Abstract
Oxidant-mediated antibacterial response systems are broadly used to control bacterial proliferation. Hypochlorite (HOCl) is an important component of the innate immune system produced in neutrophils and specific epithelia. Its antimicrobial activity is due to damaging cellular macromolecules. Little is known about how bacteria escape HOCl-inflicted damage. Recently, the transcription factor YjiE was identified that specifically protects Escherichia coli from HOCl killing. According to its function, YjiE is now renamed HypT (hypochlorite-responsive transcription factor). Here we unravel that HypT is activated by methionine oxidation to methionine sulfoxide. Interestingly, so far only inactivation of cellular proteins by methionine oxidation has been reported. Mutational analysis revealed three methionines that are essential to confer HOCl resistance. Their simultaneous substitution by glutamine, mimicking the methionine sulfoxide state, increased the viability of E. coli cells upon HOCl stress. Triple glutamine substitution generates a constitutively active HypT that regulates target genes independently of HOCl stress and permanently down-regulates intracellular iron levels. Inactivation of HypT depends on the methionine sulfoxide reductases A/B. Thus, microbial protection mechanisms have evolved along the evolution of antimicrobial control systems, allowing bacteria to survive within the host environment.
Keywords: LysR-type transcription factor, posttranslational modification, MsrA, MsrB
Reactive oxygen species (ROS) are bactericidal key effector molecules of the immune system, and are also produced by mucosal barrier epithelia (1, 2). Hypochlorite (HOCl) is a ROS with strong antimicrobial activity and oxidizing potential. It readily reacts with cellular macromolecules and particular proteins, giving rise to proteome-wide protein unfolding (3) and protein oxidation (4–10). Preferential oxidation of sulfur in cysteine (Cys) and methionine (Met) residues is a major reason for the reduced viability of cells upon HOCl exposure (5, 6). Dedicated reductases aid in restoring reduced proteins; thioredoxin and glutaredoxin reduce oxidized Cys thiols (11), and methionine sulfoxide reductases reduce Met-sulfoxide (Met-SO) back to Met (12, 13). Tissue cells are also damaged by HOCl. Neutrophil-mediated overproduction of HOCl is thought to contribute to the pathology of diseases such as chronic inflammation, cancer, and arteriosclerosis (14, 15). However, HOCl-activated protective mechanisms are only beginning to be identified. The conserved redox-regulated chaperone heat shock protein Hsp33 (16) is activated by Cys oxidation in combination with intrinsic unfolding upon HOCl stress and thus prevents the aggregation of cellular proteins (3). The Bacillus subtilis MarR-type DNA-binding transcriptional repressor hypochloric acid-specific regulator HypR is activated by disulfide-bond formation and confers protection against HOCl via the flavin oxidoreductase HypO (17). The TetR-type regulator N-ethylmaleimide reductase repressor (NemR) from Escherichia coli undergoes Cys oxidation upon exposure to HOCl and causes derepression of encoding glyoxalase I (gloA) and encoding N-ethylmaleimide reductase (nemA) that were shown to confer HOCl resistance (18). HypT (hypochlorite-responsive transcription factor) is an HOCl-specific lysine regulator (LysR)-type regulator that protects E. coli cells specifically from HOCl killing (19). It regulates many genes, most notably genes involved in Met and Cys biosynthesis and iron acquisition (19). However, how HypT is activated has remained enigmatic so far. Here we show that HOCl activates the transcription factor HypT by oxidation of Met to Met-SO, which can be reversed by methionine sulfoxide reductase (Msr)A and MsrB. Thus, HypT is a specialized transcription factor that protects cells from HOCl stress after activation by the thus far known inactivating Met oxidation.
Results
HypT Is Oxidized in Response to HOCl Stress.
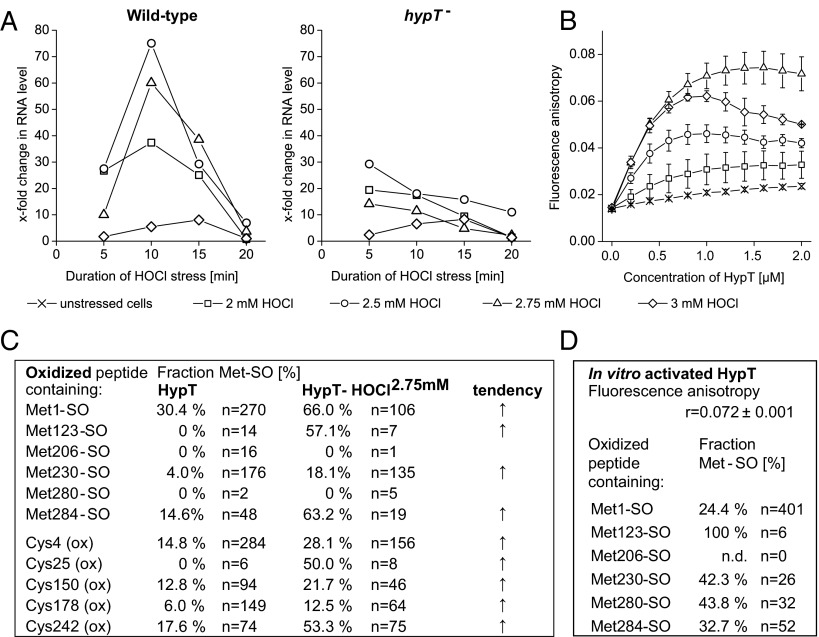
HypT protects E. coli cells from HOCl-mediated killing (19). To analyze how HypT is activated by HOCl, we purified HypT from HOCl-stressed cells and analyzed its activity and potential posttranslational modifications (PTMs). Cells growing in LB medium were stressed with 2–3 mM HOCl. There, HypT target genes such as metN, cysH, and metB are induced or repressed (fecD) (19). Gene regulation is already pronounced at 2 mM HOCl, at which cells are mildly stressed and able to recover from stress quickly. The strongest regulation occurred in wild-type cells at 10–15 min at 2.5–2.75 mM HOCl stress (Fig. 1A; shown for MetN RNA). At 3 mM HOCl, however, gene regulation is not very pronounced. Cells are 100% viable during the time course of the experiments, yet they are significantly stressed and quickly lose viability with prolonged duration of stress (Fig. S1).
Fig. 1.
HypT activation in response to HOCl stress. (A) Analysis of MetN RNA levels in HOCl-stressed C600 and C600 hypT− cells as indicated. Cells showed 100% viability during stress. Shown are the results of one representative experiment. (B) DNA binding of HypT purified from unstressed and HOCl-stressed BL21(DE3) cells as determined by FA. Protein was added stepwise to 10 nM Alexa Fluor 488–158-bp metN-promoter DNA. Results are expressed as mean ± SD (n = 3). (C) Fraction of oxidized HypT peptides relative to the total number (n) of peptides with the respective residue. Peptides were identified by MS analysis of proteolytic digests of HypT and HypT-HOCl2.75mM. For details, see SI Experimental Procedures. (D) In vitro activation of HypT. S-glutathionylated HypT was incubated with HOCl at a 6:1 molar ratio (HOCl:protein). DNA-binding activity was analyzed by FA. Results are shown as mean of anisotropy (r) ± SD (n = 3). r = 0.072 ± 0.001 for in vitro activated HypT. For comparison, r = 0.017 ± 0.013 (HypT) and r = 0.082 ± 0.013 (HypT-HOCl2.75mM). The fraction of Met-SO–containing HypT peptides was determined relative to the total number (n) of peptides with the respective Met. Met206 was not detected (n.d.).
The DNA-binding activity of HypT was analyzed by fluorescence anisotropy (FA) using fluorescent-labeled metN-promoter DNA (19). DNA binding was high when HypT was isolated from HOCl-stressed cells compared with unstressed cells. The maximum DNA-binding activity was observed after 10 min at 2.75 mM HOCl (Fig. 1B), at which strong HypT-dependent regulation of target genes was also observed (19) (Fig. 1A; compare wild-type and hypT− cells).
We subjected HypT purified from unstressed and HOCl-stressed (HypT-HOCl2.75mM) cells to proteolytic digests and analyzed the nature and localization of PTMs in HypT by LTQ Orbitrap mass spectrometry (MS). Sample preparation was performed under anaerobic conditions to eliminate spontaneous and nonspecific oxidation reactions in vitro. We found oxidation at Met and Cys residues (Fig. 1C). Met123 was oxidized to Met-SO exclusively upon HOCl stress, whereas oxidation of Met1, Met230, and Met284 increased upon HOCl stress (Fig. 1C). Met1 and Met123 also showed dioxidation to Met-sulfone, yet with a very low detection frequency (<0.2% of all detected peptides). Met206 and Met280, in contrast, were only detected in the reduced state (Fig. 1C), and with an overall very low detection frequency (<0.2% of all detected peptides), likely because they are not well-ionized. Therefore, the numbers for Met206 and Met280 given in Fig. 1C should be taken with caution. Further oxidations occurred at Cys residues preferentially after HOCl treatment (Fig. 1C; all Cys-oxidized peptides combined). We identified mono-, di-, and trioxidation at Cys, but no S-thiolation such as S-cysteinylation that was identified in the organic peroxide sensor OhrR (20). Considering that oxidized Cys was also observed in in vitro HOCl-treated HypT that showed very low DNA-binding activity (19), we assume that such oxidation is not responsible for increased HypT activity. To further assess activation of HypT, we treated S-glutathionylated HypT with HOCl in vitro, allowing us to discriminate between Cys and Met oxidation. S-glutathionylated HypT was incubated with HOCl at a molar ratio of HOCl to protein of 4:1–7:1, and then Cys was reduced and DNA-binding activity and PTMs were analyzed (Fig. 1D). HypT showed the same low DNA-binding activity as HypT isolated from unstressed cells at a 4:1 and 5:1 molar ratio and completely aggregated at a 7:1 molar ratio. At a 6:1 molar ratio, however, HypT showed an increased DNA-binding activity that was similar to that of HypT-HOCl2.75mM (Fig. 1D). Such in vitro activated HypT contained Met-SO to a somewhat different extent compared with HypT-HOCl2.75mM (Fig. 1D). Most notably, Met123 was found exclusively in the Met-SO state. The fraction of Met230-SO was much higher and the fraction of Met1-SO and Met284-SO much lower than in HypT-HOCl2.75mM. This indicates that the responsiveness of Met to HOCl oxidation is influenced by the reaction conditions, namely in vivo or in vitro oxidation, and that HypT is a very sensitive HOCl-specific regulator.
Met123, Met206, and Met230 Are Important for HypT Activity.
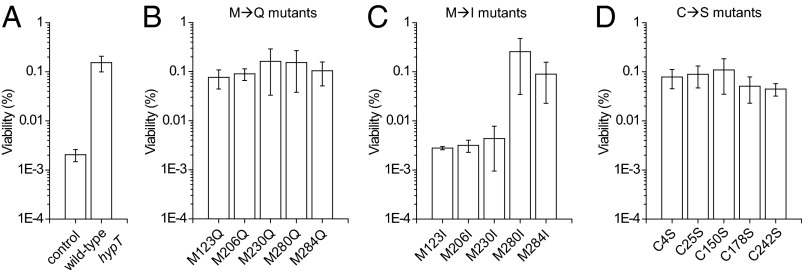
To analyze the contribution of individual Cys and Met residues to HypT activity, we generated point mutants in which Met residues were substituted by isoleucine or glutamine and Cys residues were replaced by serine. Replacing Met with glutamine mimics the Met-SO state and should result in a constitutively active HypT, whereas isoleucine substitution mimics the reduced Met and should result in inactive HypT (21, 22). Cys substitution by serine should impair their potential redox-regulatory function (23). The expression levels of all mutants were similar (M→Q/I mutants: ±10% of wild-type levels; C→S mutants: ±35% of wild-type levels), allowing comparison of viability values directly. M→Q mutants showed wild-type–like survival upon HOCl stress, indicating that the mutants are functional (compare Fig. 2, panel A with B). However, the individual substitution of Met123, Met206, or Met230 by isoleucine rendered the proteins unable to confer HOCl resistance (Fig. 2C). This is in contrast to substitution of Met280 and Met284 by isoleucine, which resulted in wild-type–like survival upon HOCl stress (Fig. 2C). This suggests that Met123, Met206, and Met230 are important, whereas Met280 and Met284 are not required, for the activity of HypT. All C→S mutants conferred wild-type–like HOCl resistance, indicating that none of the Cys is essential for activity, and therefore is not involved in HypT activation (Fig. 2D).
Fig. 2.
Met123, Met206, and Met230 are important for HypT activity. Viability assay of C600 hypT− cells carrying (A) an empty plasmid pBAD22 (control) or expressing wild-type hypT, and the hypT mutants (B) M123Q, M206Q, M230Q, M280Q, and M284Q, (C) M123I, M206I, M230I, M280I, and M284I, and (D) C4S, C25S, C150S, C178S, and C242 after treatment with 3.5 mM HOCl for 90 min. Results are expressed as mean ± SD (n ≥ 3).
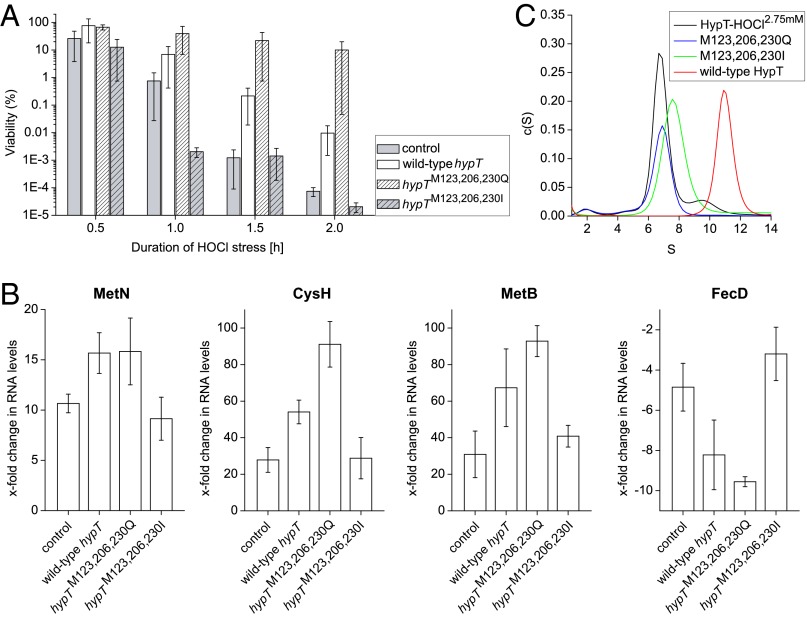
To further assess the role of Met123, Met206, and Met230 in HypT function, we generated HypT mutants in which all three residues were replaced simultaneously by either glutamine (M123,206,230Q) or isoleucine (M123,206,230I). We analyzed their ability to confer HOCl resistance and regulate target genes upon HOCl stress in comparison with wild-type HypT and the control strain harboring an empty plasmid instead of expressing hypT or a mutant. The expression level of both mutants was similar to that of wild-type hypT (+8% and +17% of wild-type levels). Expression of hypTM123,206,230Q conferred HOCl resistance and resulted in a strong regulation of MetN, CysH, MetB, and FecD RNA levels upon HOCl stress (Fig. 3 A and B), which was comparable to hypT-expressing cells and suggests that the triple M→Q mutant is functional. The triple M→I mutant, in contrast, showed control-like viability and target gene regulation (Fig. 3 A and B), demonstrating that the protein is inactive. This shows that triple M→Q substitution retains the activity of HypT upon HOCl stress, whereas M→I substitution of Met123, Met206, and Met230 renders HypT inactive.
Fig. 3.
M→Q substitution retains activity, whereas M→I substitution inactivates HypT. (A) Viability assay of C600 hypT− cells carrying an empty pBAD22 (control) or expressing hypT, hypTM123,206,230Q, and hypTM123,206,230I after treatment with 3.5 mM HOCl. Results are expressed as mean ± SD (n ≥ 4). (B) Analysis of MetN, CysH, MetB, and FecD RNA levels in cells described in A after HOCl stress (2.75 mM, 10 min) by qRT-PCR. Results are expressed as mean ± SD (n ≥ 3). (C) Analysis of the oligomerization state of HypT-HOCl2.75 mM (black), M123,206,230Q (blue), M123,206,230I (green), and wild-type HypT (red) purified from BL21(DE3) cells by aUC.
HypT is a dodecameric, ring-like protein that sediments at 10.6 S when analyzed by analytical ultracentrifugation (aUC) (19). Given that HypT dissociates in the presence of DNA to form DNA-binding active dimers and tetramers and shows a shift in oligomer distribution when analyzed in lysates from HOCl-stressed cells (19), we analyzed the oligomerization state of the triple mutants compared with HypT-HOCl2.75mM by aUC. HypT-HOCl2.75mM, M123,206,230Q, and M123,206,230I formed one main species, with 6.7–7.6 S probably corresponding to a tetramer (Fig. 3C). Thus, active HypT (i.e., HypT-HOCl2.75mM) and Met mutants comprise an oligomerization state that is different from the wild-type HypT from unstressed cells, suggesting that the Met residues are important for the quaternary structure of HypT.
Substitution of Met123, Met206, and Met230 by Glutamine Renders HypT Constitutively Active.
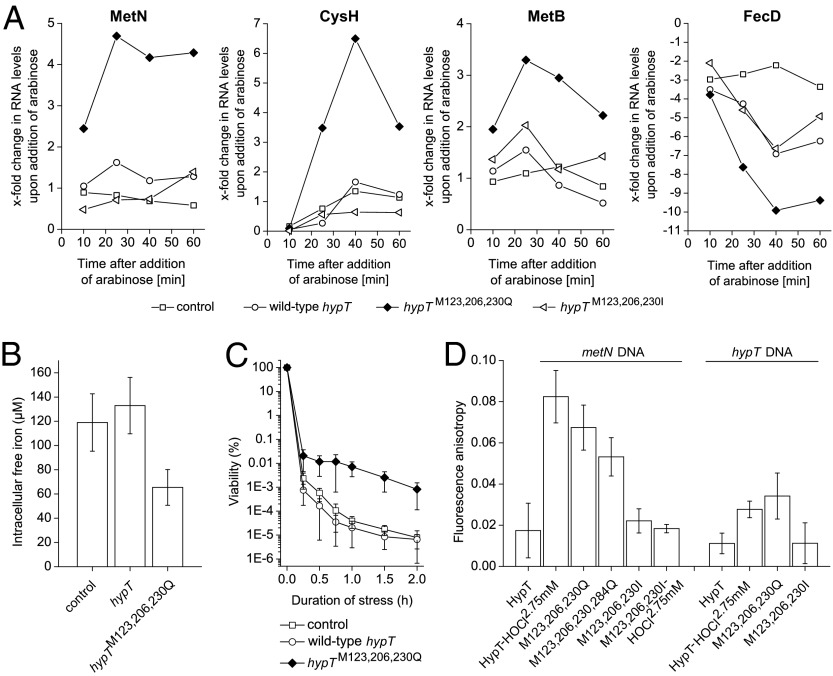
Assuming that Met oxidation activates HypT and glutamine substitution mimics the Met-SO state, a triple M→Q substitution should generate a constitutively active HypT, being able to regulate target genes independently of HOCl stress. Therefore, hypT− cells harboring an empty plasmid (control) and cells carrying a plasmid containing wild-type hypT, hypTM123,206,230Q, or hypTM123,206,230I were cultivated in the absence of arabinose. Then, arabinose was added to induce expression of plasmid-encoded hypT mutants and trigger potential HypT-dependent gene regulation (i.e., metN, cysH, metB, and fecD). Expression of hypTM123,206,230Q resulted in a strong regulation of target genes with a maximum reached at 20–40 min after arabinose induction. In contrast, the levels of target RNA were less or not affected upon expression of wild-type hypT and hypTM123,206,230I (Fig. 4A). We concluded that M123,206,230Q is constitutively active and regulates target gene expression irrespective of the presence of HOCl stress, whereas wild-type HypT and the isoleucine mutant are inactive.
Fig. 4.
hypTM123,M206,M230Q is constitutively active. (A) Analysis of MetN, CysH, MetB, and FecD RNA levels in C600 hypT− cells carrying an empty pBAD22 (control) or expressing hypT, hypTM123,206,230Q, and hypTM123,206,230I, respectively, by qRT-PCR. Expression was induced by adding arabinose at time point zero. No HOCl was added. Shown are the results of one representative experiment. (B) Intracellular free-iron levels of C600 hypT− cells carrying an empty pBAD22 (control) or expressing hypT and hypTM123,M206, M230Q, as determined by EPR. Results are expressed as mean ± SD (n ≥ 3). (C) Viability assay of C600 recA− hypT− cells carrying an empty pBAD22 (control) or expressing hypT and hypTM123,206,230Q after treatment with 2 mM H2O2. Results are expressed as mean ± SD (n ≥ 11). (D) DNA binding of HypT, HypT-HOCl2.75 mM, M123,206,230Q, M123,206,230,284Q, M123,206,230I, and M123,206,230I-HOCl2.75mM purified from BL21(DE3) cells as determined by FA. Results are expressed as mean ± SD (n ≥ 3).
To further examine the activity of M123,206,230Q, we analyzed the intracellular levels of free, unincorporated iron by electron paramagnetic resonance spectrometry (EPR) (24). We hypothesized that when M123,206,230Q constitutively down-regulates RNAs related to iron acquisition such as FecD (Fig. 4A) (19), this should concomitantly result in decreased intracellular iron levels. Confirming our considerations, cells expressing hypTM123,206,230Q showed 49 ± 13% lower unincorporated iron levels than cells expressing wild-type hypT (Fig. 4B).
Deriving from this observation, we further considered that low intracellular iron levels should generate fewer hydroxyl radicals via the Fenton reaction. To test this, we used a recA− strain (RecA is a DNA strand exchange and recombination protein) that is strongly impaired in DNA repair (25). A recA− strain suffers from increased DNA damage upon H2O2 stress (26) due to damage to iron-containing proteins and concomitantly increased hydroxyl radical formation by free iron and H2O2 (27). A recA− strain with constitutively lower iron levels should thus survive H2O2 stress better. This assay takes advantage of the fact that H2O2 does not activate HypT, as opposed to HOCl (19). We challenged recA− hypT− strains expressing wild-type hypT or hypTM123,206,230Q with H2O2 and analyzed their viability in a time-dependent manner. The hypTM123,206,230Q-expressing strain showed an up to two orders of magnitude increased viability upon H2O2 stress compared with the control and the strain expressing wild-type hypT (Fig. 4C), even though the expression level of hypTM123,206,230Q was 38% lower than that of cells expressing wild-type hypT. We also applied this assay to reevaluate the individual contribution of Met123, Met206, and Met230 to HypT activity. Expression of single or double M→Q mutants in recA− hypT− cells yielded similar levels of recombinant protein (±10% of wild-type levels) and did not confer any constitutive activity but rather a control-like viability upon H2O2 stress (Fig. S2). This demonstrates that only triple substitution of Met123, Met206, and Met230 by glutamine renders HypT constitutively active and constitutively decreases free intracellular iron levels.
Further supporting this assumption, the mutant M123,206,230Q showed a similarly high DNA-binding activity toward metN-promoter DNA as HypT-HOCl2.75mM (Fig. 4D). Additional substitution of Met284 (M123,206,230,284Q) resulted in comparable DNA-binding activity as M123,206,230Q, confirming that Met284 is not important for activity (Fig. 4D). M123,206,230I, in contrast, showed wild-type–like DNA-binding activity. Even when M123,206,230I was isolated from HOCl-stressed cells, DNA-binding activity was low, further proving that the isoleucine mutant is inactive and cannot be activated by potential additional PTMs. To analyze the specificity of DNA binding, the same experiments were performed using hypT-promoter DNA (nonspecific DNA) by taking advantage of the fact that HypT does not autoregulate its expression (19). Confirming the previous observations, the mutant M123,206,230Q showed a similarly low DNA-binding activity as HypT-HOCl2.75mM toward hypT-promoter DNA (Fig. 4D), demonstrating that HypT and M123,206,230Q are able to distinguish between target DNA (metN) and nonspecific DNA (hypT) by forming distinct DNA–HypT complexes (19).
MsrA and MsrB Reverse Met Oxidation and Inactivate HypT.
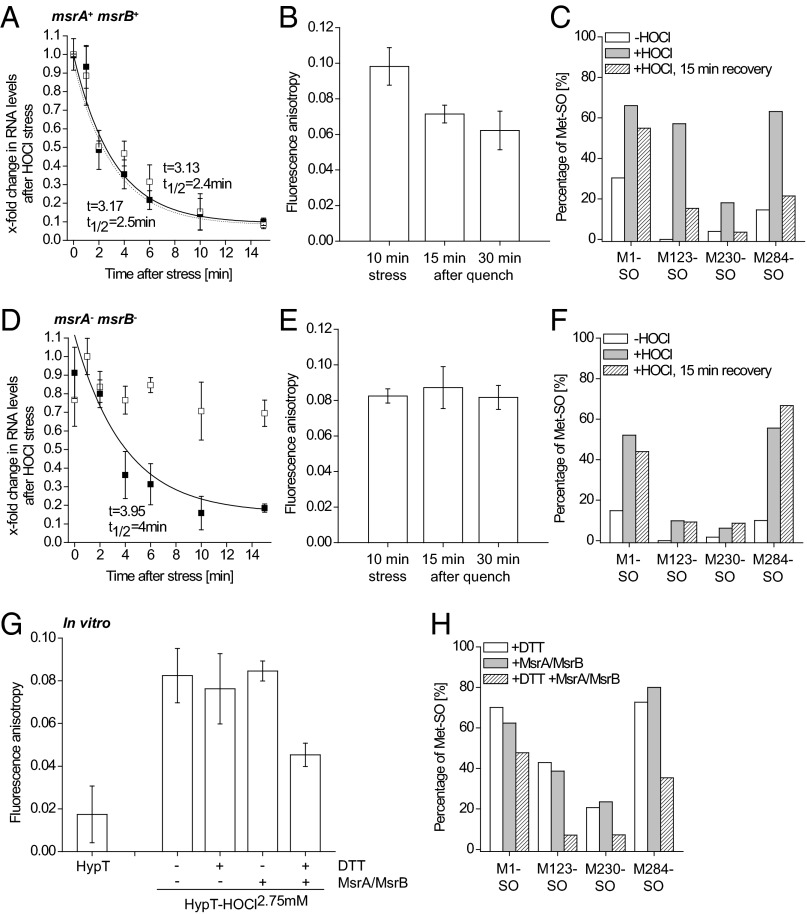
PTMs involved in redox regulation are mostly reversible. MsrA and MsrB reduce Met-SO back to Met (12, 13). Therefore, we analyzed the inactivation of HypT in E. coli msrA+ msrB+ (Fig. 5 A–C) or msrA− msrB− cells expressing hypT from a plasmid (Fig. 5 D–F). Samples were removed during HOCl stress (2.75 mM, 10 min) and then HOCl was quenched and cells were allowed to recover (Fig. 5 A–F and Fig. S3A) or cells remained HOCl-stressed (no quenching of HOCl; Fig. S3 B–E). Of note, MsrB protein levels are similar ±HOCl (Fig. S3F). Then, either RNA was isolated for quantitative (q)RT-PCR or HypT was purified for DNA binding and MS analysis. Inactivation of HypT in msrA+ msrB+ cells occurred quickly after the HOCl stress was quenched (Fig. 5 A–C). HypT-dependent regulation of MetN RNA levels was relieved quickly after HOCl stress and paralleled the levels observed for the control strain carrying an empty plasmid (Fig. 5A, time constant t = 3.13 and 3.17 min). The DNA-binding activity of active HypT decreased with t = 14 min (Fig. 5B), which was paralleled by the reduction of Met1-SO, Met123-SO, Met230-SO, and Met284-SO (Fig. 5C). Differences in the kinetics of decreases in RNA levels and inactivation of HypT may be derived from strongly different hypT expression levels in C600 and BL21(DE3) cells (SI Experimental Procedures). In sharp contrast, no inactivation of HypT was observed in msrA− msrB− cells. Relief of MetN RNA regulation in msrA− msrB− control cells was similar to the wild-type strain (Fig. 5D, t = 4 min). This indicates that the decrease in target RNA levels is partially independent of HypT, similar to the regulation of target genes upon HOCl stress that is also partially independent of HypT (Figs. 1A and 3B). However, a protracted decrease in MetN RNA levels was observed in msrA− msrB− cells expressing hypT, and RNA levels remained at about 70% of this during stress (Fig. 5D). Further, the DNA-binding activity remained similar during recovery (Fig. 5E) and no Met-SO reduction was observed (Fig. 5F). Thus, we conclude that MsrA and MsrB are vital for the inactivation of HypT in vivo and thus its reversible activation.
Fig. 5.
HypT is inactivated by MsrA/MsrB. (A–F) E. coli msrA+ msrB+ cells (A–C) and msrA− msrB− cells (D–F) were treated with HOCl (2.75 mM, 10 min). Then, fivefold concentrated LB medium (20% of the volume of cells) was added to quench HOCl and relieve the stress immediately and to replenish nutrients. Samples were removed at the indicated time points. (A and D) MetN RNA levels in stressed and quenched cells (A: C600 msrA+ msrB+ hypT−; D: C600 msrA− msrB− hypT−) either carrying an empty pBAD22 (control, closed symbol) or expressing hypT from pBAD22 (open symbol) by qRT-PCR. Shown are mean ± SD (n = 3). Shown are relative values; the highest value in one set of samples was set to 1. MetN RNA levels decreased with (A) time constants of ca. 3.1 min and t1/2 ca. 2.5 min and (D) t = 3.95 min and t1/2 = 4 min (control). No reliable kinetic parameters for MetN RNA levels could be calculated for hypT-expressing cells in D because they remained at ca. 70% of the stress level. (B and E) DNA binding of HypT purified from stressed and quenched (15 and 30 min after addition of LB medium) BL21(DE3) cells (B: msrA+ msrB+; E: msrA− msrB−) as determined by FA. Results are expressed as mean ± SD (n = 3). FA values in B decreased with t = 14 min (single exponential fit). (C and F) Fraction of Met-SO–containing peptides relative to the total number of peptides with the respective Met and Met-SO. Peptides were identified by MS analysis of proteolytic digests of HypT purified from unstressed (white bar), stressed (gray), and quenched (striped) BL21(DE3) cells (C: msrA+ msrB+; F: msrA− msrB−). (G) DNA binding of HypT, HypT-HOCl2.75 mM, and HypT-HOCl2.75 mM treated with DTT, and HypT-HOCl2.75 mM [all purified from BL21(DE3) cells] after DTT and MsrA/MsrB treatment as determined by FA. Results are expressed as mean ± SD (n ≥ 3). (H) Fraction of Met-SO–containing peptides relative to the total number of peptides with the respective Met and Met-SO. Peptides were identified by MS analysis of proteolytic digests of HypT-HOCl2.75 mM purified from BL21(DE3) cells and treated with DTT (white), MsrA/MsrB (gray), and combined DTT and MsrA/MsrB (striped).
When HOCl was not quenched and cells remained HOCl-stressed, MetN RNA levels decreased much more slowly than in quenched samples and the DNA-binding activity of HypT remained at the activated level (Fig. S3 B–E). In hypT− msrA+ msrB+ cells, MetN RNA levels remained high for 4 min before decreasing with t = 5.65 min and t1/2 = 5 min (control) and t = 6.84 min and t1/2 = 15.8 min (+hypT) (Fig. S3B). This protracted decrease of MetN RNA levels in hypT-expressing cells likely reflects the time cells require to reestablish redox homeostasis and recover catalytically active Msr that can inactivate HypT. In hypT− msrA− msrB− cells, in contrast, MetN RNA levels remained at about 60–70% of the stress level (Fig. S3D), indicating that functional MsrA/MsrB are required for complete relief of HOCl-induced MetN RNA regulation.
Additionally, HypT purified from HOCl-stressed cells was incubated with MsrA/MsrB and DTT in vitro and DNA-binding activity and Met oxidation were analyzed. As a control, samples were treated with DTT alone or with MsrA/MsrB in the absence of DTT. MsrA/MsrB require reductive conditions for catalytic activity and, thus, lack of reductive conditions inhibits Met-SO reduction in a substrate (28). DTT alone and MsrA/MsrB treatment alone altered neither the DNA-binding activity nor the Met-SO levels of HypT-HOCl2.75mM (Fig. 5H). MsrA/MsrB and DTT treatment of HypT-HOCl2.75mM, however, resulted in decreased DNA-binding activity (Fig. 5G) and reduction of Met-SO to Met (Fig. 5H). However, not all Met was completely reduced. This may be explained by limited access of MsrA/MsrB to Met-SO at the chosen conditions, and may explain why the DNA-binding activity after MsrA/MsrB treatment was still higher than that of HypT purified from unstressed cells. Taken together, we show that Met123, Met206, and Met230 of HypT function as oxidative stress sensors and become oxidized upon HOCl treatment to activate the transcription factor.
Discussion
HypT is a redox-regulated transcription factor and is suited perfectly to protecting cells against the consequences of HOCl stress. Even though HOCl is highly reactive and causes extensive inactivation and aggregation of proteins (3, 5, 29), it activates HypT. HypT is particularly sensitive to HOCl-dependent oxidation. In vitro activation of HypT requires as little as a 6:1 molar ratio of HOCl to HypT. This makes HypT similarly sensitive to activation by HOCl as the chaperone Hsp33, which is activated in vitro at a 10-fold molar excess of HOCl to protein (3). HOCl activation of HypT involves defined oxidative modifications of Met residues, which are reversible by MsrA/MsrB upon return to nonstress conditions. The requirement for simultaneous oxidation of three Met residues may be the reason why H2O2 was unable to activate HypT; the oxidizing power of H2O2 is apparently not high enough for HypT activation. The reactivity of HOCl and H2O2 with Met in a protein differs tremendously [3.8 × 107 M−1⋅s−1 and 3 × 10−2 M−1⋅s−1, respectively (30)], suggesting that the ability of HOCl to oxidize and activate HypT quickly is derived from its high activity. It should be noted that Met can indeed serve as an H2O2 sensor and become oxidized to regulate protein function. H2O2-mediated Met oxidation inactivates calmodulin (21, 31), whereas it activates calmodulin kinase II in mammalian cells (32). Thus, Met apparently functions as a specific HOCl sensor in HypT, responding to a defined range of HOCl that is capable of oxidizing all three Met residues at the same time.
Simultaneous substitution of Met123, Met206, and Met230 by glutamine generates a constitutively active HypT. The M123,206,230Q mutant regulates target genes independently of HOCl stress. In contrast, M123,206,230I neither conferred HOCl resistance nor regulated target genes. Even when M123,206,230I was purified from HOCl-stressed cells (M123,206,230I2.75mM; Fig. 4D), it showed the same low DNA-binding activity as HypT purified from unstressed cells. This shows that Cys oxidation in HypT, which occurs in HOCl-stressed cells, is not involved in HypT activation. M123,206,230Q decreased intracellular free-iron levels, providing further evidence for its constitutive activity. The tight regulation of intracellular free-iron levels appears to be a key to limiting formation and accumulation of reactive and destructive species such as hydroxyl radicals, which are formed in a Fenton-type reaction.
Amino acids involved in a defined activity regulation scheme would be expected to be conserved [e.g., the active-site Cys in Hsp33, oxidative stress regulator OxyR, NemR, and HypR (16–18, 33), and the Met in calmodulin kinase II (32)]. However, only M230 and M206 of HypT are located in a highly conserved region; M230 is conserved, whereas M206 is partially conserved and M123 is fairly variable (see alignment in ref. 19). The structure of HypT is not known. To assess the potential localization of the Met residues in HypT, we performed a secondary structure and disorder prediction and aligned the HypT sequence with that of the LysR-type transcription factor OxyR from E. coli. Further, we modeled the HypT structure using the structure prediction software Phyre 2 (34) and compared it with the structure of reduced OxyR from E. coli (33) (Fig. S4). According to the secondary prediction (Fig. S4A) and the structural model (Fig. S4C), M123 in HypT would be located at the end of an α-helix. M206 in HypT would be located in the redox loop in OxyR where the active-site Cys208 is located, and M230 in HypT would be located next to a β-sheet in OxyR. Both the redox loop and the β-sheet undergo major conformational changes upon OxyR oxidation (33). The β-sheet forms a pseudohelical loop after OxyR oxidation and is important for dimer interaction (33). M205 in the redox loop of OxyR corresponds to M206 in HypT (Fig. S4B) and is important for tetrameric interactions in reduced and oxidized OxyR and thus in oxidation-mediated changes in DNA binding (33). One could speculate that, if HypT and OxyR are indeed structurally similar, oxidation-mediated conformational rearrangements in HypT alter contacts within the dimer/tetramer. This may alter HypT binding to target DNA and activate HypT. Support for this consideration may be derived from the oligomerization state of HypT-HOCl2.75mM and M123,206,230Q. Both proteins form tetramers instead of dodecamers, as observed for wild-type HypT (19). It appears that the active HypT protein needs to be present in a tetrameric state to be able to bind efficiently to target DNA. Thus, shifting the equilibrium toward the active oligomeric state may be beneficial for activity. However, this shift alone is not sufficient for activity because it was also observed for M123,206,230I.
Taken together, we provide evidence that bacteria have evolved a transcription factor well-suited to protect them from HOCl killing during the innate immune response. This indicates a unique concept of redox regulation and reversible activation of a response molecule by HOCl-derived Met oxidation.
Experimental Procedures
Deletion strains and HypT mutants were generated via standard procedures (Table S1). Cultivation of strains, viability assays, protein purification, analysis of DNA binding using Alexa Fluor 488–158-bp DNA and unlabeled HypT variants, aUC analysis, and qRT-PCR were performed exactly as described (19). Modified residues were identified by MS analysis after proteolytic digest under anaerobic conditions using an LTQ Orbitrap XL (Thermo Scientific). In vitro HOCl treatment of HypT was performed using S-glutathionylated HypT at a molar ratio of HOCl to HypT of 4:1–7:1. For the analysis of constitutive HypT activity in vivo, arabinose was added to induce expression of the desired mutant gene and RNA levels were analyzed by qRT-PCR at various time points. To analyze the effect of MsrA/MsrB on HypT in vivo, msrA+ msrB+ and msrA− msrB− cells expressing hypT were HOCl-stressed (2.75 mM, 15 min), and then fivefold concentrated LB medium was added to quench HOCl and allow recovery of cells. Samples were removed at different time points and RNA was isolated for qRT-PCR or HypT was purified for FA and MS analysis. MsrA/MsrB treatment with purified HypT was performed under reducing conditions and DTT, MsrA, and MsrB were removed by gel filtration. Detailed materials and methods can be found in SI Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Drs. Johannes Buchner, James Imlay, and Stefan Gleiter for discussions; Dr. Johannes Graumann and Helmut Krause for initial MS analysis; Dr. Katharina Gebendorfer for generating the KMG232 strain, Amelie Tsoutsoulopoulos for assistance; Dr. Jean-Francois Collet for providing the anti-MsrB antibody; and Dr. Hauke Lilie and Maike Krause for aUC experiments. EPR analysis was performed at the Illinois EPR Research Center. A.D. was supported by the Elitenetzwerk Bayern, J.P. by the Studienstiftung des Deutschen Volkes, Y.L. by the Boehringer Ingelheim Fonds, and J.W. by the Emmy-Noether Program of the Deutsche Forschungsgemeinschaft.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300578110/-/DCSupplemental.
References
- 1.Ha EM, Oh CT, Bae YS, Lee WJ. A direct role for dual oxidase in Drosophila gut immunity. Science. 2005;310(5749):847–850. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- 2.Roos D, Winterbourn CC. Immunology. Lethal weapons. Science. 2002;296(5568):669–671. doi: 10.1126/science.1071271. [DOI] [PubMed] [Google Scholar]
- 3.Winter J, Ilbert M, Graf PC, Ozcelik D, Jakob U. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell. 2008;135(4):691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghesquière B, et al. Redox proteomics of protein-bound methionine oxidation. Mol Cell Proteomics. 2011;10(5):M110.006866. doi: 10.1074/mcp.M110.006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosen H, et al. Methionine oxidation contributes to bacterial killing by the myeloperoxidase system of neutrophils. Proc Natl Acad Sci USA. 2009;106(44):18686–18691. doi: 10.1073/pnas.0909464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leichert LI, et al. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci USA. 2008;105(24):8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chi BK, et al. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol Cell Proteomics. 2011;10(11):M111.009506. doi: 10.1074/mcp.M111.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawkins CL, Pattison DI, Davies MJ. Hypochlorite-induced oxidation of amino acids, peptides and proteins. Amino Acids. 2003;25(3-4):259–274. doi: 10.1007/s00726-003-0016-x. [DOI] [PubMed] [Google Scholar]
- 9.Robison AJ, Winder DG, Colbran RJ, Bartlett RK. Oxidation of calmodulin alters activation and regulation of CaMKII. Biochem Biophys Res Commun. 2007;356(1):97–101. doi: 10.1016/j.bbrc.2007.02.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szuchman-Sapir AJ, et al. Hypochlorous acid oxidizes methionine and tryptophan residues in myoglobin. Free Radic Biol Med. 2008;45(6):789–798. doi: 10.1016/j.freeradbiomed.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 11.Berndt C, Lillig CH, Holmgren A. Thioredoxins and glutaredoxins as facilitators of protein folding. Biochim Biophys Acta. 2008;1783(4):641–650. doi: 10.1016/j.bbamcr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Ejiri SI, Weissbach H, Brot N. Reduction of methionine sulfoxide to methionine by Escherichia coli. J Bacteriol. 1979;139(1):161–164. doi: 10.1128/jb.139.1.161-164.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimaud R, et al. Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J Biol Chem. 2001;276(52):48915–48920. doi: 10.1074/jbc.M105509200. [DOI] [PubMed] [Google Scholar]
- 14.Cook NL, et al. Myeloperoxidase-derived oxidants inhibit sarco/endoplasmic reticulum Ca2+-ATPase activity and perturb Ca2+ homeostasis in human coronary artery endothelial cells. Free Radic Biol Med. 2012;52(5):951–961. doi: 10.1016/j.freeradbiomed.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Güngör N, et al. Genotoxic effects of neutrophils and hypochlorous acid. Mutagenesis. 2010;25(2):149–154. doi: 10.1093/mutage/gep053. [DOI] [PubMed] [Google Scholar]
- 16.Jakob U, Muse W, Eser M, Bardwell JC. Chaperone activity with a redox switch. Cell. 1999;96(3):341–352. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- 17.Palm GJ, et al. Structural insights into the redox-switch mechanism of the MarR/DUF24-type regulator HypR. Nucleic Acids Res. 2012;40(9):4178–4192. doi: 10.1093/nar/gkr1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray MJ, Wholey WY, Parker BW, Kim M, Jakob U. NemR is a bleach-sensing transcription factor. J Biol Chem. March 27, 2013 doi: 10.1074/jbc.M113.454421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gebendorfer KM, et al. Identification of a hypochlorite-specific transcription factor from Escherichia coli. J Biol Chem. 2012;287(9):6892–6903. doi: 10.1074/jbc.M111.287219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JW, Soonsanga S, Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci USA. 2007;104(21):8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bigelow DJ, Squier TC. Thioredoxin-dependent redox regulation of cellular signaling and stress response through reversible oxidation of methionines. Mol Biosyst. 2011;7(7):2101–2109. doi: 10.1039/c1mb05081h. [DOI] [PubMed] [Google Scholar]
- 22.Vogt W. Oxidation of methionyl residues in proteins: Tools, targets, and reversal. Free Radic Biol Med. 1995;18(1):93–105. doi: 10.1016/0891-5849(94)00158-g. [DOI] [PubMed] [Google Scholar]
- 23.Klomsiri C, Karplus PA, Poole LB. Cysteine-based redox switches in enzymes. Antioxid Redox Signal. 2011;14(6):1065–1077. doi: 10.1089/ars.2010.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keyer K, Imlay JA. Inactivation of dehydratase [4Fe-4S] clusters and disruption of iron homeostasis upon cell exposure to peroxynitrite. J Biol Chem. 1997;272(44):27652–27659. doi: 10.1074/jbc.272.44.27652. [DOI] [PubMed] [Google Scholar]
- 25.Courcelle J, Hanawalt PC. RecA-dependent recovery of arrested DNA replication forks. Annu Rev Genet. 2003;37:611–646. doi: 10.1146/annurev.genet.37.110801.142616. [DOI] [PubMed] [Google Scholar]
- 26.Imlay JA, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240(4857):1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 27.Jang S, Imlay JA. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J Biol Chem. 2007;282(2):929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boschi-Muller S, et al. A sulfenic acid enzyme intermediate is involved in the catalytic mechanism of peptide methionine sulfoxide reductase from Escherichia coli. J Biol Chem. 2000;275(46):35908–35913. doi: 10.1074/jbc.M006137200. [DOI] [PubMed] [Google Scholar]
- 29.Davies MJ. The oxidative environment and protein damage. Biochim Biophys Acta. 2005;1703(2):93–109. doi: 10.1016/j.bbapap.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 30.Pan B, et al. Comparative oxidation studies of methionine residues reflect a structural effect on chemical kinetics in rhG-CSF. Biochemistry. 2006;45(51):15430–15443. doi: 10.1021/bi061855c. [DOI] [PubMed] [Google Scholar]
- 31.Chin D, Means AR. Methionine to glutamine substitutions in the C-terminal domain of calmodulin impair the activation of three protein kinases. J Biol Chem. 1996;271(48):30465–30471. doi: 10.1074/jbc.271.48.30465. [DOI] [PubMed] [Google Scholar]
- 32.Erickson JR, et al. A dynamic pathway for calcium-independent activation of CaMKII by methionine oxidation. Cell. 2008;133(3):462–474. doi: 10.1016/j.cell.2008.02.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi H, et al. Structural basis of the redox switch in the OxyR transcription factor. Cell. 2001;105(1):103–113. doi: 10.1016/s0092-8674(01)00300-2. [DOI] [PubMed] [Google Scholar]
- 34.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: A case study using the Phyre server. Nat Protoc. 2009;4(3):363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.