to short-term hematopoiesis (P = 0.008 and P < 0.001, respectively) (Figure 3a) as well as the reconstitution of the KLS progenitor pool (P = 0.04 and P < 0.001, respectively) (Figure 3b). Unlabeled CD34− KLS cells did not reduce the recovery of GFP+ hematopoiesis (P = 0.13) (Figure 3a), consistent with the abundance of mature precursors in unfractionated marrow and the relative paucity of such progenitors among CD34− KLS cells. By contrast, osteopoietic engraftment of GFP+ cells was strikingly diminished by cotransplantation with only 300 CD34− KLS cells (P < 0.001) (Figure 3c). These stem/primitive progenitor cells, identified by the CD34− KLS phenotype, demonstrated a 100- to 200-fold greater capacity to inhibit osteopoietic engraftment than did unsubfractionated KLS cells.
Figure 3.
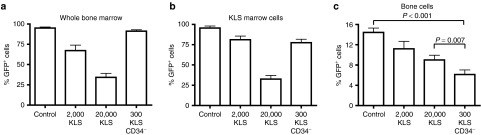
Competitive inhibition by primitive hematopoietic cells. (a) Reconstitution of whole bone marrow, (b) KLS hematopoietic marrow compartment, or (c) bone cell compartment, after transplantation of 2 × 105 GFP-expressing marrow cells alone (control) or with 2 × 103 or 2 × 104 unlabeled KLS cells, or 300 CD34− KLS competitor cells. Mean (± SD) percentages of GFP-expressing cells were determined by flow cytometry (in a,b) or immunohistochemical staining (in c). GFP, green fluorescent protein.
LTR-HSC have dual hematopoietic-osteopoietic differentiation capacity
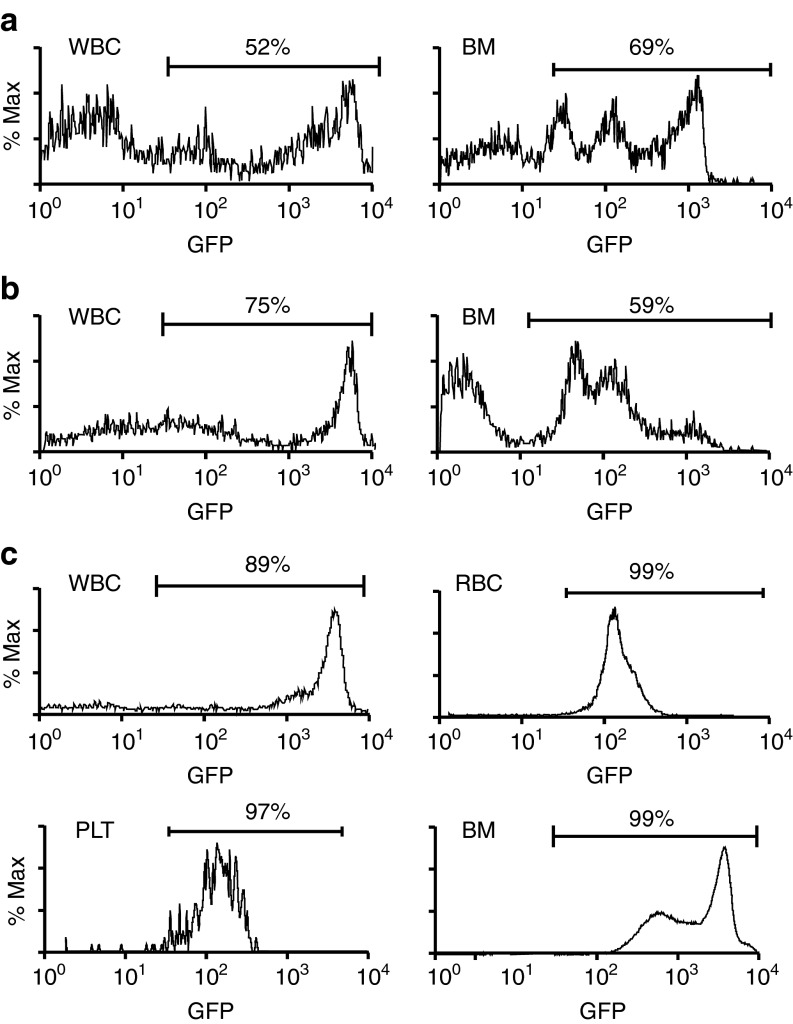
Our data suggest a dual hematopoietic-osteopoietic differentiation capacity for marrow cells exhibiting the CD34− KLS phenotype. Although not strictly homogeneous, CD34− KLS cells are considered to represent LTR-HSC.15 Collectively, then, our data raise the possibility that the LTR-HSC possess an osteopoietic differentiation potential. To rigorously test this idea, we transplanted single FACS-sorted GFP+ KLS cells into lethally irradiated mice (n = 60) that were also given 1 × 105 Sca-1− wild-type marrow cells to support short-term hematopoietic recovery. At 16 weeks post-transplantation, four of the mice showed a contribution to blood leukocytes (3–52%) and the BM (1.3–69%) from the single infused GFP+ cell (Figure 4a). Unfractionated BM cells (3 × 106) from the three mice with 26–69% GFP+ marrow reconstitution were then transplanted into lethally irradiated secondary recipients (n = 4 mice per primary recipient). At 3 weeks after secondary transplantation, all mice showed GFP+ hematopoiesis in blood (leukocytes, 56–75%, Figure 4b; erythrocytes, 32–42%; platelets, 40–72%), with 38–59% GFP+ engraftment in the BM (Figure 4b) of killed mice (n = 2 per primary recipient) demonstrating a contribution to short-term hematopoiesis. Evidence for self-renewal and LT hematopoietic repopulating capacity of the single KLS cells was provided in the secondary recipients (n = 2 per primary recipient transplanted), which were maintained for 18 weeks following transplantation. At that time, blood showed 69–89% GFP+ leukocytes, 52–99% GFP+ erythrocytes, and 45–97% GFP+ platelets; GFP+ donor engraftment was also observed in the BM (31–99%) (Figure 4c).
Figure 4.
Dual hematopoietic-osteopoietic engraftment of a single KLS cell. (a) Flow cytometric analysis showing high-level green fluorescent protein (GFP) expression among blood leukocytes (WBC) and unfractionated marrow cells (BM) from the primary recipient. (b) Flow cytometric analysis for GFP positivity among blood leukocytes and unfractionated marrow cells from a secondary recipient at 3 weeks post-transplantation of marrow from the primary recipient studied in a. (c) Flow cytometric analysis showing high level of GFP expression among blood leukocytes (WBC), blood erythrocytes (RBC), blood platelets (PLT), and unfractionated marrow cells (BM), collected from a secondary recipient at 18 weeks post-transplantation of marrow from the primary recipient studied in a. BM, bone marrow.
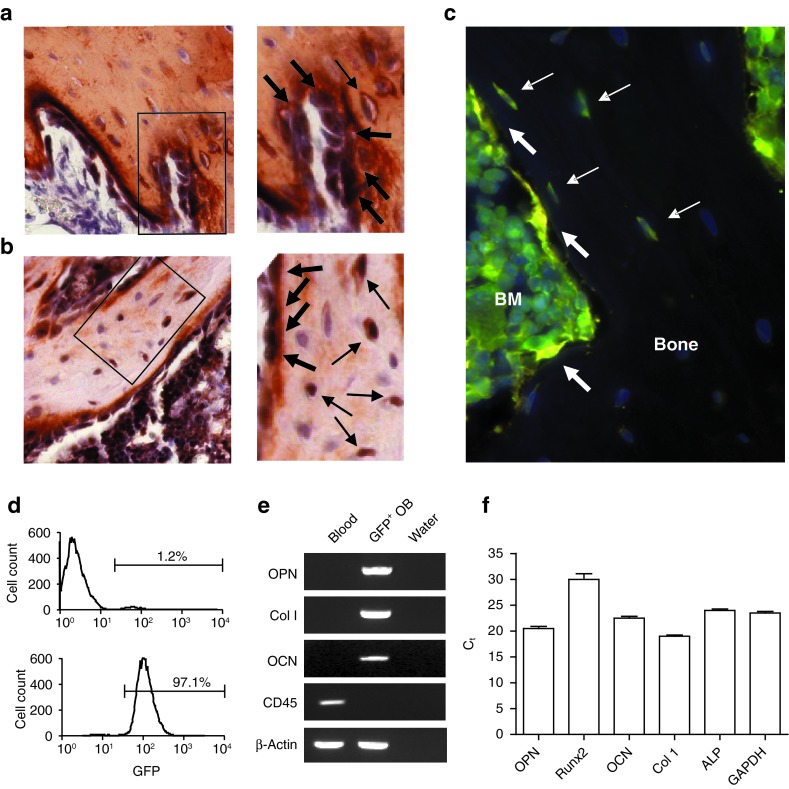
Strikingly, donor-derived (GFP+) osteopoiesis was evident within the epiphysis and metaphysis of the long bones of these secondary recipients (5.1–8.0%) at 3 weeks after transplantation, the optimal time to assess osteopoietic engraftment.16 Double immunohistochemical staining for collagen I and GFP (Figure 5a) and for osteocalcin and GFP (Figure 5b) identified the GFP+ cells as osteoblasts and osteocytes. Immunofluorescence microscopy shows donor-derived osteoblasts and osteocytes (Figure 5c). Both microscopy studies demonstrated donor-derived osteopoietic cells in clusters, a pattern consistent with our previous observations after conventional transplantation of unfractionated BM.16
Figure 5.
Engrafted cells express osteoblast markers. (a,b) Double immunohistochemical staining of bone taken from a secondary recipient 3 weeks post-transplantation was positive for (a) both collagen I (red) and GFP (black) and (b) both osteocalcin (red) and GFP (black). Bar = 50 μm. The panels to the right, are digitally enlarged images of the respective boxed regions in a and b. Osteoblasts and osteocytes are indicated by thick and thin arrows, respectively. (c) Immunofluorescence microscopy of bone from a secondary recipient strongly stained for GFP (green) and lightly stained for osteocalcin (red) to minimize background fluorescence. Three yellow osteoblasts (thick arrows) and four green-yellow osteocytes (thin arrows) are indicated. (d) Flow cytometric analyses of culture-expanded osteoblasts before (top) and after (bottom) FACS enrichment for GFP expression. (e) Reverse transcriptase-PCR analysis showing expression of osteopontin (OPN), collagen I (Col I), and osteocalcin (OCN), but not CD45, in the GFP-enriched osteoblasts from d. (f) Quantitative reverse transcriptase-PCR showing the expression of five osteoblast genes compared with the GAPDH expression in the same cell population as in e. BM, bone marrow; Ct, threshold cycle; FACS, fluorescence-activated cell sorting; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; GFP, green fluorescent protein.
To further validate GFP expression in osteoblasts, we FACS-sorted culture-expanded cells obtained from the bone of a secondary recipient to enrich for GFP expression, attaining a population that was >97% GFP-positive (Figure 5d). Reverse transcriptase-PCR demonstrated that these plastic-adherent GFP+ cells expressed osteopontin, collagen I, and osteocalcin, but not CD45, indicative of an osteoblast phenotype (Figure 5e). Quantitative reverse transcriptase-PCR demonstrated comparable expression of five bone-specific genes to the ubiquitously expressed GAPDH (glyceraldehyde 3-phosphate dehydrogenase) in this cell population confirming the osteoblast phenotype of these GFP+cells, which must be progeny of the originally transplanted, single GFP-expressing cell (Figure 5f).
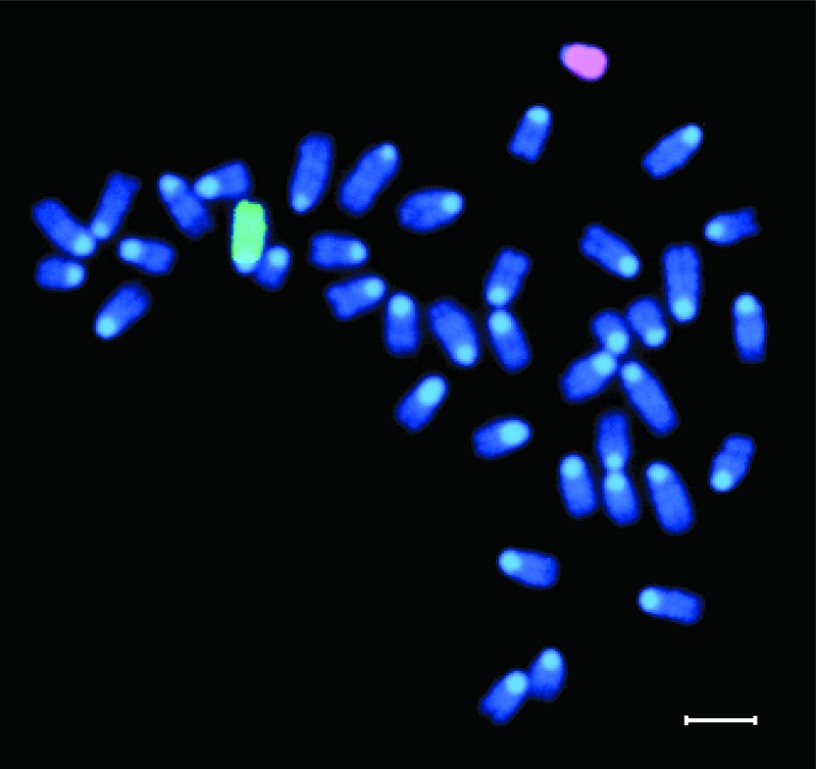
Our data demonstrate individual cells expressing markers of LTR-HSC origin (GFP) and osteoblasts/osteocytes suggesting differentiation of LTR-HSC to bone cells. Fusion of donor-derived hematopoietic cells with host osteoblasts in the secondary recipients was excluded by fluorescence in situ hybridization in culture-expanded, donor-derived (Y chromosome) osteoblasts (Figure 6).
Figure 6.
Representative cytogenetic analysis of cultured osteoblasts obtained from secondary recipients. The X (green) and Y (red) chromosomes were identified by fluorescence in situ hybridization, using chromosome-specific probes. Bar = 10 μm.
Discussion
In contrast to purported rare instances of HSC plasticity which have been effectively refuted,17 our data indicate a reproducible, robust osteopoietic differentiation of the LTR-HSC. This study was founded on several prior observations. First, we2,18 and others3,4,5,6 have shown that transplantation of BM cells (>99.9% hematopoietic cells) leads to donor-derived osteopoiesis in mice. Second, while MSCs are thought to be marrow osteoprogenitors, we have shown that non(plastic)-adherent marrow cells are far more robust osteoprogenitors than MSCs after intravenous transplantation.2 Third, the level of donor-derived osteopoiesis is saturable18 suggesting the transplantable osteoprogenitor engrafts at distinct, saturable sites in the marrow microenvironment. Finally, after transplantation, early hematopoietic engraftment seems to occur at discrete sites within the marrow space,19,20,21 and donor-derived osteopoiesis invariably mirrors the clusters of donor hematopoietic engraftment16,19 suggesting a common niche for both regenerating activities.
Our pilot experiment showing that transplanted Lin− Sca-1+ cells (2 × 105) resulted in a level of osteopoietic engraftment similar to what we observed after transplantation of tenfold more (2 × 106) unfractionated BM cells18 supported our hypothesis that primitive hematopoietic cells have dual hematopoietic-osteopoietic differentiation capacity. Had the observed engraftment been due to a contaminating adherent MSC then transplantation of an equal number of purified MSCs should have resulted in a similar amount of engraftment rather than the nearly tenfold less that was observed.
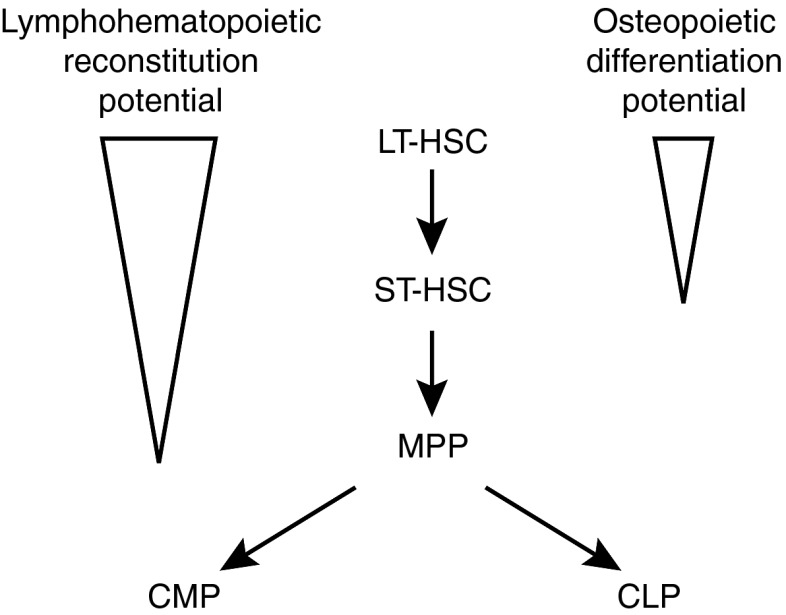
We next refined the phenotype of the transplantable osteoprogenitor to the population of KLS cells, which are thought to be solely hematopoietic progenitors further supporting the notion of a dual hematopoietic-osteopoietic activity among primitive marrow cells. These data, together with our previous observation that osteopoietic engraftment in mice is quite robust early after transplantation, but decreases over time,16 led us to consider that the putative bipotent progenitor cell might be a short-term repopulating HSC. While transplantation with CD34+ KLS cells, the marrow population containing the short-term repopulating HSC, gave low levels of osteopoietic engraftment, transplantation with CD34− KLS cells resulted in almost four times as much. Nonetheless, the low level of engraftment by the CD34+ cells was significantly greater than in the negative control. Although the latter finding could reflect contamination of the CD34+ fraction with CD34− cells, the more likely interpretation is that the osteopoietic capacity of CD34− KLS cells decreases with maturation. Hence, the most primitive stem cells in the marrow compartment, marked by the lack of CD34 expression, would have the greatest osteoprogenitor activity with a gradual loss of this potential as the cells mature, marked by increasing CD34 expression (progressively brighter fluorescence as measured by flow cytometry). Once the cells have committed to hematopoietic differentiation within a specific lineage, all of their osteopoietic capacity should be lost. This interpretation is supported by the lack of donor-derived osteopoiesis from transplanted multipotent progenitor, common myeloid progenitor or common lymphoid progenitor cells. These findings implicate the population of CD34− KLS cells, which are highly enriched for LTR-HSC,15 as the most robust transplantable osteoprogenitors within the BM and suggest that these primitive cells lose their osteopoietic differentiation capacity as they mature (Figure 7).
Figure 7.
Proposed model for relative hematopoietic and osteopoietic contributions of primitive hematopoietic progenitors after transplantation. Triangles indicate decreasing differentiation potential, with LT-HSCs having the strongest osteopoietic activity. ST-HSCs have low (but detectable) activity, whereas MPPs, CMPs, and CLPs lack osteopoietic potential altogether. CLP, common lymphoid progenitor; CMP, common myeloid progenitor; LT-HSC, long-term hematopoietic stem cell; MPP, multipotent progenitor; ST-HSC, short-term hematopoietic stem cell.
If our working hypothesis is correct, that primitive hematopoietic stem/progenitor cells identified by CD34− KLS expression engraft at discrete, saturable sites in the marrow space and generate local osteopoietic cells, then donor-derived osteopoiesis after BMT should be inhibited by cotransplantation of purified CD34− KLS cells, as these cells should compete with marrow CD34− KLS cells for engraftment. Indeed, cotransplantation of unlabeled CD34− KLS cells with GFP-expressing marrow cells dramatically reduced the contribution of unfractionated GFP+ marrow cells to osteopoiesis further substantiating the notion that the pool of CD34− KLS cells is the source of transplantable osteopoiesis and engrafts at defined, finite sites in the marrow microenvironment.
Although our previous report of a common retroviral integration site in clonogenic hematopoietic and osteopoietic cells after transplantation of unfractionated, gene-marked marrow cells provided compelling evidence of a common hematopoietic-osteopoietic primitive progenitor,2 our single cell secondary transplantation data are the sine qua non to establish dual hematopoietic-osteopoietic differentiation activity of LTR-HSC. We showed short-term and LT hematopoietic engraftment and the osteopoietic engraftment in mice transplanted with the BM from a mouse reconstituted from a single GFP+ KLS cell. While it is generally accepted that all GFP-expressing blood cells in secondary recipients, measured by flow cytometry, were derived from the single GFP+ cell transplanted into primary recipients, the potential pitfall of these studies is inadequate evidence of multi-tissue differentiation. In our study, we demonstrated that the GFP+ cells in bone were osteoblasts/osteocytes by histologic localization, bright field microcopy of double immunohistochemically stained bone, immunofluorescence microscopy of doubly stained bone, and gene expression analysis of GFP-enriched, culture-expanded cells (osteoblasts). Moreover, we excluded fusion as an alternative explanation by karyotype analysis. Thus, an LTR-HSC, defined by its capacity to generate GFP+ blood and marrow at 16 weeks post-transplantation and to undergo self-renewal in primary recipients, retained sufficient potency to contribute robustly to both hematopoiesis and osteopoiesis in secondary recipients, thereby satisfying the criteria for “stemness” as defined by Lagasse et al.22
Notably, donor-derived osteopoiesis ranged from 5 to 8% in our secondary recipients; however, we would stress that the maximal contribution of donor cells to the osteopoietic compartment after infusion of very high doses of marrow cells is about 15% of the bone cells in the metaphysis and epiphysis.18 The observed engraftment in bone after secondary transplantation, therefore, represents about a third to a half of the maximal level of engraftment. Since the donor origin of transplanted cells is identified by GFP expression and the transplanted marrow of the primary recipient was 26–69% GFP+, the magnitude of engraftment we observed in the secondary recipients was precisely what one would predict.
If single LTR-HSC contribute significantly to the osteopoietic compartment after transplantation into lethally irradiated mice, what might account for the relatively rapid decrease in donor-derived osteopoiesis following BMT?16 One explanation is that osteopoiesis in the immediate post-transplantation setting reflects a physiological response of donor HSC that have newly engrafted in endosteal niches. After restoration of the homeostatic condition of the niche, where HSC are maintained in a quiescent state, osteopoietic differentiation ceases. This interpretation argues that the osteopoietic activity of HSC is a genetically programmed, physiologically relevant response evident in the setting of hematopoietic cell transplantation, rather than a rare instance of developmental plasticity. Consistent with this view is the expression pattern of critical transcription factors. For example, Runx2, the master regulator of osteoblast differentiation from pluripotent mesenchymal progenitors,23 is highly expressed in HSCs but rapidly decreases with myeloid differentiation,24 analogous to the marked osteopoietic differentiation potential of primitive LT-HSC that decreases with maturation (Figures 2c and 3a,b). Similarly, Runx1, a key hematopoietic transcription factor25 essential for fetal liver hematopoiesis,26 also contributes to fetal skeletal development.27,28 In addition, after clinical hematopoietic cell transplantation, circulating collagen I-positive osteoprogenitors coexpress CD34 and/or CD45 and are derived from transplanted donor hematopoietic cells.29 Moreover, Singh et al.6 recently reported that BMT leads to engraftment of functional donor-derived osteoblasts as well as mesenchymal progenitors consistent with our current findings and confirming our prior observations.2,16,18 Efforts to harness this osteopoietic activity for the treatment of bone disorders or bone injury should be directed toward understanding the specific molecular cues that allow HSC to choose an osteoblastic fate.
Materials and Methods
Transplantation. All animal studies were approved by the Institutional Animal Use and Care Committees of The Children's Hospital of Philadelphia. BM was harvested from transgenic mice (FVB/N genetic background) expressing GFP under the control of the H2K promoter.11 The unfractionated or FACS-sorted (FACSVantage; BD Biosciences, San Jose, CA) marrow cells were transplanted into 4–6 week old lethally irradiated (1,125 cGy) FVB/N mice by tail vein injection within 2 hours after irradiation, as previously described.2 For the single cell transplant studies, marrow cells from our GFP-expressing transgenic (FVB/N) mice were sorted on a FACSVantage at 1 cell/well in a 96-well plate containing 200 μl Hank's balanced salt solution with 2% fetal bovine serum (Mediatech, Manassas, VA). Individual wells were screened by light and fluorescence microscopy to verify that they contained a single GFP-expressing cell. To each well, we added 1 × 105 wild-type Sca-1− cells, and then the entire cell graft was transplanted into lethally irradiated recipients as described above.
Immunohistochemical staining and analysis by microscopy. The immunohistochemical staining of bone sections (4-μm thickness) and microscopic evaluation of the metaphysis and epiphysis of femurs were performed as previously described.2,16 Three sections from each femur were studied and the mean (± SD) percentage of donor cells calculated. The number of animals evaluated in each experimental group is given in the text. Stained slides were examined on a Nikon E800 microscope (Nikon Instruments, Melville, NY) with either a 40x/0.95 N.A. or a 60x/0.95 N.A. dry objective. Photomicrographs were produced with the attached Nikon DXM 1200 color camera and Nikon ACT-1 Version 2.11 software (Nikon Instruments). Images were cropped and labeled using Photoshop CS4 and Illustrator CS4 (Adobe Systems, San Jose, CA).
Immunofluorescence staining and analysis by microscopy. Formalin-fixed specimens were decalcified, paraffin embedded and 3 μm thickness section were prepared for double immunofluorescence assays. The sections were deparaffinized, rehydrated, and permeabilized with phosphate-buffered saline (PBS) 1X containing 0.12% triton X-100. Antigen retrieval was performed with incubation in a chondroitinase ABC solution (0.35 IU/ml; Sigma-Aldrich, St Louis, MO) for 25 minutes. To block nonspecific binding of the antibodies, the sections were incubated for 45 minutes with 10% blocking reagent (Roche Diagnostics, Indianapolis, IN) dissolved in maleic buffer and fetal calf serum. Bone sections were incubated in a humidified chamber overnight at 4 °C with a solution of PBS 1X with 0.1% bovine serum albumin, 10% donkey serum (Jackson ImmunoResearch Laboratories, West Grove, PA), and 0.4% triton X-100. In this solution, were dissolved the two primary antibodies: rabbit anti-GFP (1:200, cat. no. ab290; Abcam, Cambridge, MA) and goat anti osteocalcin (1:100, cat. no. 7060-18-15; AbD Serotec, Raleigh, NC). After three washes in high salt PBS 1M, slides were incubated for 2 hours in the dark in the mixture of two secondary antibodies, with two different fluorochromes, composed by PBS 1X, 10% donkey serum, goat anti-rabbit (1:200, cat. no. 35552; Thermo Scientific, Waltham, MA) and donkey anti goat (1:200, cat. no. ab96933; Abcam) antibodies. After three washes in high salt PBS 1M, slides were mounted with a drop of mounting medium with DAPI (4′,6-diamidino-2-phenylindole) for DNA staining (Sigma). Slides were stored at 4 °C in dark. Stained slides were examined on an inverted microscope, Zeiss Axio Observer.A1 (Carl Zeiss MicroImaging, Thornwood, NY) with epi-fluorescence and a 40x/0.75 NA dry objective in a darkened room. Photomicrographs were acquired with AxioCam MRc5 color camera (Carl Zeiss MicroImaging) and AxioVision software. Images were adjusted for brightness, contrast, color balance by AxioVision LE.
Osteoblast isolation. Osteoblasts were isolated from freshly obtained bone and expanded in tissue culture as previously described.2
Flow cytometry. Blood or marrow cells were analyzed with a FACSAria flow cytometer (BD Biosciences), using commercially available antibodies (BD Biosciences or eBioscience, San Diego, CA). The data were analyzed with Cell Quest Pro Software, version 5.2.1 (BD Biosciences).
Reverse transcriptase-PCR. Total RNA was isolated from culture-expanded osteoblast using TRIzol Reagent (Invitrogen, Carlsbad, CA). First-strand cDNA was synthesized from 10 μg total RNA using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Foster City, CA), according to the manufacturer's protocol. PCR was performed with the HotStarTaq Master Mix (Qiagen, Valencia, CA). Conditions for amplification were 35 cycles at 95 °C for 1 minute, 53 °C (collagen I and osteocalcin), 55 °C (β-actin and osteopontin), or 57 °C (CD45) for 1 minute, and 72 °C for 1 minute. Primer sequences were as follows: osteopontin, 5′-TCACCATTCGGATGAGTCTG-3′ and 5′-ACTTGTGGCTCTGATGTTCC-3′ collagen I, 5′-AGCGGAGGTGGCTATGACTT-3′ and 5′-GCGCGGCTGTATGAGTTCTT-3′ osteocalcin, 5′-CTGACCTCACAGATGCCAAG-3′ and 5′-GGAGCTGCTGTGACATCC-3′ CD45, 5′-CTTCGACGGAGAGTTAATGC-3′ and 5′-GTCGCCTTAGCTTGACAACA-3′ β-actin, 5′-CATTGTGATGGACTCCGGAGACGG-3′ and 5′-CATCTCCTGCTCGAAGTCTAGAGC-3′. Amplified products were then identified by 2% agarose gel electrophoresis and visualized with ethidium bromide and a ultraviolet transilluminator.
Quantitative reverse transcriptase-PCR. Total RNA was isolated as above. First-strand cDNA was synthesized from 1 μg of total RNA using a revertAid H minus first-strand cDNA synthesis kit (Fermentas, Waltham, MA) according to the manufacturer's instructions. Quantitative real-time PCR was performed using the Applied Biosystems StepOne Real-Time PCR System and the Fast SYBR Green Master Mix reagent. Forward and reverse primers were designed using http://test.idtdna.com/home/home.aspx web site, specifying mRNA rather than genomic DNA by ensuring the selected primers spanned an intron sequence. Primer sequences were as follows: osteopontin, 5′-GTG ATT TGC TTT TGC CTG TTT G-3′ and 5′-GAG ATT CTG CTT CTG AGA TGG G-3′ runx2, 5′-GTA GCC AGG TTC AAC GAT CTG-3′ and 5′-CCG TCC ACT GTC ACT TTA ATA GC-3′ osteocalcin, 5′-CTGACCTCACAGATGCCAAG-3′ and 5′-GGAGCTGCTGTGACATCC-3′ collagen I, 5′-AAG GAT ACA GTG GAT TGC AGG-3′ and 5′-TCT ACC ATC TTT GCC AAC GG-3′ alkaline phosphatase, 5′-CTC CAA AAG CTC AAC ACC AAT G-3′ and 5′-ATT TGT CCA TCT CCA GCC G-3′ GAPDH, 5′-GCC TTC CGT GTT CCT ACC-3′ and 5′-CCT CAG TGT AGC CCA AGA TG-3′. Relative expression of the target gene was normalized to the endogenous β-actin gene as reference. The 2−ΔΔCt (cycle threshold) method,30 was used to calculate relative expression levels of the target genes.
Fluorescence in situ hybridization. Culture-expanded osteoblasts from the bone of secondary recipient mice were incubated for 4 hours in colcemid and harvested by standard methods. Directly labeled X and Y chromosome-specific paint probes were purchased from Applied Spectral Imaging (Vista, CA) and used in hybridization studies, according to the manufacturer's instructions. Chromosomes were analyzed as described previously.2
Statistical analysis. Engraftment rates were compared by Student's t-test (two-tailed) and one-way analysis of variance using Prism version 4 software (GraphPad, San Diego, CA), with a P value of <0.05 considered statistically significant.
Acknowledgments
The authors gratefully acknowledge Virginia Valentine for assistance with the fluorescence in situ hybridization studies. This work was supported in part by grant R01 HL077643 from the National Institutes of Health, St. Jude Children's Research Hospital Cancer Center Support (CORE) grant P30 CA21765, and grants from the Ministero Istruzione Università e Ricerca (PRIN 2006), the Regione Emilia Romagna, the Associazione per il Sostegno dell'Ematologia e dell'Oncologia Pediatrica (ASEOP), and Fondazione Cassa di Risparmio di Modena. The authors declared no conflict of interest.
References
- Scadden DT. The stem cell niche in health and leukemic disease. Best Pract Res Clin Haematol. 2007;20:19–27. doi: 10.1016/j.beha.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici M, Pritchard C, Garlits JE, Hofmann TJ, Persons DA, Horwitz EM. Hematopoietic cells and osteoblasts are derived from a common marrow progenitor after bone marrow transplantation. Proc Natl Acad Sci USA. 2004;101:11761–11766. doi: 10.1073/pnas.0404626101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson SK, Dooner MS, Weier HU, Frenkel B, Lian JB, Stein GS, et al. Cells capable of bone production engraft from whole bone marrow transplants in nonablated mice. J Exp Med. 1999;189:729–734. doi: 10.1084/jem.189.4.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmsted-Davis EA, Gugala Z, Camargo F, Gannon FH, Jackson K, Kienstra KA, et al. Primitive adult hematopoietic stem cells can function as osteoblast precursors. Proc Natl Acad Sci USA. 2003;100:15877–15882. doi: 10.1073/pnas.2632959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra M, Rosol M, Ogawa M, Larue AC. Amelioration of a mouse model of osteogenesis imperfecta with hematopoietic stem cell transplantation: microcomputed tomography studies. Exp Hematol. 2010;38:593–602. doi: 10.1016/j.exphem.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh L, Brennan TA, Kim JH, Egan KP, McMillan EA, Chen Q, et al. Long-term functional engraftment of mesenchymal progenitor cells in a mouse model of accelerated aging. Stem cells. 2012;31:607–611. doi: 10.1002/stem.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz EM, Prockop DJ, Fitzpatrick LA, Koo WW, Gordon PL, Neel M, et al. Transplantability and therapeutic effects of bone marrow-derived mesenchymal cells in children with osteogenesis imperfecta. Nat Med. 1999;5:309–313. doi: 10.1038/6529. [DOI] [PubMed] [Google Scholar]
- Horwitz EM, Prockop DJ, Gordon PL, Koo WW, Fitzpatrick LA, Neel MD, et al. Clinical responses to bone marrow transplantation in children with severe osteogenesis imperfecta. Blood. 2001;97:1227–1231. doi: 10.1182/blood.v97.5.1227. [DOI] [PubMed] [Google Scholar]
- Marini JC. Osteogenesis imperfecta calls for caution. Nat Med. 1999;5:466–467. doi: 10.1038/8326. [DOI] [PubMed] [Google Scholar]
- Horwitz EM, Gordon PL, Koo WK, Marx JC, Neel MD, McNall RY, et al. Isolated allogeneic bone marrow-derived mesenchymal cells engraft and stimulate growth in children with osteogenesis imperfecta: Implications for cell therapy of bone. Proc Natl Acad Sci USA. 2002;99:8932–8937. doi: 10.1073/pnas.132252399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici M, Tadjali M, Kepes S, Allay ER, Boyd K, Ney PA, et al. Transgenic mice with pancellular enhanced green fluorescent protein expression in primitive hematopoietic cells and all blood cell progeny. Genesis. 2005;42:17–22. doi: 10.1002/gene.20121. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- Dominici M, Marino R, Rasini V, Spano C, Paolucci P, Conte P, et al. Donor cell-derived osteopoiesis originates from a self-renewing stem cell with a limited regenerative contribution after transplantation. Blood. 2008;111:4386–4391. doi: 10.1182/blood-2007-10-115725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagers AJ, Sherwood RI, Christensen JL, Weissman IL. Little evidence for developmental plasticity of adult hematopoietic stem cells. Science. 2002;297:2256–2259. doi: 10.1126/science.1074807. [DOI] [PubMed] [Google Scholar]
- Marino R, Martinez C, Boyd K, Dominici M, Hofmann TJ, Horwitz EM. Transplantable marrow osteoprogenitors engraft in discrete saturable sites in the marrow microenvironment. Exp Hematol. 2008;36:360–368. doi: 10.1016/j.exphem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici M, Rasini V, Bussolari R, Chen X, Hofmann TJ, Spano C, et al. Restoration and reversible expansion of the osteoblastic hematopoietic stem cell niche after marrow radioablation. Blood. 2009;114:2333–2343. doi: 10.1182/blood-2008-10-183459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askenasy N, Zorina T, Farkas DL, Shalit I. Transplanted hematopoietic cells seed in clusters in recipient bone marrow in vivo. Stem Cells. 2002;20:301–310. doi: 10.1634/stemcells.20-4-301. [DOI] [PubMed] [Google Scholar]
- Askenasy N, Stein J, Yaniv I, Farkas DL. The topologic and chronologic patterns of hematopoietic cell seeding in host femoral bone marrow after transplantation. Biol Blood Marrow Transplant. 2003;9:496–504. doi: 10.1016/s1083-8791(03)00150-2. [DOI] [PubMed] [Google Scholar]
- Lagasse E, Shizuru JA, Uchida N, Tsukamoto A, Weissman IL. Toward regenerative medicine. Immunity. 2001;14:425–436. doi: 10.1016/s1074-7613(01)00123-6. [DOI] [PubMed] [Google Scholar]
- Komori T. Regulation of osteoblast differentiation by Runx2. Adv Exp Med Biol. 2010;658:43–49. doi: 10.1007/978-1-4419-1050-9_5. [DOI] [PubMed] [Google Scholar]
- Kuo YH, Zaidi SK, Gornostaeva S, Komori T, Stein GS, Castilla LH. Runx2 induces acute myeloid leukemia in cooperation with Cbfbeta-SMMHC in mice. Blood. 2009;113:3323–3332. doi: 10.1182/blood-2008-06-162248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano K, Kurokawa M. The role of Runx1/AML1 and Evi-1 in the regulation of hematopoietic stem cells. J Cell Physiol. 2010;222:282–285. doi: 10.1002/jcp.21953. [DOI] [PubMed] [Google Scholar]
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- Lian JB, Balint E, Javed A, Drissi H, Vitti R, Quinlan EJ, et al. Runx1/AML1 hematopoietic transcription factor contributes to skeletal development in vivo. J Cell Physiol. 2003;196:301–311. doi: 10.1002/jcp.10316. [DOI] [PubMed] [Google Scholar]
- Liakhovitskaia A, Lana-Elola E, Stamateris E, Rice DP, van ‘t Hof RJ, Medvinsky A. The essential requirement for Runx1 in the development of the sternum. Dev Biol. 2010;340:539–546. doi: 10.1016/j.ydbio.2010.02.005. [DOI] [PubMed] [Google Scholar]
- Suda RK, Billings PC, Egan KP, Kim JH, McCarrick-Walmsley R, Glaser DL, et al. Circulating osteogenic precursor cells in heterotopic bone formation. Stem Cells. 2009;27:2209–2219. doi: 10.1002/stem.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan JS, Wang D, Stewart CN., Jr Statistical methods for efficiency adjusted real-time PCR quantification. Biotechnol J. 2008;3:112–123. doi: 10.1002/biot.200700169. [DOI] [PubMed] [Google Scholar]