Abstract
Understanding the evolutionary history of microbial pathogens is critical for mitigating the impacts of emerging infectious diseases on economically and ecologically important host species. We used a genome resequencing approach to resolve the evolutionary history of an important microbial pathogen, the chytrid Batrachochytrium dendrobatidis (Bd), which has been implicated in amphibian declines worldwide. We sequenced the genomes of 29 isolates of Bd from around the world, with an emphasis on North, Central, and South America because of the devastating effect that Bd has had on amphibian populations in the New World. We found a substantial amount of evolutionary complexity in Bd with deep phylogenetic diversity that predates observed global amphibian declines. By investigating the entire genome, we found that even the most recently evolved Bd clade (termed the global panzootic lineage) contained more genetic variation than previously reported. We also found dramatic differences among isolates and among genomic regions in chromosomal copy number and patterns of heterozygosity, suggesting complex and heterogeneous genome dynamics. Finally, we report evidence for selection acting on the Bd genome, supporting the hypothesis that protease genes are important in evolutionary transitions in this group. Bd is considered an emerging pathogen because of its recent effects on amphibians, but our data indicate that it has a complex evolutionary history that predates recent disease outbreaks. Therefore, it is important to consider the contemporary effects of Bd in a broader evolutionary context and identify specific mechanisms that may have led to shifts in virulence in this system.
Emerging infectious diseases (EIDs) pose significant challenges for human health, agricultural crops, and economically and ecologically important populations in nature (1–4). The incidence of EIDs has been steadily rising over the last several decades (5, 6), and EIDs are of particular concern in an increasingly globalized world. For example, the majority of human EIDs is zoonoses that originate in wildlife (5) and subsequently, create a significant burden for global economies and public health (7, 8). Therefore, scientific efforts to understand and respond to EIDs are critical in diverse fields from biomedicine to conservation biology.
Although EIDs result from a complex interplay of factors, many studies focus primarily on the emergence of novel microbial pathogens. There are, in fact, high-profile examples of EIDs caused by the rapid appearance of novel, hypervirulent, or host-switching strains (9–11), but EIDs are not always caused by rapid or recent evolution of the pathogen itself. Virulence itself is an emergent property of microbe–host–environment interactions (12). Thus, EIDs can result from shifts in any factor—or combination of factors—in the microbe–host–environment epidemiological triangle (13). Characterizing the evolutionary history of emerging pathogens is, thus, critical, allowing us to determine whether observed EIDs result from rapid, recent shifts in organisms with pathogenic potential.
Chytridiomycosis is an EID responsible for declines in amphibian species around the world. The chytrid fungus Batrachochytrium dendrobatidis (Bd) was discovered and linked to amphibian declines in 1998 (14, 15). Chytridiomycosis is caused by Bd and kills amphibians by disrupting the integrity of their skin, a physiologically important organ that is involved in gas exchange, electrolyte balance, hydration, and protection from other pathogens (16, 17). Bd infects hundreds of species of amphibians, is found on all continents where amphibians occur, and is responsible for declines and extirpations in a diversity of amphibian hosts (18).
Soon after Bd was discovered, researchers proposed two competing hypotheses for the emergence of chytridiomycosis. The emerging pathogen hypothesis posited that a novel disease agent caused chytridiomycosis, and the endemic pathogen hypothesis proposed that an environmental shift disrupted a previously benign microbe–host interaction (19). Over the years, spatiotemporal and genetic data have supported the emerging pathogen hypothesis (reviewed in refs. 20 and 21). Spatiotemporal data provided clear evidence that Bd arrived and spread through geographic regions where it was not present historically (22–24). Early genetic studies also found very little genetic differentiation in Bd with no geographic signal, consistent with a recent, rapid spread of a novel disease agent (25–27). Recently, genetic and genomic data have been used to describe a geographically widespread Bd lineage [termed the global panzootic lineage (GPL)] (28) and several putatively endemic Bd lineages (28–30). However, different studies have used different methods and focused sampling in different parts of the world, precluding integration across studies to determine the evolutionary history leading to the emergence of Bd as a global threat to amphibians.
Here, we present whole-genome sequencing from a global panel of Bd isolates to show that Bd has a historically deeper and more complex evolutionary history than previously appreciated. We sequenced Bd genomes from around the world and also, a non-Bd chytrid outgroup that does not attack amphibians [Homolaphlyctis polyrhiza (Hp)] (31). Our focus was primarily on the evolutionary dynamics of Bd in the Americas, because many of the most devastating outbreaks have occurred in the New World. We address outstanding questions about the origins, genetic diversity, and genome structure of Bd that can be resolved using whole-genome data. We also integrate our genomic data with those data from a previous study with complementary geographic sampling (28). Our results reveal that the evolutionary history of Bd is complex, with multiple divergent lineages, heterogeneous patterns of genomic evolution, and no simple link between a single evolutionary event and observed amphibian declines.
Results and Discussion
Bd-induced amphibian declines are of general concern given the important role that amphibians play in food webs, the potential medicinal value of amphibian compounds, and the aesthetic value of amphibian biological diversity (32, 33). Studying the evolutionary history of Bd is also critically important for understanding the origin and spread of EIDs and effectively focusing disease mitigation efforts. Here, we synthesize our results and their implications for key questions about the evolutionary history of Bd based on genomic sequencing of a global panel of isolates (Table S1).
Is Bd Comprised of Multiple Lineages?
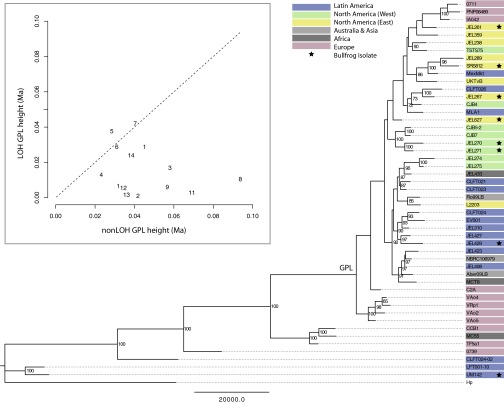
Our results show deep phylogenetic structure in the Bd tree (Fig. 1). We inferred the phylogenetic relationships among the 29 isolates sequenced in this study based on an average of 24× sequencing depth per isolate and 101,931 nuclear SNPs (Fig. S1). Additionally, we inferred the relationships among the 49 global isolates based on 76,515 nuclear SNPs from an integrated analysis of our data and the data from Farrer et al. (28) (Fig. 1). Both analyses were rooted with the closest known non-Bd chytrid (Hp), allowing us to infer the order of branching events. We recovered unbalanced topologies with less diversity in the early branching lineages and a more diverse crown group. The crown group has been referred to as the GPL, because much of the global Bd diversity described thus far is contained in this group and there is an apparent association with the onset of epizootics (28).
Fig. 1.
Rooted Bd phylogeny for 49 isolates based on 76,515 SNPs. The phylogeny for 29 focal isolates based on 101,931 SNPs is presented as Fig. S1. Maximum parsimony tree is shown with nodal support values generated from 200 bootstrap replicates. (Inset) Estimated divergence dates for the GPL based on different partitions of the largest supercontigs of the Bd genome. The x and y axes show GPL node height in millions of years for regions of the genome that did and did not exhibit pervasive LOH, respectively. Supercontig numbers are shown on the plot. Supercontig 1 contained two nonoverlapping LOH regions and thus, is represented two times on the plot. Supercontigs 10, 15, 16, and 17 did not contain clear LOH blocks and are not included in the plot. The average mean divergence date for the GPL was significantly younger (P < 0.001) for LOH regions (9,843 y ago) compared with non-LOH regions (41,805 y ago).
The tree topologies indicate far greater genetic and evolutionary diversity in Bd than has previously been recognized. In the combined tree, we identify several distinct clades (Fig. 1), including one clade composed of Bd isolates from Brazil, one clade previously identified by Farrer et al. (28) from southern Africa and Spain, and one clade restricted to Switzerland. Our genomic sequencing also revealed additional diversity within the GPL. The analysis of our 29 focal isolates indicated two well-supported clades within the GPL (Fig. S1). Our meta-analysis of 49 isolates did not show high bootstrap support for this primary split within the GPL (possibly because of reduced power from fewer SNPs) but did contain other well-supported clusters within the GPL (Fig. 1). Without additional sampling, it is premature to consider whether the diversity within the Bd tree represents variation within a single species or deeper taxonomic variation. In all probability, with more sampling, additional deeply branching lineages will be uncovered. We conclude that Bd is composed of multiple divergent lineages and that the emergence of the globally distributed GPL occurred within the context of deeper ancestral diversity, much of which may yet be discovered.
Is Bd a Novel or Endemic Pathogen?
The novel and endemic pathogen hypotheses generate different predictions for genetic variation within Bd. If Bd was largely endemic, we would expect deep phylogenetic structure in the Bd tree corresponding to geographic locality and/or host association. If Bd was entirely novel, we would expect a shallow and comb-like tree topology. Previous genetic data have largely supported the novel pathogen hypothesis (25–28), but many of these datasets have relied on fewer variable markers or more limited geographic sampling, resulting in tree topologies that appear more shallow and comb-like than those topologies presented here.
Overall, our genomic data suggest that we should consider a less binary classification rather than attempting to reconcile the diversity of Bd isolates as purely novel or purely endemic. On one end of the spectrum, our data were not broadly consistent with the endemic pathogen hypothesis. Generally, isolates that shared a common geographic or host association did not cluster within clades. Some closely related isolates did share a common geography (e.g., the pairing of JEL270 and JEL271 isolated from Bullfrogs in Point Reyes, California; the pairing of CJB5 and CJB7 isolated from Mountain Yellow-Legged Frogs in the Sierra Nevada Mountains, California; and the clade containing VAo2, VAo4, VAo5, and VRp1 from Valencia, Spain). This microgeographic signal does not reflect ancient geographic structure or long-term host associations but instead, is likely explained by the invasion and subsequent evolution of clones in particular geographic regions. On the other end of the spectrum, the presence of lineages that diverged before the GPL indicates that the evolutionary history of Bd is more ancient than suggested by the novel pathogen hypothesis. Even the GPL, which contains closely related and globally dispersed isolates, is not genetically homogenous (Fig. 1). Therefore, our data reveal that Bd is likely endemic in some parts of its range but novel in others.
Bd certainly functions as an invasive species in many parts of the world, and the novel pathogen hypothesis is supported in particular locations by unambiguous spatiotemporal evidence. For example, there are well-documented examples of recent introduction and spread of Bd in many regions, including Panama (24, 34), California (23), and Australia (18, 35). However, these recent introductions should be viewed from a backdrop of Bd diversity. One plausible scenario supported by our data is that recent Bd introductions occurred from an ancestral pool of variation that is deeper than previously considered.
Where Did Bd Originate?
Because our genomics dataset included a non-Bd chytrid (Hp), we were able to root our phylogenetic analysis. Thus, we could identify lineages that were more basal in the Bd tree and determine whether these lineages showed an association with particular hosts or particular regions of the world. The basal lineage in our tree was from Brazil (Fig. 1). One of the Brazilian Bd isolates was collected from Hylodes ornatus from São Paulo, Brazil, and one was collected from a bullfrog in a US market that likely traces back to Brazil (30). Brazil seems to contain ancestral variation, and Bd dynamics in natural ecosystems in Brazil may be largely consistent with an endemic scenario [i.e., Bd has been reported in a number of Brazilian frog species, but mass die offs have not been observed (36–38)]. In addition to the more basal Bd-Brazil clade, a number of isolates from Brazil were contained in the GPL. A previous study suggested that novelty could arise by hybridization of Bd-Brazil with GPL isolates and subsequently spread through the international wildlife trade (30). However, both bullfrog and Brazilian isolates were scattered throughout our tree (Fig. 1), and we have no direct evidence for any specific region or host species as an evolutionary source.
Our results underscore that it is premature to conclude a geographic location for the origin of Bd or any of its subclades. Our analyses revealed deeply branching lineages from Brazil and Africa but no close sister relationship between either of these clades and the GPL. Previous studies have promoted various possibilities for the origin of Bd (29, 30, 39, 40). However, sampling for genomic studies thus far has focused primarily on Europe (28) and the Americas (this study). Additional genomic sampling in other regions is critical. However, undiscovered isolates may fall in key positions in the tree (e.g., base of Bd tree, sister to GPL) and clarify the geographic and host associations at important transition points in the evolutionary history of Bd. However, the true origin of Bd may never be resolved, even with exhaustive sampling of contemporary Bd isolates, given historical dynamics of extinction and migration.
When Did Bd Originate?
Our dating analysis suggests the possibility that some Bd lineages are more ancient than previously appreciated. We applied a molecular clock to estimate the age of key nodes within our tree. Time calibration of phylogenetic trees must be done with caution, particularly for analyses that cannot rely on fossil evidence and trees with incomplete taxon sampling. We, therefore, do not advocate that our inferred node depths be upheld as absolute dates but only relative estimates to distinguish between competing hypotheses. Below, we discuss our results for two key nodes: the base of the Bd tree and the common ancestor of the GPL. The dates provided are dependent on an assumed rate of evolution under a strict clock (i.e., 0.0081 substitutions per site per million years) (41) and a tree prior derived from the constant population-size coalescent (42). Although we report results for these model assumptions, results were quantitatively similar with alternative assumptions (e.g., exponentially increasing population size prior, relaxed clock).
In the analysis including 76,515 SNPs for all 49 isolates, the root for the entire Bd tree was estimated to be 104,700 y B.P., and an age for the most recent common ancestor of the GPL was 26,400 y B.P. To refine our estimates of the age of the GPL, we also conducted separate analyses for different partitions of the Bd genome for this group. We looked at loss of heterozygosity (LOH) and non-LOH regions separately for each supercontig (Fig. 1). Considering all 49 isolates in a common analysis, the non-LOH data partition revealed significantly earlier divergence dates than the LOH data partition (Welch t testdf = 15.635, t = 4.3849, P < 0.001). Specifically, the average divergence depths recovered for the GPL from the non-LOH regions were 41,805 y and the LOH regions were 9,843 y. The youngest depth estimate for the GPL was from an LOH region on supercontig 2 (1,024 y), and the oldest depth estimate was from a non-LOH region on supercontig 8 (90,000 y).
It is not unusual for different data partitions to recover different divergence depths given among-locus variation in core processes that influence genetic variation (e.g., mutation hotspots, regions under selection, incomplete lineage sorting, recombination, and gene conversion) (43). Furthermore, among-locus discordance can be informative for distinguishing between alternative demographic scenarios. For example, our data suggest that the GPL may be older than previously proposed. The most consistent signal in our data suggests an origin of the GPL in the 10,000- to 40,000-y range. If the GPL is, in fact, tens of thousands of years old, the reduced variation in the LOH region on supercontig 2 (where a large chromosomal region is homozygous and nearly monomorphic throughout the GPL) could be caused by purifying selection, ongoing gene conversion, or a selective sweep propagated through the GPL by strong selection. Alternatively, it is possible that the LOH event on supercontig 2 predated the emergence of the GPL and was inherited asexually during the spread of GPL. This alternative is supported by the fact that the LOH boundary is largely identical among isolates (Fig. S2). If the age of the GPL is estimated solely by the LOH region on supercontig 2, the GPL seems much younger (1,024 y with a confidence range of 500–1,500 y). The young date estimated from this genomic segment is still older than—but within one order of magnitude of—the divergence date presented by Farrer et al. (28). If the GPL is, indeed, this young, we must explain why there is so much sequence variation among isolates in other regions of the genome. If this pervasive variation represents sorting of ancestral variants, it would suggest a highly diverse ancestor to the GPL. It is also important to note that even the youngest divergence dates obtained predate contemporary observed amphibian declines.
What Is the Role of Hybridization and Sexual Reproduction in Bd?
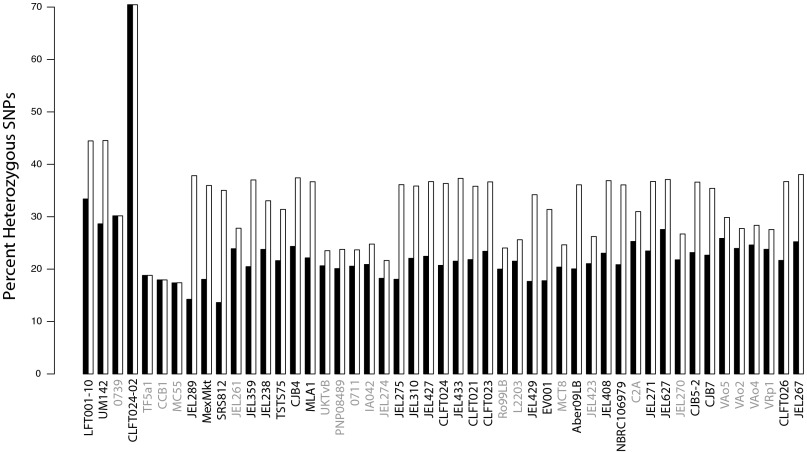
Although sexual reproduction has never been observed in Bd, sexual recombination and hybridization have been proposed as important mechanisms in the evolutionary history of Bd. For example, a recent discovery of a hybrid between the GPL and a divergent Bd lineage from Brazil suggests that genetically distinct isolates can hybridize (isolate CLFT024-02) (30). Moreover, hybridization has been proposed as a potential explanatory mechanism for the origin of the GPL clade as a whole (28). Our genome-scale data are consistent with the hypothesis that some isolates may have a hybrid history. Specifically, we show that isolate CLFT024-02 has elevated heterozygosity with nearly double the percentage of heterozygous SNPs than the other sampled isolates (Fig. 2).
Fig. 2.
Percentage of heterozygous SNPs. Black bars represent all data from the largest 16 supercontigs, and white bars represent only non-LOH regions. As expected, the percentage of heterozygous SNPs is elevated when LOH regions are excluded. Also highlighted is elevated heterozygosity in the putative hybrid isolate CLFT024-02. Isolates from the study by Farrer et al. (28) are shown with gray isolate names and generally show lower heterozygosity because of lower sequencing coverage.
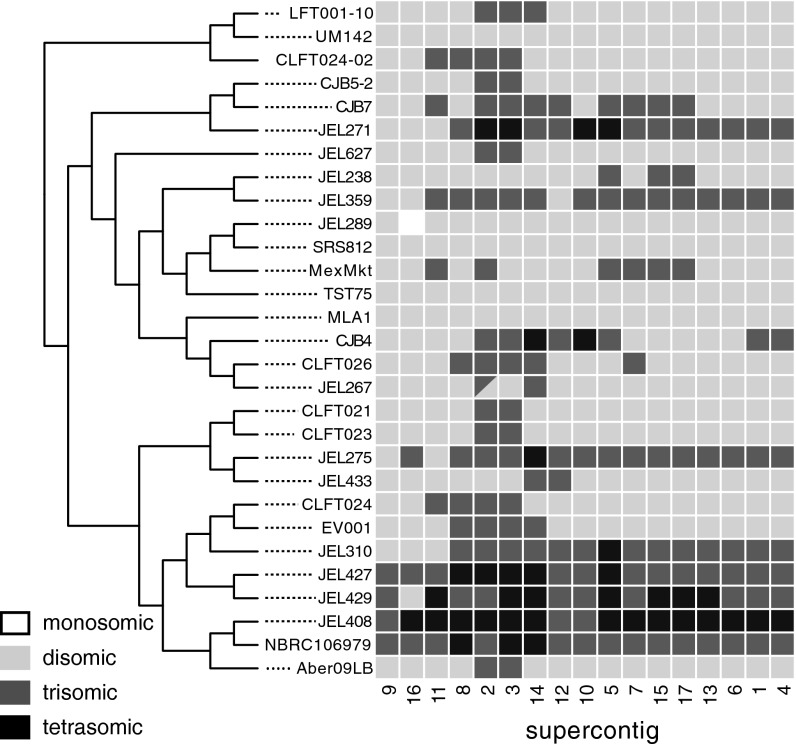
However, our data do not support the hypothesis of a hybrid origin for the GPL as a whole. Recent hybridization between two lineages, such as observed in CLFT024-02, should be observed as increased heterozygosity or ploidy relative to nonhybrid isolates. However, we did not observe elevated heterozygosity in the GPL, with no significant difference between the percentage of heterozygous SNPs comparing GPL with non-GPL isolates (t test: P = 0.56, excluding the hybrid CLFT024-02 from analyses and focusing only on SNPs outside LOH regions). We also did not observe a shared pattern of increased ploidy in the GPL (Fig. 3). Thus, although hybridization potentially plays a role in generating and sorting genetic and functional variation in Bd, our genomic data do not provide evidence that a specific historical hybridization event led to the emergence of the GPL.
Fig. 3.
Evidence for variation in chromosomal copy numbers. Copy numbers shown for each isolate across all major supercontigs. Supercontigs have been rearranged to form clusters according to Euclidean distances.
We also looked for genomic evidence of recurrent sexual reproduction in Bd. Recurrent sexual reproduction will lead to an inbreeding coefficient (FIS) (44) of zero, but the mean FIS observed among all SNPs in the GPL was −0.319 (SE ± 0.002; SD ± 0.473). The high variance in FIS and the large range of heterozygosity values across isolates (Fig. 2) could be caused by either rare sexual reproduction or mitotic recombination (27, 30). Meiotic recombination and mitotic recombination leave different genomic signatures and thus, can be distinguished with resequencing data. With sexual recombination within the GPL (equivalent to selfing), we expect a positive correlation between chromosome size and heterozygosity, because per base pair recombination rate increases with chromosome size (45, 46). With mitotic recombination, we expect a negative correlation between heterozygosity and chromosome size, because randomly placed mitotic cross-overs affect a larger number of base pairs as chromosome sizes increase. We found that FIS was negatively correlated with supercontig size (r2 = 0.60; P < 0.001) with all supercontigs except the largest, having an overall negative FIS average (Fig. S3). Therefore, although we cannot exclude the possibility that some Bd clades undergo sexual reproduction, genome-wide patterns are more consistent with pervasive mitotic rather than meiotic recombination.
What Are the Dynamics of Genome Evolution in Bd?
Our data identified other dynamic genome processes in Bd, including dramatic chromosomal aneuploidy, copy number variation (CNV), and LOH. We documented extensive aneuploidy (i.e., changes in chromosome copy number without changes in ploidy of the entire chromosomal set) among isolates and among chromosomal segments within isolates (Fig. 3). Chromosomal copy number varied among our samples from monosomic to tetrasomic (Fig. 3). In fact, only a handful of isolates showed consistent patterns of disomy across their entire genomes (e.g., UM142, TST75, MLA1, and SRS812). Some isolates showed elevated copy numbers at most chromosomal segments (e.g., JEL408, JEL427, JEL429, JEL271, and NCRC106979) but not consistently across the genome. Although some of the isolates with higher chromosomal copy number were found in the GPL, chromosomal copy number was a poor indicator of relatedness in general (i.e., ploidy patterns were not clustered in the tree). We found evidence of monosomy in only one isolate (JEL289) at supercontig 16. In addition to patterns across isolates, we also evaluated patterns across chromosomal segments. Several segments, such as supercontigs 3 and 5, tended to have increased copy numbers, but we did not find a strong pattern of shared aneuploidy in particular genomic regions across a majority of isolates. Estimates of mean copy number of supercontigs inferred from our genomic resequencing correlated (r2 = 0.70; P < 0.01) with estimates of DNA content from flow cytometry (30) for eight isolates where both types of data were available. Therefore, CNV patterns described from resequencing data can be used to give an accurate picture of underlying genomic processes.
Our data also showed striking patterns of LOH throughout the Bd tree. LOH by mitotic recombination has been proposed as a mechanism for the uneven distribution of genetic variation within and among Bd isolates (27). Our genomic sequencing revealed widespread LOH and consolidated blocks of LOH in Bd genomes. Among isolates, the average total size of LOH blocks across the genome was 7.7 ± 3.7 megabases (Mb), spanning a large proportion of the ∼25-Mb genome. Some LOH breakpoints were shared across isolates (Fig. S2). Shared ancestry seems the most likely explanation, because breakpoints are nearly identical across isolates, a pattern difficult to explain with selection and recombination. LOH regions, therefore, characterize groups of isolates (for example, the LOH region on supercontig 2) that is found in the GPL. We also identified LOH regions that are shared among subgroups of the GPL and those LOH regions that occur only in single isolates.
Ploidy changes and LOH events seem to occur relatively rapidly and affect large regions of the genome (47). Therefore these dynamic processes could play a role in rapid functional shifts. Future work is needed to determine whether specific triggers (i.e., recombination hotspots, biased gene conversion, or natural selection) lead to CNV and LOH and how quickly these processes can occur in Bd. For example, if these genomic changes happen over short timescales or in response to specific conditions, they could be a mechanism for rapid changes in response to environmental shifts or invasion of new habitats. One fruitful avenue would be to determine whether specific genomic processes like LOH can be linked to attenuation of virulence [which may occur in some laboratory isolates over rounds of passaging (48)].
What Regions of the Genome Are Under Selection?
We examined the Bd genomes for signatures of adaptive genomic evolution. We identified genes with an excess of nonsynonymous relative to synonymous substitutions and tested for enrichment of particular Gene Ontology (GO) functional groups in those genes with dN/dS ratios greater than one. We found enrichment of molecular functions related to protein binding (P = 1.7E-17), ATP binding (P = 4.3E-7), and RNA nucleotide binding (P = 3E-5) and enrichment for biological functions in regulation of response to stimulus (P = 6.7E-6) and signaling (P = 6.7E-6). We identified specific genes that showed high dN/dS ratios, including some protease genes in expanded gene families such as BDEG_05170 (fungalysin metallopeptidase), BDEG_02225, and BDEG_03861 (aspartyl proteases). We also conducted GO enrichment analyses to determine whether LOH or CNV gene regions disproportionately contained genes that could play a role in Bd pathogenesis. In LOH blocks, we found a significant enrichment of genes encoding proteins in processes of reactive oxygen metabolism (P < 0.002), l-serine metabolism (P < 0.003), and superoxide dismutase and oxidoreductase activities (P < 0.002). Most intriguingly, in CNV regions, we found a significant enrichment of genes encoding serine and aspartic peptidases (P = 6.1E-9), gene families that have been discussed previously as potential Bd pathogenicity factors (49).
A number of lines of evidence now point to the role of protease genes as potentially important in Bd infection of amphibian skin. Several protease gene families are dramatically expanded in the Bd genome relative to other closely related fungi (49), and many of these genes have increased expression when Bd is exposed to amphibian skin (50). Estimates of the age of the gene duplication events in these gene families range from tens of thousands to tens of millions of years (49), suggesting that the protease gene family expansions occurred well before the spread of the GPL. Other nonprotease gene family expansions have also recently been identified that could affect Bd cell wall and adhesion properties (51). Comparative and functional studies of these candidate loci are now needed to determine whether specific alleles in these gene families are associated with key Bd phenotypes or evolutionary transitions.
Priorities for Future Research.
Our results indicate that Bd lineages are older, more diverse, and exhibit more heterogeneous and dynamic genomic architecture than previously documented, and these results have specific implications for our understanding of Bd-related amphibian declines. First, additional efforts to describe the evolutionary diversity in the lineages currently grouped as Bd are essential. Genetic data from additional isolates will be imperative to further reveal phylogenetic structure at the base of the Bd tree. Not only must we focus our effort on less-sampled parts of the world but also, other host species and environmental samples. The diversity of chytrids in many environments is underappreciated (52). Understanding the origins of Bd will require a focus not only on the GPL but also, the discovery and description of more deeply branching lineages, including non-Bd relatives.
Second, linking phylogenetic and functional variation in Bd is challenging but necessary. Evidence from laboratory studies suggests that isolates differ in virulence (48, 53), with evidence that some GPL isolates may be more deadly than the early-diverging isolate from South Africa (28). However, comparing virulence traits among isolates is inherently challenging given that virulence is an emergent property of the host–microbe–environment interaction. Controlled experiments with factorial designs (multiple isolates tested against multiple host species under multiple environmental conditions) are needed. Additionally, we must develop consistent assays for the Bd phenotypes that vary across isolates and affect the ability of Bd to infect frogs. Mapping these phenotypes onto the Bd tree will provide insight into how the evolutionary diversity described here is linked to functional diversity.
Third, our data indicate that we must consider both ecological and evolutionary shifts that may have led to the observed disease-related amphibian declines. Contemporary changes, such as increased global trade, are almost certainly responsible for Bd introductions to new localities with naïve hosts. However, our data show that Bd has a deep and complex evolutionary history and that key nodes in the Bd tree are older than observed amphibian die offs. Therefore, our analyses underscore an important unanswered question in the emergence of Bd as a wildlife pathogen: why now? Our research efforts now need to focus on understanding genetic and environmental control of disease dynamics in parts of the world where Bd is endemic and areas devastated by the GPL. Close comparative examination of Bd isolates will determine whether particular genomic regions can be unambiguously linked to the spread of the GPL and whether the dynamic genomic processes described here could play a role in functional shifts in Bd. We also need to renew efforts to understand cofactors that may help explain the recent impacts of Bd on amphibians and maintain an integrative focus on microbe–host–environment interactions. Overall our data emphasize the importance of considering the evolutionary origins of seemingly novel disease agents and help focus our Bd research agenda on the evaluation and implications of a more complex and ancient history for a contemporary ecological crisis.
Methods
Complete methodological details are presented in SI Methods. Briefly, we selected 29 Bd isolates for de novo sequencing using Illumina GA IIx and Illumina HiSeq platforms (Table S1). Genomic data are accessioned in the National Center for Biotechnology Information Short Read Archive (accession no. SRA062886). We quality-filtered Illumina reads and generated a stringent SNP dataset by applying a strict minimum coverage filter. We also integrated our data with published data from an additional geographically complementary 20 Bd isolates (28). We estimated rooted phylogenies using Hp as an outgroup. We used BEAST (v 1.7.3; 40) to sample from the posterior density of time-calibrated trees. We also conducted divergence–time analyses using datasets subsampled at the level of the supercontig and compared estimated heights of the GPL between LOH and non-LOH regions of each supercontig. We also identified regions of the genome exhibiting LOH and chromosome CNV. Finally, we conducted a genomic scan for selection and an analysis of GO enrichment function for genes showing elevated nonsynonymous SNP counts.
Supplementary Material
Acknowledgments
We acknowledge Peter Murphy, Ché Weldon, Victoria Vasquez, Cinthya Mendoza-Almeralla, and Lisa Schloegel for isolating Bd samples that were included in this study. We thank Ana V. Longo Berríos and David Vallejo for assistance with laboratory work, Scott Cashins for collection of tadpoles for Bd isolation, and Jamie Voyles and two anonymous reviewers for comments on earlier versions of the manuscript. This project was funded by National Science Foundation Grants EF-0723871 and IOS-0825355 (to E.B.R.) and grants from the Cornell Center for Comparative Population Genetics (3CPG) and the Atkinson Center for a Sustainable Future (ACSF; to K.R.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The genomic data reported in this paper have been deposited in the NCBI Sequence Read Archive (accession no. SRA062886).
See Commentary on page 9193.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300130110/-/DCSupplemental.
References
- 1.Collins JP. Amphibian decline and extinction: What we know and what we need to learn. Dis Aquat Organ. 2010;92(2–3):93–99. doi: 10.3354/dao02307. [DOI] [PubMed] [Google Scholar]
- 2.Blehert DS, et al. Bat white-nose syndrome: An emerging fungal pathogen? Science. 2009;323(5911):227. doi: 10.1126/science.1163874. [DOI] [PubMed] [Google Scholar]
- 3.McCallum H. Tasmanian devil facial tumour disease: Lessons for conservation biology. Trends Ecol Evol. 2008;23(11):631–637. doi: 10.1016/j.tree.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 4.Woolhouse M, Gaunt E. Ecological origins of novel human pathogens. Crit Rev Microbiol. 2007;33(4):231–242. doi: 10.1080/10408410701647560. [DOI] [PubMed] [Google Scholar]
- 5.Jones KE, et al. Global trends in emerging infectious diseases. Nature. 2008;451(7181):990–993. doi: 10.1038/nature06536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher MC, et al. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2012;484(7393):186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binder S, Levitt AM, Sacks JJ, Hughes JM. Emerging infectious diseases: Public health issues for the 21st century. Science. 1999;284(5418):1311–1313. doi: 10.1126/science.284.5418.1311. [DOI] [PubMed] [Google Scholar]
- 8.Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430(6996):242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith GJD, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459(7250):1122–1125. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- 10.Lipsitch M, et al. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003;300(5627):1966–1970. doi: 10.1126/science.1086616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LaDeau SL, Kilpatrick AM, Marra PP. West Nile virus emergence and large-scale declines of North American bird populations. Nature. 2007;447(7145):710–713. doi: 10.1038/nature05829. [DOI] [PubMed] [Google Scholar]
- 12.Casadevall A, Fang FC, Pirofski LA. Microbial virulence as an emergent property: Consequences and opportunities. PLoS Pathog. 2011;7(7):e1002136. doi: 10.1371/journal.ppat.1002136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scholthof KBG. The disease triangle: Pathogens, the environment and society. Nat Rev Microbiol. 2007;5(2):152–156. doi: 10.1038/nrmicro1596. [DOI] [PubMed] [Google Scholar]
- 14.Berger L, et al. Chytridiomycosis causes amphibian mortality associated with population declines in the rain forests of Australia and Central America. Proc Natl Acad Sci USA. 1998;95(15):9031–9036. doi: 10.1073/pnas.95.15.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Longcore JE, Pessier AP, Nichols DK. Batrachochytrium dendrobatidis gen et sp nov, a chytrid pathogenic to amphibian. Mycologia. 1999;91(2):219–227. [Google Scholar]
- 16.Voyles J, et al. Pathogenesis of chytridiomycosis, a cause of catastrophic amphibian declines. Science. 2009;326(5952):582–585. doi: 10.1126/science.1176765. [DOI] [PubMed] [Google Scholar]
- 17.Rosenblum EB, Poorten TJ, Settles M, Murdoch GK. Only skin deep: Shared genetic response to the deadly chytrid fungus in susceptible frog species. Mol Ecol. 2012;21(13):3110–3120. doi: 10.1111/j.1365-294X.2012.05481.x. [DOI] [PubMed] [Google Scholar]
- 18.Skerratt LF, et al. Spread of chytridiomycosis has caused the rapid global decline and extinction of frogs. EcoHealth. 2007;4(2):125–134. [Google Scholar]
- 19.Rachowicz LJ, et al. The novel and endemic pathogen hypotheses: Competing explanations for the origin of emerging infectious diseases of wildlife. Conserv Biol. 2005;19(5):1441–1448. [Google Scholar]
- 20.Kilpatrick AM, Briggs CJ, Daszak P. The ecology and impact of chytridiomycosis: An emerging disease of amphibians. Trends Ecol Evol. 2010;25(2):109–118. doi: 10.1016/j.tree.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 21.Rosenblum EB, et al. A molecular perspective: Biology of the emerging pathogen Batrachochytrium dendrobatidis. Dis Aquat Organ. 2010;92(2–3):131–147. doi: 10.3354/dao02179. [DOI] [PubMed] [Google Scholar]
- 22.Lips KR, Diffendorfer J, Mendelson JR, Sears MW. Riding the wave: Reconciling the roles of disease and climate change in amphibian declines. PLoS Biol. 2008;6(3):e72. doi: 10.1371/journal.pbio.0060072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vredenburg VT, Knapp RA, Tunstall TS, Briggs CJ. Dynamics of an emerging disease drive large-scale amphibian population extinctions. Proc Natl Acad Sci USA. 2010;107(21):9689–9694. doi: 10.1073/pnas.0914111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng TL, Rovito SM, Wake DB, Vredenburg VT. Coincident mass extirpation of neotropical amphibians with the emergence of the infectious fungal pathogen Batrachochytrium dendrobatidis. Proc Natl Acad Sci USA. 2011;108(23):9502–9507. doi: 10.1073/pnas.1105538108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morehouse EA, et al. Multilocus sequence typing suggests the chytrid pathogen of amphibians is a recently emerged clone. Mol Ecol. 2003;12(2):395–403. doi: 10.1046/j.1365-294x.2003.01732.x. [DOI] [PubMed] [Google Scholar]
- 26.Morgan JAT, et al. Population genetics of the frog-killing fungus Batrachochytrium dendrobatidis. Proc Natl Acad Sci USA. 2007;104(34):13845–13850. doi: 10.1073/pnas.0701838104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.James TY, et al. Rapid global expansion of the fungal disease chytridiomycosis into declining and healthy amphibian populations. PLoS Pathog. 2009;5(5):e1000458. doi: 10.1371/journal.ppat.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farrer RA, et al. Multiple emergences of genetically diverse amphibian-infecting chytrids include a globalized hypervirulent recombinant lineage. Proc Natl Acad Sci USA. 2011;108(46):18732–18736. doi: 10.1073/pnas.1111915108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goka K, et al. Amphibian chytridiomycosis in Japan: Distribution, haplotypes and possible route of entry into Japan. Mol Ecol. 2009;18(23):4757–4774. doi: 10.1111/j.1365-294X.2009.04384.x. [DOI] [PubMed] [Google Scholar]
- 30.Schloegel LM, et al. Novel, panzootic and hybrid genotypes of amphibian chytridiomycosis associated with the bullfrog trade. Mol Ecol. 2012;21(21):5162–5177. doi: 10.1111/j.1365-294X.2012.05710.x. [DOI] [PubMed] [Google Scholar]
- 31.Longcore JE, Letcher PM, James TY. Homolaphlyctis polyrhiza gen. et sp nov., a species in the Rhizophydiales (Chytridiomycetes) with multiple rhizoidal axes. Mycotaxon. 2011;118:433–440. [Google Scholar]
- 32.Gomes A, et al. Bioactive molecules from amphibian skin: Their biological activities with reference to therapeutic potentials for possible drug development. Indian J Exp Biol. 2007;45(7):579–593. [PubMed] [Google Scholar]
- 33.Whiles M, et al. Disease-driven amphibian declines alter ecosystem processes in a tropical stream. Ecosystems. 2012 doi: 10.1007/s10021-10012-19602-10027. [DOI] [Google Scholar]
- 34.Velo-Anton G, et al. Amphibian-killing fungus loses genetic diversity as it spreads across the New World. Biol Conserv. 2012;146(1):213–218. [Google Scholar]
- 35.Murray K, et al. The distribution and host range of the pandemic disease chytridiomycosis in Australia, spanning surveys from 1956–2007. Ecology. 2010;91(4):E091–E108. [Google Scholar]
- 36.Carnaval AC, et al. Amphibian chytrid fungus broadly distributed in the Brazilian Atlantic rain forest. EcoHealth. 2006;3(1):41–48. [Google Scholar]
- 37.Toledo LF, Britto FB, Araujo OGS, Giasson LMO, Haddad CFB. The occurance of Batrachochytrium dendrobatidis in Brazil and the inclusion of 17 new cases of infection. South Am J Herpetol. 2006;1(3):185–191. [Google Scholar]
- 38.Gründler MC, et al. Interaction between breeding habitat and elevation affects prevalence but not infection intensity of Batrachochytrium dendrobatidis in Brazilian anuran assemblages. Dis Aquat Organ. 2012;97(3):173–184. doi: 10.3354/dao02413. [DOI] [PubMed] [Google Scholar]
- 39.Bai C, et al. Global and endemic Asian lineages of the emerging pathogenic fungus Batrachochytrium dendrobatidis widely infect amphibians in China. Divers Distrib. 2012;18(3):307–318. [Google Scholar]
- 40.Soto-Azat C, Clarke BT, Poynton JC, Cunningham AA. Widespread historical presence of Batrachochytrium dendrobatidis in African pipid frogs. Divers Distrib. 2010;16(1):126–131. [Google Scholar]
- 41.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290(5494):1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 42.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ballard JWO, Whitlock MC. The incomplete natural history of mitochondria. Mol Ecol. 2004;13(4):729–744. doi: 10.1046/j.1365-294x.2003.02063.x. [DOI] [PubMed] [Google Scholar]
- 44.Weir BS. Genetic Data Analysis II. Sunderland, MA: Sinauer; 1996. [Google Scholar]
- 45.Riles L, et al. Physical maps of the six smallest chromosomes of Saccharomyces cerevisiae at a resolution of 2.6 kilobase pairs. Genetics. 1993;134(1):81–150. doi: 10.1093/genetics/134.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jensen-Seaman MI, et al. Comparative recombination rates in the rat, mouse, and human genomes. Genome Res. 2004;14(4):528–538. doi: 10.1101/gr.1970304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Forche A, et al. Stress alters rates and types of loss of heterozygosity in Candida albicans. mBio. 2011;2(4):e00129-11. doi: 10.1128/mBio.00129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berger L, Marantelli G, Skerratt LF, Speare R. Virulence of the amphibian chytrid fungus Batrachochytium dendrobatidis varies with the strain. Dis Aquat Organ. 2005;68(1):47–50. doi: 10.3354/dao068047. [DOI] [PubMed] [Google Scholar]
- 49.Joneson S, Stajich JE, Shiu SH, Rosenblum EB. Genomic transition to pathogenicity in chytrid fungi. PLoS Pathog. 2011;7(11):e1002338. doi: 10.1371/journal.ppat.1002338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosenblum EB, Poorten TJ, Joneson S, Settles M. Substrate-specific gene expression in Batrachochytrium dendrobatidis, the chytrid pathogen of amphibians. PLoS One. 2012;7(11):e49924. doi: 10.1371/journal.pone.0049924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abramyan J, Stajich JE. Species-specific chitin-binding module 18 expansion in the amphibian pathogen Batrachochytrium dendrobatidis. mBio. 2012;3(3):e00150-12. doi: 10.1128/mBio.00150-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Freeman KR, et al. Evidence that chytrids dominate fungal communities in high-elevation soils. Proc Natl Acad Sci USA. 2009;106(43):18315–18320. doi: 10.1073/pnas.0907303106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gahl MK, Longcore JE, Houlahan JE. Varying responses of northeastern North American amphibians to the chytrid pathogen Batrachochytrium dendrobatidis. Conserv Biol. 2012;26(1):135–141. doi: 10.1111/j.1523-1739.2011.01801.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.