SUMMARY
De novo DNA methylation is an essential aspect of the epigenetic reprogramming that takes place during early development, yet factors responsible for its instatement at particular genomic loci are poorly defined. Here, we demonstrate that the KRAB-ZFP-mediated recruitment of KAP1 to DNA in embryonic stem cells (ESCs) induces cytosine methylation. This process is preceded by H3K9 trimethylation, and genome-wide analyses reveal that it spreads over short distances from KAP1-binding sites so as to involve nearby CpG islands. In sharp contrast, in differentiated cells, KRAB/KAP1-induced heterochromatin formation does not lead to DNA methylation. Correspondingly, the methylation status of CpG islands in the adult mouse liver correlates with their proximity to KAP1-binding sites in ESCs, not in hepatocytes. Therefore, KRAB-ZFPs and their cofactor KAP1 are in part responsible for the establishment during early embryogenesis of site-specific DNA methylation patterns that are maintained through development.
INTRODUCTION
The genome-wide site-specific methylation of cytosine residues is a key epigenetic contributor to the identity, fate, and transcriptional activity of a cell. This process is tightly regulated during development (Reik, 2007; Saitou et al., 2012). A set of enzymes partakes in the deposition and maintenance of the 5-methylcytosine (5mC) mark. DNA methyltransferase (DNMT) 3a and DNMT3b, in conjunction with DNMT3L, recognize nonmethylated DNA and catalyze de novo cytosine methylation, while DNMT1, recruited at the replication fork by PCNA and NP95 (also known as UHRF1), is responsible for the maintenance of this modification during cell division (Sharif et al., 2007). In contrast, what tethers de novo DNMTs to specific genomic loci remains ill defined, although an association with chromatin changes, notably methylation of histone 3 at lysine 9 (H3K9), has been noted in some circumstances. For instance, the H3K9 mono- and dimethyltransferase G9a appears to be important for de novo DNA methylation of the OCT4 promoter (Feldman et al., 2006) and of murine leukemia virus (MLV)-derived retroviral vectors in embryonic stem cells (ESCs) (Leung et al., 2011), the H3K9 trimethyltransferase SETDB1 was found to recruit DNMT3a at tumor suppressor genes in cancer cells (Li et al., 2006), and the H3K36me3 mark was observed to foster DNA methylation (Dhayalan et al., 2010). This is reminiscent of the interplay between chromatin changes and DNA methylation that has been documented in plants and fungi (Jackson et al., 2002; Tamaru and Selker, 2001).
The first four days of embryonic development are characterized by a genome-wide wave of demethylation. However, this reprogramming spares imprinted loci and is incomplete over sequences derived from retrotransposons such as endogenous retroviruses (ERVs) and long interspersed nuclear elements (LINEs). DNA methylation is then rapidly re-established by the time of implantation. During this window of epigenetic instability, both DNA demethylating and de novo methylating activities are expressed. How their opposite influences lead to specific methylation patterns remains unclear, although the role of cis-acting elements that autonomously determine DNA methylation states during this period was recently revealed (Lienert et al., 2011b; Saitou et al., 2012).
Recent evidence implicates the KRAB/KAP1 epigenetic regulation system in the early embryonic control of DNA methylation. With almost 400 members in both human and mouse, KRAB-containing zinc finger proteins (KRAB-ZFPs) constitute the largest family of transcriptional regulators encoded by higher vertebrates (Emerson and Thomas, 2009). Using their C-terminal array of C2H2 zinc fingers for DNA binding, KRAB-ZFPs recruit their universal cofactor KAP1 (also known as TRIM28 or TIF1-beta) via their N-terminal KRAB domain. KAP1 then acts as a scaffold for chromatin-modifying complexes that, notably, comprise histone deacetylases and the histone methyltransferase SETDB1, leading to histone deacetylation, deposition of H3K9me3, binding of HP1, formation of heterochromatin, and transcriptional silencing (Friedman et al., 1996; Moosmann et al., 1997). KAP1 and KRAB-ZFPs were recently shown to silence ERVs and murine leukemia virus in ESCs (Rowe et al., 2010; Wolf and Goff, 2007), and this system was implicated in DNA methylation maintenance of imprints during early development. (Li et al., 2008; Quenneville et al., 2011). Finally, artificial tethering of KAP1 to the body of a lentiviral vector leads to DNA methylation and permanent silencing of an adjacent promoter when allowed to occur during the first few days of embryogenesis (Wiznerowicz et al., 2007).
In this study, we provide formal evidence that the KRAB/KAP1-mediated sequence-specific recognition of thousands of genomic loci in ESCs leads to their DNA methylation, thus ensuring the genome-wide establishment of site-specific epigenetic marks that are subsequently maintained during development.
RESULTS
KRAB/KAP1-Induced De Novo DNA Methylation in ESCs
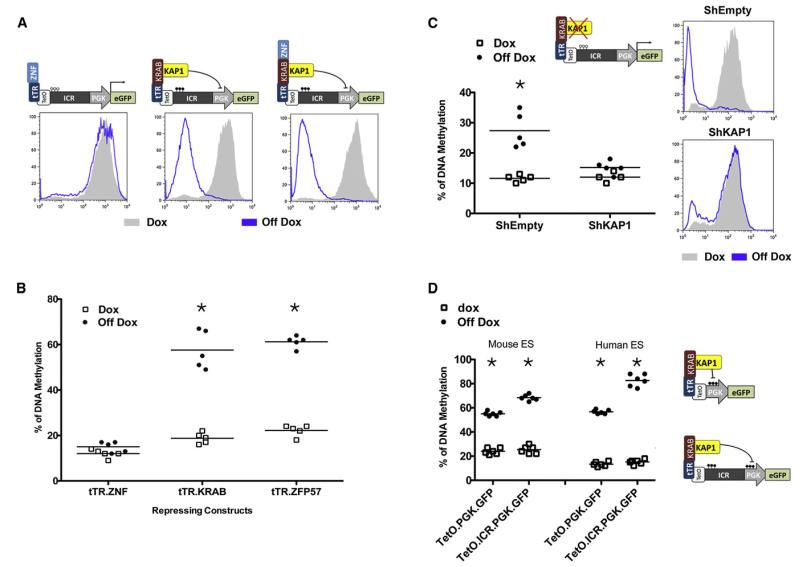
In order to investigate the role of KRAB-ZFPs and KAP1 in de novo methylation, we first used a lentiviral vector-based system suitable for monitoring both DNA methylation and epigenetic silencing in either mouse or human ESCs. A first set of vectors expressed a Tet repressor (tTR) fused to different parts of ZFP57, the KRAB-ZFP involved in the maintenance of imprinting (Li et al., 2008; Quenneville et al., 2011). The second set contained a PGK.GFP expression cassette downstream of TetO repeats, with or without a 2 kb KvDMR1 imprinted control region (ICR) as intervening sequence. We previously demonstrated that, in this configuration, tTR fusion proteins bind TetO in a doxycycline-preventable manner (Wiznerowicz and Trono, 2003). We first engineered murine ESC lines to produce the various tTR derivatives, before transduction with the TetO.ICR.PGK.GFP vector in the presence or absence of Dox. Three weeks later, we examined GFP expression and ICR DNA methylation, respectively, by fluorescence-activated cell sorting (FACS) (Figure 1A) and pyrosequencing (Figure 1B). In the presence of Dox, all cell lines exhibited high levels of GFP and the ICR displayed low rates (<20%) of CpG methylation. In contrast, when doxycycline was omitted; hence, when tTR.ZFP57 or tTR.KRAB was allowed to bind its TetO target, GFP production was silenced and the ICR was methylated to about 60%. Both GFP repression and tTR-induced ICR methylation were absent in tTR.ZNF-expressing cells, irrespective of Dox exposure, consistent with the KAP1-docking dependence of these processes.
Figure 1. KRAB/KAP1-Induced De Novo DNA Methylation in ESCs.
(A) Murine ESC expressing indicated tTR derivatives were transduced with a lentivector containing TetO repeats upstream of an ICR and a PGK.GFP cassette and examined by FACS after 2 days of culture with or without doxycycline.
(B) Methylation of ICR DNA, measured by pyrosequencing 3 weeks after introduction of the lentivector into murine ESCs expressing tTR fusion proteins (full-length ZFP57 [tTR.ZFP57], KRAB-deleted [tTR.ZNF], KRAB-domain [tTR.KRAB]).
(C) TetO.ICR.PGK.GFP and tTR.KRAB vectors were introduced in control (sh-empty) or KAP1-depleted (shKAP1) human ESCs, subsequently analyzed as in (A) and (B). Transcriptional repression and ICR DNA methylation are both abrogated in Kap1 KD cells.
(D) The PGK promoter contained in indicated vectors becomes methylated in both mouse and human ESCs, with a slight increase when the ICR is inserted upstream.
(B–D) *Statistically significant (p < 0.01).
In mouse ESCs, KAP1 deletion leads to growth arrest, differentiation, and ultimately cell death (Rowe et al., 2010). However, human ESCs can maintain pluripotent self renewal after shRNA-mediated knockdown of the master regulator (D.T., unpublished data). We thus tranduced a human ESC line expressing tTR.KRAB with either a control (shEMPTY) or a Kap1-specific (shKAP1) shRNA lentiviral vector that reduced the Kap1 mRNA level to less than 10% that of wild-type (data not shown). We then introduced TetO.ICR.PGK.GFP in these cells, in the presence or absence of Dox. Without the drug, shEMPTY-transduced human ESCs efficiently repressed GFP expression, while this phenotype was almost completely abrogated in the Kap1 knockdown cells (Figure 1C). As a corollary, methylation of the lentivector-contained ICR occurred in control cells, but was prevented by KAP1 depletion (Figure 1C). Together, these results demonstrate that human ESCs support KRAB-induced de novo DNA methylation and confirm the KAP1 dependence of this process on the PGK promoter and KvDMR1 ICR.
To assess the possible impacts of the ICR sequence and of the distance between the KAP1-docking site and the promoter, we examined in parallel an ICR-devoid TetO.PGK.GFP vector, in which the transcription start site is about 2 kb closer to the TetO motif. Pyrosequencing-based examination of six CpGs situated within the PGK promoter revealed that these readily underwent KAP1-recruitment-dependent de novo DNA methylation, whether or not the ICR was present (Figure 1D). If anything, 5mC deposition at the PGK promoter was higher in the presence of the intervening sequence, demonstrating not only that DNA methylation could spread at some distance from the KAP1-docking site, but also that the KvDMR1 ICR may contain methylation-promoting elements.
Differential Kinetics of KRAB/KAP1-Induced Transcriptional Repression and De Novo DNA Methylation
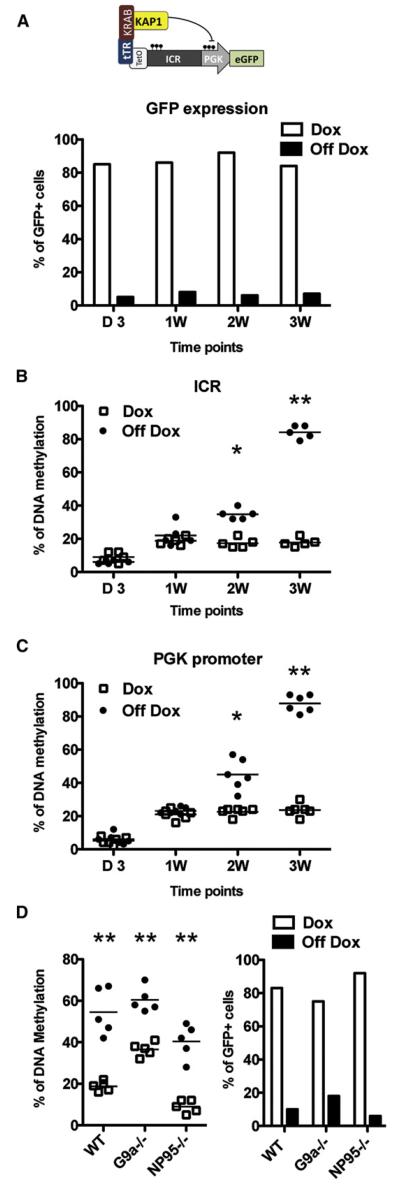
With a similar system, we previously demonstrated that transcriptional repression is driven by the deposition of repressive histone marks at the promoter and initially does not reflect DNA methylation, which only later establishes a secondary state of irreversible silencing (Wiznerowicz et al., 2007; Wiznerowicz and Trono, 2003). KAP1-mediated GFP silencing was immediately apparent in our system (Figure 2A), confirming that histone-mediated transcriptional repression proceeds very rapidly. To determine the underlying kinetics of KRAB/KAP1-induced DNA methylation, we examined 5mC deposition at the CpG dinucleotides contained in both the ICR and PGK promoter in murine ESCs at a series of time points after transduction with the TetO.ICR.PGK.GFP vector (Figures 2B and 2C). At day 3, levels of DNA methylation were extremely low over both sequences. By 1 week, they raised to about 20%, and they stayed at this level if Dox was present in the culture. If tTR.KRAB was allowed to bind the TetO motif, CpG methylation progressively increased to reach more than 80% at both the ICR and the PGK promoter within 3 weeks.
Figure 2. De Novo Methylation Is Progressive and Independent from G9a and NP95.
(A–C) In the absence of Dox, GFP expression is repressed rapidly and stays low (A), whereas DNA methylation at ICR (B) and PGK promoters (C) slowly increases over 3 weeks.
(D) DNA methylation at ICR is significantly increased upon KAP1 docking (off Dox) in both wild-type and G9a or NP95 KO murine ESCs (left), correlating GFP repression (right). *p < 0.05, * *p < 0.01.
G9a and NP95 Independence of KRAB/KAP1-Induced De Novo DNA Methylation
It has been reported that G9a histone methyltransferase is needed for methylation of an MLV-derived retroviral vector in murine ESCs (Leung et al., 2011), and it was proposed that NP95 plays a similar role for the CMV immediate early promoter (Meilinger et al., 2009). To find out whether these findings related to KRAB/KAP1-dependent events, we stably expressed tTR.KRAB into murine ESCs knocked out for either one of these factors (Dong et al., 2008; Sharif et al., 2007) and transduced the resulting cell lines with the TetO.ICR.PGK.GFP lentiviral vector. In both settings, KAP1-induced GFP repression was fully preserved and Dox-controllable ICR methylation was maintained. Indeed, both baseline and induced ICR methylation levels were higher in G9a knockout than in control ESCs, whereas both were lower in NP95−/− cells, as anticipated from a defect in the maintenance of this modification (Figure 2D). Because the deletion of SETDB1 rapidly leads to ESC growth arrest and death, we could not test the importance of this known partner of KAP1 (Matsui et al., 2010) in KAP1-induced DNA methylation.
Spreading of KAP1-Induced DNA Methylation to Nearby CpG Islands
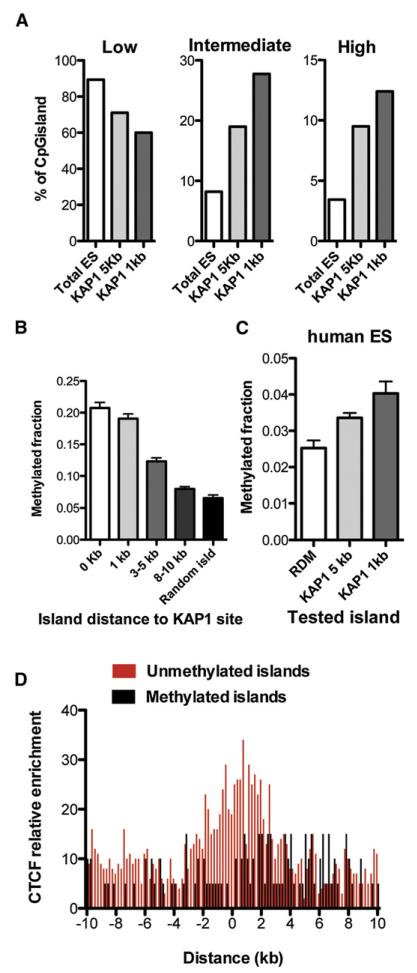
In order to assess the impact of KAP1 genomic recruitment and patterns of DNA methylation in ESCs, we used a combination of publically available and in-house ChiP-seq and CpG methylation data, for both murine and human ESCs (Lienert et al., 2011a; Meissner et al., 2008; Stadler et al., 2011). Considering the very strong levels of overall DNA methylation of the genome, we concentrated our analysis on so-called CpG islands, which are highly enriched in CpG dinucleotides and are typically hypomethylated. We first classified CpG islands in three subsets according to their DNA methylation status in murine ESCs: low (less than 20% methylation), intermediate (between 20% and 80%) and high (more than 80%) (Figure 3A). Of the total CpG island pool, 89% was found in the low methylation subset, whereas only 8% and 3% exhibited intermediate and high levels of this modification, respectively. However, when CpG islands located less than 5 kb (591) or less than 1 kb (324) from a KAP1-binding site were examined, the fraction with low methylation levels decreased to 70% and 60%, respectively Correspondingly, the proportion of CpG islands with intermediate and high levels of DNA methylation increased, reaching 27% in intermediate and 12% in highly methylated categories for islands situated within 1 kb of a KAP1-binding site (p < 10−15). To further examine the distance at which KAP1 can promote the methylation of a CpG island, we performed a reverse analysis, in which the average methylation levels of CpG islands located at given distances from a KAP1-binding site were determined (Figure 3B). CpG islands coinciding precisely with a KAP1 site (0 kb) had the highest average methylation score, while a relatively rapid decrease was noted afterward, CpG islands situated between 8 and 10 kb from a KAP1 site displaying profiles no different from a random pool of these sequence elements. These results, obtained on the basis of the analysis of CpG-rich sequences, were confirmed when elements identified as CpG islands were examined in the mouse genome through biochemical purification (Figure S1). In human ESCs, we used reduced representation bisulfite sequencing (RRBS) data (Meissner et al., 2008) and similarly found that levels of methylation of CpG islands were inversely proportional to their distance to a KAP1-binding site (Figure 3C). Remarkably, in both murine and human ESCs, a strong enrichment in CTCF-binding sites was seen in the space separating KAP1 landing sites and unmethylated KAP1-close CpG islands, compared with their methylated counterparts (Figure 3D). In this setting, CTCF may thus act as a barrier element, or directly counteract the deposition of methylation marks, as previously suggested (Stadler et al., 2011).
Figure 3. Short-Range Spreading of KRAB/KAP1-Associated DNA Methylation.
(A) Distribution of CpG islands among groups exhibiting low (<20%), intermediate (>20%, <80%) and high (>80%) levels of methylation, their entire pool (Total ES) compared with the subsets located 5 kb and 1 kb from KAP1-binding sites in murine ESCs.
(B) Methylation levels of CpG islands in mouse ESCs, according to distance from KAP1-binding sites. Fischer’s exact test shows significance (p < 0.001) until 8–10 kb (225–532 islands considered per group).
(C) KAP1-close CpG islands (n = 758 for 5 kb and n = 224 for 1 kb) also exhibit higher methylation levels in human ESCs (RDM, randomly selected islands). Measured methylation levels are lower than in murine ESCs due to the use of RRBS (as opposed to full-genome sequencing).
(D) Anticorrelation of CTCF enrichment and DNA methylation on KAP1-close CpG islands in murine ESCs. The 0 kb point represents the middle of the distance between KAP1-binding sites and CpG islands, restricting this in silico analysis to islands situated 5 kb or less from a KAP1 site.
KRAB/KAP1-Induced Repression and DNA Methylation Patterns in Adult Tissues
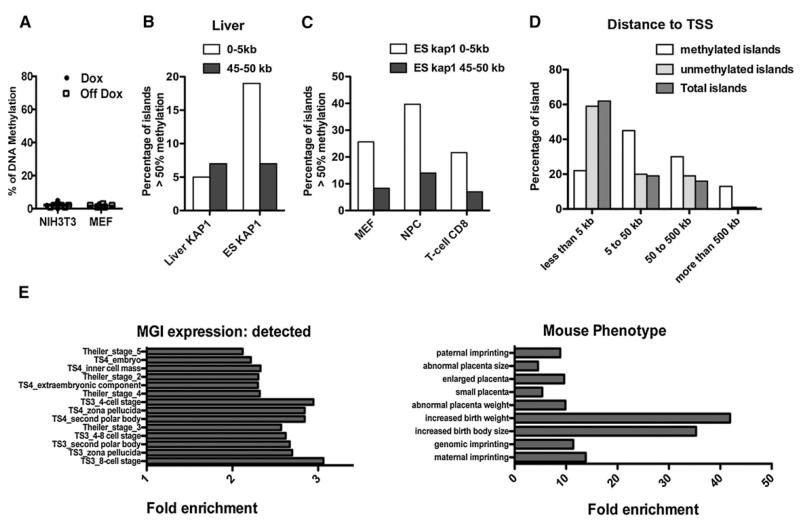
We previously observed that KRAB/KAP1-induced transcriptional repression is reversible when triggered in somatic cells (Wiznerowicz and Trono, 2003). This suggested that, in this setting, docking of the silencing complex does not lead to DNA methylation. To ascertain this point, we introduced the TetO.ICR.PGK.GFP vector in tTR.KRAB-expressing NIH 3T3 cells and primary mouse embryonic fibroblasts (MEFs). In either cell type, Dox-controllable GFP repression was fully operational, but no significant methylation of the ICR could be detected (Figure 4A). DNMT3a, DNMT3b, or DNMT3L overexpression in NIH 3T3 (Figure S2) did not trigger any increase in KRAB/KAP1-induced DNA methylation, indicating that other factors account for the ESC specificity of this process. To confirm the general implications of this finding, we examined a possible relationship between the methylation status of CpG islands in the mouse liver, as previously determined by RRBS (Meissner et al., 2008), with that of KAP1-binding sites mapped in this organ by ChIP-seq (Bojkowska et al., 2012). There was no correlation between these two parameters (Figure 4B). Instead, the likelihood that a CpG island was found to be methylated in the liver was conditioned by its proximity to a KAP1-binding site in ESCs, a relationship that was also observed in three other somatic cell types (Figure 4C). Thus, whereas KRAB and KAP1 do not govern DNA methylation in differentiated tissues, site-specific methylation marks established by this system in early embryogenesis are maintained through development.
Figure 4. KRAB/KAP1-Mediated De Novo Methylation Induces the Early Embryonic Setting of Permanent Epigenetic Marks.
(A) Same experiment as in Figures 1A and 1B, in indicated mouse somatic cells. KAP1 recruitment does not induce DNA methylation.
(B) DNA methylation in the mouse liver of CpG islands close (0–5 kb) or far (45–50 kb) from liver KAP1 sites (liver KAP1, n = 492 and 552, respectively) or in ESCs (ES KAP1, n = 591 and 429). A t test (Mann-Whitney U) reveals a difference (p < 0.001) between ES KAP1 groups but not between liver KAP1 groups.
(C) Higher levels of DNA methylation in somatic cells (mouse embryonic fibroblasts, neural progenitor cells, and CD8-positive T cells) for islands located close to KAP1-binding sites in ESCs. All groups are of similar size (n between 538 and 655), and a t test (Mann-Whitney U) reveals a difference (p < 0.001) between close and distant sites.
(D) The distribution of KAP1-close (<5 kb) methylated, unmethylated, and total CpG islands demonstrates that methylated islands are usually remote from total Refseq TSSs.
(E) Gene ontology analyses (GREAT) of transcriptional units situated near ESCs and KAP1-close methylated CpG islands identifies genes expressed in preimplantation development and in extraembryonic tissues (left) and genes, the inactivation of which results notably in imprinting and early developmental phenotypes (right).
While KRAB/KAP1-induced DNA methylation is expected to be functionally beneficial at ERVs and ICRs, the resulting permanent inactivation of its other targets, such as promoters or enhancers of cellular genes, could be problematic. In that respect, our genome-wide exploration revealed interesting features. Methylated CpG islands were on average positioned farther from transcriptional start sites (TSSs) than their unmethylated counterparts, suggesting that they functioned in their majority as enhancers rather than promoters (Figure 4D), while the few TSSs (26) close to these CpG islands were transcriptionally inactive (Figure S3). Also, KAP1-close CpG islands were often linked to genes active during the earliest phases of embryogenesis and in extraembryonic tissues; that is, not likely to suffer from the functional inactivation of cis-acting transcriptional activators in inner cell-mass- or epiblast-derived ESCs (Figure 4E).
DISCUSSION
The present study demonstrates that the site-specific KRAB/KAP1-mediated induction of heterochromatin in ESCs can lead to de novo DNA methylation and suggests a mechanism for the establishment of this epigenetic mark during early embryogenesis at many genomic loci found to be methylated in adult tissues. Our data support a model in which, at KRAB-ZFP-targeted sites, KAP1 and associated chromatin modifiers, including DNA methyltransferases, directly counter the DNA-demethylating forces that are active across the genome during this period. This process is particularly relevant for endogenous retroviruses, which constitute primary targets of KRAB/KAP1-mediated silencing and most likely drove the positive selection undergone by KRAB-ZFP genes during evolution. Our data suggest that the sequence-specific recognition of ERVs by KRAB-ZFPs, most of which are expressed in ESCs but shut down shortly thereafter (unpublished data), leads to their permanent silencing by DNA methylation, alleviating the need for continuous expression of their cognate trans-repressors. Although emanating from the same general mechanism, the control of imprinted loci represents an interesting variant, since the responsible KRAB-ZFP, ZFP57, recognizes only the methylated version of a motif present in all ICRs (Quenneville et al., 2011). This elegantly ensures that methyl marks are preserved on the imprinted allele during the wave of reprogramming that takes place after fertilization, while avoiding their instatement on the nonimprinted allele.
On the basis of the analysis of CpG islands at least, the spreading of KAP1-induced DNA methylation was limited in ESCs, averaging no more than 3 to 5 kb from the KAP1-docking sites. While this could reflect the rapid exhaustion of a putative heterochromatin-propagating activity, the local recruitment of counteracting factors such as CTCF may play a role. Our observation of limited KAP1-induced heterochromatin spreading is also consistent with the description of DNA methylation occurring in mouse ESCs over the 5′ long terminal repeat and genomic flanking sequences of a retroviral vector, but not over its 3′ regions (Berwin and Barklis, 1993). Such short-range propagation also explains how KRAB/KAP1 might be involved in controlling tens of thousands of endogenous retroelements during early embryogenesis without triggering the complete methylation-mediated blockade of the host genome.
In 3T3 or primary murine embryonic fibroblasts, KRAB/KAP1-mediated induction of heterochromatin was not followed by DNA methylation. This is in agreement with our previous observation that the repression of a promoter through the doxycycline-controllable nearby docking of KRAB/KAP1 is fully reversible in a wide variety of somatic cells, whether in tissue culture or in vivo (Wiznerowicz and Trono, 2003). These data are also consistent with our finding that the methylation status of CpG islands in the mouse liver is conditioned by their proximity to KAP1-binding sites in ESCs, not in hepatocytes. That epigenetic changes triggered in differentiated tissues by the recruitment of KRAB/KAP1 at specific loci be histone-rather than DNA-based explains how this system can regulate such highly dynamic processes as lymphohematopoietic differentiation, B cell activation, hepatic metabolism, and management of behavioral stress (Bojkowska et al., 2012; Santoni de Sio et al., 2012; Jakobsson et al., 2008). Nevertheless, our finding of such a fundamental difference in the outcome of KRAB/KAP1-mediated repression between ESCs and differentiated tissues warrants a characterization of the molecular complexes recruited at KAP1-binding sites in either setting. This effort may not only shed light on the mechanisms involved in conditioning the establishment of permanent epigenetic marks at specific loci under physiological circumstances, but also point to the kind of dysregulation driving their aberrant instatement in pathologies such as cancer.
EXPERIMENTAL PROCEDURES
Cell-Based Assays
All cell lines were cultured under standard conditions. Lentiviral vector was used with a mutiplicity of infection (MOI) of 50 and 10, respectively, for tTR-fusion expression vectors and 36 and 10 for TetO-containing reporter vectors, into mouse (ES, MEF, and NIH 3T3) and human (ES) cells. Human ESCs transduced with control or Kap1 shRNA lentiviral vectors were selected with hygromycin (see Extended Experimental Procedures). DNA was extracted and bisulfite conversion was performed, followed by PCR (primers are available in Extended Experimental Procedures) and pyrosequencing performed according to standard protocols.
In Silico Analyses
Data from different public data sets (GEO IDs GSE28247 GSE11034 GSE30280 GSE31183) were used to examine whole-genome bisulfite sequencing, RRBS-, and CTCF-binding sites in mouse ESCs. Mouse and human CpG islands and human RRBS data were obtained from the ENCODE project (ENCODE Project Consortium, 2011). KAP1 ChIP-seq data were from our previous work (Bojkowska et al., 2012; Rowe et al., 2010). Data were analyzed with the use of the “ChIP-seq on-line analysis tools” package (http://ccg.vital-it.ch/chipseq/), the GREAT website (http://bejerano.stanford.edu/great/public/html/) (McLean et al., 2010), and in-house shell scripts using h19 and mm9 assemblies. Fisher’s exact test was used to determine statistical significance.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dirk Schuebeler (Friedrich Miescher Institute) for helpful discussions, and Matthew Lorincz (University of British Columbia) and Haruhiko Koseki (Riken Center for Allergy and Immunology), for the gift of the G9a and NP95 knockout ES cells, respectively. Part of the computations were performed in the Vital-IT facility (www.vital-it.ch) of the Swiss Institute of Bioinformatics. This work was supported by grants from the Swiss national Science Foundation and the European Research Council.
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes Extended Experimental Procedures and three figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2012.08.043.
Publisher's Disclaimer: This is an open-access article distributed under the terms of the Creative Commons Attribution-Noncommercial-No Derivative Works 3.0 Unported License (CC-BY-NC-ND; http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode).
REFERENCES
- Berwin B, Barklis E. Retrovirus-mediated insertion of expressed and non-expressed genes at identical chromosomal locations. Nucleic Acids Res. 1993;21:2399–2407. doi: 10.1093/nar/21.10.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkowska K, Aloisio F, Cassano M, Kapopoulou A, de Sio FS, Zangger N, Offner S, Cartoni C, Thomas C, Quenneville S, et al. Liver-specific ablation of KRAB associated protein 1 in mice leads to male-predominant hepatosteatosis and development of liver adenoma. Hepatology. 2012 doi: 10.1002/hep.25767. Published online June 8, 2012. http://dx.doi.org/10.1002/hep.25767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J. Biol. Chem. 2010;285:26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, Yang HW, Lam LL, Mager DL, Schübeler D, et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 2008;27:2691–2701. doi: 10.1038/emboj.2008.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson RO, Thomas JH. Adaptive evolution in zinc finger transcription factors. PLoS Genet. 2009;5:e1000325. doi: 10.1371/journal.pgen.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- Friedman JR, Fredericks WJ, Jensen DE, Speicher DW, Huang XP, Neilson EG, Rauscher FJ., 3rd KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- Jakobsson J, Cordero MI, Bisaz R, Groner AC, Busskamp V, Bensadoun JC, Cammas F, Losson R, Mansuy IM, Sandi C, Trono D. KAP1-mediated epigenetic repression in the forebrain modulates behavioral vulnerability to stress. Neuron. 2008;60:818–831. doi: 10.1016/j.neuron.2008.09.036. [DOI] [PubMed] [Google Scholar]
- Leung DC, Dong KB, Maksakova IA, Goyal P, Appanah R, Lee S, Tachibana M, Shinkai Y, Lehnertz B, Mager DL, et al. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc. Natl. Acad. Sci. USA. 2011;108:5718–5723. doi: 10.1073/pnas.1014660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Rauch T, Chen ZX, Szabó PE, Riggs AD, Pfeifer GP. The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J. Biol. Chem. 2006;281:19489–19500. doi: 10.1074/jbc.M513249200. [DOI] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell. 2008;15:547–557. doi: 10.1016/j.devcel.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienert F, Mohn F, Tiwari VK, Baubec T, Roloff TC, Gaidatzis D, Stadler MB, Schübeler D. Genomic prevalence of heterochromatic H3K9me2 and transcription do not discriminate pluripotent from terminally differentiated cells. PLoS Genet. 2011a;7:e1002090. doi: 10.1371/journal.pgen.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienert F, Wirbelauer C, Som I, Dean A, Mohn F, Schübeler D. Identification of genetic elements that autonomously determine DNA methylation states. Nat. Genet. 2011b;43:1091–1097. doi: 10.1038/ng.946. [DOI] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meilinger D, Fellinger K, Bultmann S, Rothbauer U, Bonapace IM, Klinkert WE, Spada F, Leonhardt H. Np95 interacts with de novo DNA methyltransferases, Dnmt3a and Dnmt3b, and mediates epigenetic silencing of the viral CMV promoter in embryonic stem cells. EMBO Rep. 2009;10:1259–1264. doi: 10.1038/embor.2009.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosmann P, Georgiev O, Thiesen HJ, Hagmann M, Schaffner W. Silencing of RNA polymerases II and III-dependent transcription by the KRAB protein domain of KOX1, a Krüppel-type zinc finger factor. Biol. Chem. 1997;378:669–677. doi: 10.1515/bchm.1997.378.7.669. [DOI] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, Trono D. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol. Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Saitou M, Kagiwada S, Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development. 2012;139:15–31. doi: 10.1242/dev.050849. [DOI] [PubMed] [Google Scholar]
- Santoni de Sio FR, Massacand J, Barde I, Offner S, Corsinotti A, Kapopoulou A, Bojkowska K, Dagkis A, et al. KAP1 regulates gene networks controlling B lymphoid differentiation and function. Blood. 2012;119:4675–4685. doi: 10.1182/blood-2011-12-401117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiznerowicz M, Jakobsson J, Szulc J, Liao S, Quazzola A, Beermann F, Aebischer P, Trono D. The Kruppel-associated box repressor domain can trigger de novo promoter methylation during mouse early embryogenesis. J. Biol. Chem. 2007;282:34535–34541. doi: 10.1074/jbc.M705898200. [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.