Abstract
A long-held dogma posits that strong presentation to the immune system of the dominant influenza virus glycoprotein antigens neuraminidase (NA) and hemagglutinin (HA) is paramount for inducing protective immunity against influenza virus infection. We have deliberately violated this dogma by constructing a recombinant influenza virus strain of A/PR8/34 (H1N1) in which expression of NA and HA genes was suppressed. We down-regulated NA and HA expression by recoding the respective genes with suboptimal codon pair bias, thereby introducing hundreds of nucleotide changes while preserving their codon use and protein sequence. The variants PR8-NAMin, PR8-HAMin, and PR8-(NA+HA)Min (Min, minimal expression) were used to assess the contribution of reduced glycoprotein expression to growth in tissue culture and pathogenesis in BALB/c mice. All three variants proliferated in Madin–Darby canine kidney cells to nearly the degree as WT PR8. In mice, however, they expressed explicit attenuation phenotypes, as revealed by their LD50 values: PR8, 32 plaque-forming units (PFU); HAMin, 1.7 × 103 PFU; NAMin, 2.4 × 105 PFU; (NA+HA)Min, ≥3.16 × 106 PFU. Remarkably, (NA+HA)Min was attenuated >100,000-fold, with NAMin the major contributor to attenuation. In vaccinated mice (NA+HA)Min was highly effective in providing long-lasting protective immunity against lethal WT challenge at a median protective dose (PD50) of 2.4 PFU. Moreover, at a PD50 of only 147 or 237, (NA+HA)Min conferred protection against heterologous lethal challenges with two mouse-adapted H3N2 viruses. We conclude that the suppression of HA and NA is a unique strategy in live vaccine development.
Keywords: chemical synthesis, computer-aided design, margin of safety, Protective dose 50, hetero-subtypic immunity
Influenza virus is a human disease agent that leads to >30,000 deaths in the United States and several hundred thousand deaths globally each year (1). Why is influenza not better controlled? There are four reasons. First, the major neutralization antigenic proteins providing protective immunity on the virion surface, hemagglutinin (HA) and neuraminidase (NA), undergo yearly genetic variation by point mutations (i.e., genetic drift). This renders the viruses resistant to population immunity, thereby setting the stage for “seasonal” epidemics (2). Second, influenza virus may acquire a new antigenic makeup (i.e., reassortment of heterologous genes, referred to as genetic shift), leading to pandemics (3). Third, because of the seasonal flu, new vaccines must be produced every year. This is a complex undertaking, considering that more than one type or strain of influenza virus cocirculates in any flu season, possibly requiring the development of multiple new vaccines each year. Fourth, only two major types of vaccines are currently licensed: intramuscularly administered trivalent inactivated vaccine (TIV) and (cold-adapted) live attenuated vaccine (LAIV), given intranasally. The efficacy of these two vaccines is suboptimal.
The injectable TIVs, which require a large quantity of starting material, the equivalent of ∼1010 plaque-forming units (PFU) per dose, are incapable of inducing significant cell-mediated immunity, which is now recognized as an important determinant of protection (4). Moreover, the overall efficacy of TIVs in the US adult population aged 18–65 y is only 59% (5). In contrast, LAIVs induce both humoral and cellular immunity, but are restricted for use in persons aged 2–49 y (6, 7). Moreover, recurrent administration of LAIVs, which use the same attenuating viral backbone, could result in tolerance in repeat recipients (8).
Influenza viruses, classified as types A, B, and C, are enveloped, negative-strand RNA viruses of Orthomyxoviridae, of which subtypes of type A are the major culprit in human disease (9). The viruses transcribe and replicate their multipartite genomes in the cell nucleus, with each segment encoding one or two polypeptides. Of these, the most important antigenic molecules are the glycoproteins HA and NA.
A basic premise in influenza vaccination is adequate delivery of HA and NA to vaccine recipients, assuming that a very high dose (TIV) or a dose corresponding to live viral infection (LAIV) of these traditionally dominant antigenic polypeptides alone is sufficient for adequate vaccine efficacy. Here we present data supporting the opposite, where deliberate reduction of influenza HA and NA expression leads to an ultraprotective live vaccine. Infection of BALB/c mice with a live subtype A H1N1 (PR8) virus designed to significantly reduce HA and NA synthesis led to a vaccine candidate with excellent growth properties in tissue culture cells but with an extraordinary safety profile, as shown by a favorable median protective dose (PD50). The results obtained with this counterintuitive strategy may lead to an entirely new class of live influenza virus vaccines.
To modify the expression of HA and NA in the H1N1 vaccine candidate, we made use of a little-known phenomenon known as codon pair bias. In this bias, pairs of synonymous codons do not appear in protein-encoding sequences of a species at the frequency that would be predicted based on the genome-wide frequency (or “codon usage”) of the individual codons (10–12). Certain pairs are statistically overrepresented (preferred) in the ORFeome, whereas other pairs are underrepresented (disliked); the difference can be quantitated by calculating a codon pair score that is more positive for the preferred codon pairs and more negative for the disliked codon pairs (13, 14). The differences in biological function between overrepresented and underrepresented codon pairs are small, but if a gene is recoded with a large number of underrepresented (“deoptimized”) codon pairs (leading to hundreds of point mutations), the effect on expression of the corresponding protein can be substantial (13, 14). Importantly, neither codon use nor protein sequences are changed during these manipulations.
Using a specific algorithm and chemical synthesis (9), we constructed virus PR8 (A/Puerto Rico/8/1934; H1N1), containing an excess of underrepresented codon pairs (“deoptimized”) in NA and HA genes [variant (NA+HA)Min; Min, minimal gene expression]. Variant (NA+HA)Min has surpassed our expectations of safety and PD50 in the mouse system. Moreover, mice vaccinated with 1,000 PFU of (NA+HA)Min variant were fully protected against lethal challenge with two different heterologous, mouse-adapted H3N2 influenza viruses.
Results and Discussion
Construction and Characterization of an HA and NA Codon Pair-Deoptimized Influenza Virus in Tissue Culture.
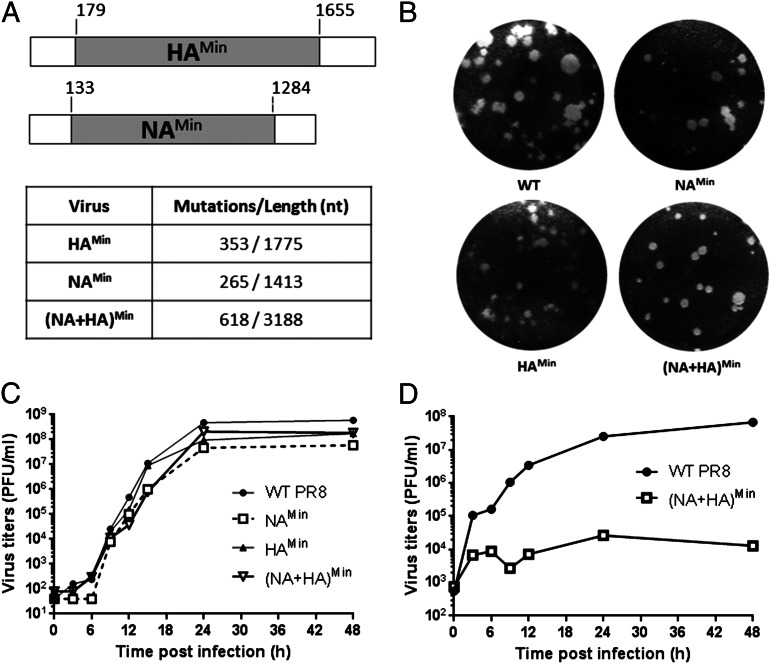
To achieve attenuation of influenza virus PR8, we deoptimzed codon pair bias (introducing underrepresented codon pairs) in viral genes HA and NA according to computer algorithms (13, 14) and chemical syntheses (15), aiming to reduce the expression level of the targeted viral genes. The NA and HA glycoproteins are considered the major dominant antigenic determinants of influenza viruses. Our seemingly counterintuitive approach of targeting the expression level of the two most important antigens was based on the assumption that neither the HA gene product nor the NA gene product is directly involved in the intracellular replicative cycle (i.e, transcription, translation, and RNA replication). Thus, an influenza virus expressing reduced amounts of HA and NA protein might still replicate well up to the steps of virus budding, whereas maturation and cell-to-cell spread may be impaired. We have previously shown that PR8 with only deoptimized HA (HAMin; 353 synonymous mutations in 1,775 nucleotides) replicated well in Madin–Darby canine kidney (MDCK) epithelial cells (14). Similarly, a variant with a deoptimized NA gene (NAMin; 265 of 1,413 synonymous mutations; Fig. 1A) also replicated well in MDCK cells (Fig. 1C), and expressed a phenotype with only a slightly smaller plaque size than WT PR8 (Fig. 1B). Interestingly, variant (NA+HA)Min (618 of 3,188 nucleotide changes), combining the HAMin and NAMin genes, expressed growth and plaque phenotypes in MDCK cells comparable to those of the individual HAMin and NAMin variants (Fig. 1 B and C). Apparently, in MDCK cells, the codon pair deoptimization of (NA+HA)Min does not involve an “additive” relationship of the effect of deoptimized genomes, with significantly more negative codon pair scores in HAMin and NAMin, as was found with poliovirus (13).
Fig. 1.
Construction of codon pair-deoptimized variants and phenotypes in tissue cultures. (A) NAMin and HAMin were designed as described previously (12), leaving 120–200 nt of WT sequences at the 5′ and 3′ ends (3), and constructed by chemical synthesis. They were then used to replace one or two corresponding genes of WT PR8 by reverse genetics (14). The number of synonymous mutations is shown. (B) Recovered viruses were analyzed for plaque size on MDCK monolayers. (C) Growth kinetics of WT PR8 and codon pair-deoptimized variants were analyzed in MDCK cells after infection at an MOI of 0.01. At every 3 h p.i., cell supernatants were collected and analyzed for virus titers by plaque assays. (D) Growth kinetics of WT PR8 and (NA+HA)Min virus were analyzed in A549 cells after infection at an MOI of 1.
However, in adenocarcinomic human alveolar basal epithelial (A549) cells, the (NA+HA)Min variant was highly attenuated (Fig. 1D), growing to a final titer three to four orders of magnitude lower than WT PR8. A549 cells retain a complex signaling network that is related to the innate host response (16, 17). Our results suggest that the innate immune response may have a greater impact on (NA+HA)Min than on WT PR8, with the latter growing to similar titers in MDCK and A549 cells.
Reduced NA mRNA and HA Protein Levels in (NA+HA)Min-Infected Cells.
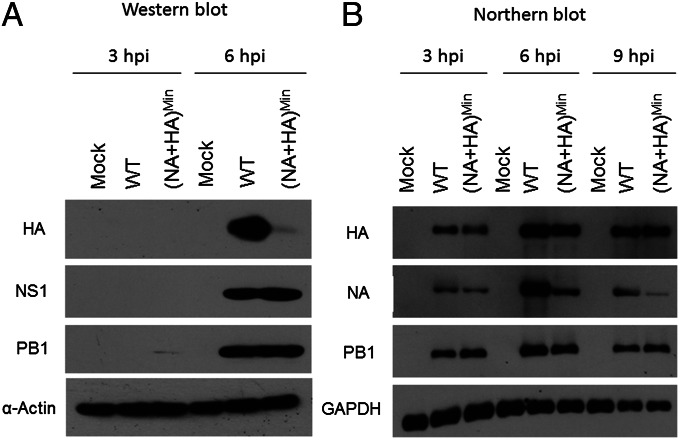
The adequate growth properties of (NA+HA)Min in MDCK cells raised questions as to the extent of HA and NA synthesis from its codon pair-deoptimized genes. We first tested the apparent yield of HA polypeptide by Western blot analysis in MDCK cells at 3 h and 6 h postinfection (p.i.) with WT virus or (NA+HA)Min at a multiplicity of infection (MOI) of 5. At 6 h p.i., expression of HA protein was significantly reduced in (NA+HA)Min-infected cells compared with PR8-infected cells, whereas polymerase 1 (PB1) and nonstructural protein 1 (NS1) were synthesized to equal levels by viruses (Fig. 2A). Unfortunately, we were unable to carry out the same analysis for NA, owing to a lack of PR8 NA-specific antibodies.
Fig. 2.
Protein expression and mRNA levels in (NA+HA)Min-infected tissue culture cells. MDCK cells were infected with (NA+HA)Min or WT PR8 at an MOI of 5. (A) Western blot analyses were performed to determine the viral protein expression in infected cells at 3 h and 6 h p.i. (B) Northern blot analyses were performed to determine mRNA levels of HA, NA, PB1, and GAPDH in (NA+HA)Min- or WT PR8-infected MDCK cells (Materials and Methods). At 3, 6, and 9 h p.i., cytoplasmic mRNAs were collected and analyzed. For HAMin and HAWT transcript probes, we used the same 150 nucleotides that recognized the common 3′ end of the respective genes. Similarly, the probes for NAMin and NAWT have the same 150-nt sequence corresponding to the common 3′ end of the NA genes.
There are two possible explanations for the reduced amount of HA protein in (NA+HA)Min-infected cells. First, codon pair deoptimization of the HA gene might render the corresponding mRNA ineffective in translation, a scenario fitting with our hypothesis of translation of mRNAs with significantly negative codon pair scores (13). Second, there might be faster turnover of mRNA transcripts of the modified HA gene. However, using the levels of PB1 and GAPDH mRNAs as controls, Northern blot analyses of mRNA levels in (NA+HA)Min-infected cells revealed only slight reductions of HAMin mRNA at 3 h and 6 h (Fig. 2B). We suggest that the small extent of this reduction cannot explain the greatly diminished amount of HA protein (Fig. 2A). This result supports our hypothesis of reduced protein translation of the recoded HA mRNA (13). In contrast, Northern blot analyses indicated extensive reduction of the recoded NAMin mRNA after 6 h p.i. and particularly after 9 h p.i. (Fig. 2B). The reason for the decreased recoded NAMin mRNA signal is not known. The underrepresentation of NAMin mRNA is not likely related to inadequate transcription, considering that the sequences in the nontranslated region and terminal sequences of the ORF gene were kept WT (Fig. 1A).
Early in the course of infection (3 h p.i.), the level of NAMin mRNA was slightly reduced, yet on progression of replication, a turnover of the recoded NAMin mRNA seemed possible. The turnover of NAMin mRNA may be caused by a ribosome surveillance mechanism (e.g., no-go-decay) (18). How (NA+HA)Min can proliferate well in MDCK cells despite the reduced level of HA and presumably also of NA is unclear. Apparently, those reduced levels are not yet below the threshold of protein requirement in MDCK cells, where cell-to-cell spread of the virus is virtually unimpeded by host immune surveillance or physical barriers (extracellular matrix).
Characterization of the Codon Pair-Deoptimized Variants as Vaccine Candidates in Mice.
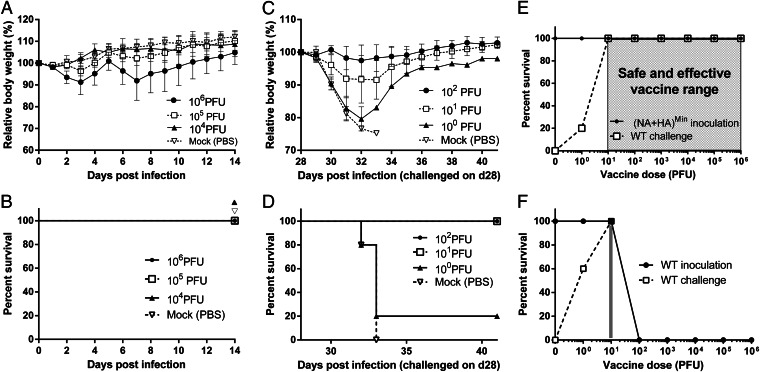
Given that the (NA+HA)Min variant grew well in MDCK cells but poorly in A594 cells, we were interested in examining its growth phenotype and pathogenesis in an animal model. Groups of five BALB/c mice received (NA+HA)Min at a dose of 104, 105, or 106 PFU intranasally, and body weight and survival was monitored continuously for 14 d p.i. (Fig. 3 A and B). Interestingly, the (NA+HA)Min variant did not induce apparent disease after a dose of up to 105 PFU. Even at 106 PFU, mice sustained transient weight loss, but all survived. Thus, the theoretical LD50 of the (NA+HA)Min variant was calculated to be ≥3.16 × 106 PFU, which exceeds that of WT PR8 by a factor of at least 100,000.
Fig. 3.
Pathogenicity in mice. (A and B) Measurements of LD50. Groups of five male Balb/C mice were intranasally infected with the (NA+HA)Min variant at 104, 105, or 106 PFU, and their relative body weight (A) and survival rate (B) were monitored for 14 d p.i.. Mice that lost 25% of their body weight were euthanized. LD50 was calculated based on the Reed–Muench method (23). (C and D) Measurement of PD50. Groups of five male Balb/C mice were intranasally vaccinated with 102, 101, or 100 PFU of (NA+HA)Min on day 0. On day 28 postvaccination, all mice were challenged with 105 PFU of WT PR8 virus, and their relative body weight (C) and survival rate (D) after challenge were monitored. PD50 was calculated based on the Reed–Muench method (23). (E and F) Safe and effective vaccine ranges of the (NA+HA)Min (open box) and WT PR8 virus (gray zone) were plotted. Any vaccine dose within these regions was associated with survival, as well as complete protection from lethal homologous challenge. Error bars represent SD.
To what extent do the recoded HAMin and NAMin genes contribute to the attenuated phenotype of the double mutant (NA+HA)Min? Whereas the (NA+HA)Min, HAMin, and NAMin variants replicated with nearly equal efficiency and similar kinetics as WT PR8 in MDCK cells (Fig. 1C), the LD50 of the variants were by orders of magnitude different: PR8 = 32 PFU, HAMin = 1.7 × 103 PFU (14), NAMin = 2.4 × 105 PFU (Fig. S1), and (NA+HA)Min ≥3.16 × 106. Because individually, the NAMin is about 100-fold more attenuated than the HAMin, it appears that, surprisingly, the recoding of the NA gene is the major contributor to (NA+HA)Min attenuation. We hypothesize that this observation is related to the important biological role of neuraminidase in aiding virus spread by destroying virus receptors on the cells from which newly synthesized virions are being released (19, 20). Another proposed function of NA is facilitating virus access to the respiratory epithelial target cells, by enzymatically degrading the protective barrier of mucin-like glycoconjugates covering the respiratory tract (21, 22). Such a function may be more critical for infectivity of the virus in an intact host organisms compared with tissue culture cells, where few such barriers exist.
If (NA+HA)Min were to be used as a vaccine candidate it must be capable of providing, at low dose, long-term protection from challenge with a lethal dose of WT virus. Therefore, we determined the dose of (NA+HA)Min required to protect 50% of vaccinated animals from subsequent lethal WT challenge (PD50). Groups of five BALB/c mice were vaccinated with a single dose of 100, 101, or 102 PFU of (NA+HA)Min intranasally. 28 d after vaccination, the animals were challenged with 105 PFU (3,000 × LD50) of WT PR8 virus. As with the original infections, we monitored body weight and survival of the animals for 14 d after the challenge. Remarkably, although (NA+HA)Min was highly attenuated in mice, it was also highly proficient at protecting against lethal challenge with the WT virus. As little as 10 PFU of (NA+HA)Min protected all five mice from lethal challenge (Fig. 3 C and D). The PD50 value calculated by the Reed–Muench method (23) was only 2.4 PFU. To our knowledge, this is the lowest reported protective dose of an experimental vaccine in a mouse model.
As a measure of safety versus efficacy, we determined vaccine safety and protective range with various doses of either (NA+HA)Min variant or WT PR8. A zone of five orders of magnitude (from 10 to 106 PFU) can be considered the “region of safety” of (NA+HA)Min vaccination (Fig. 3E), given that all mice receiving increasing doses of “vaccine” within this region were protected from lethal challenge with the WT virus. In contrast, the safe and effective region for WT PR8 was extremely limited (Fig. 3F).
Greatly Reduced Growth of (NA+HA)Min in the Lungs of Vaccinated Mice.
To determine parameters of the (NA+HA)Min pathogenicity in vivo, we infected BALB/c mice with 104 PFU of WT PR8 or (NA+HA)Min. On days 1, 3, 5, 7, 9, and 11, three mice each from the WT PR8 and (NA+HA)Min groups were euthanized, after which their lungs were homogenized and virus titers in the homogenates were determined by plaque assays. As expected, WT PR8 replicated well, but even (NA+HA)Min replicated noticeably in lungs of the vaccinated animals. Both PR8 and the variant achieved maximum titers at around day 3 (Fig. 4A) although there was an ∼100-fold difference in the titers between the two viruses. All WT PR8-infected mice died on day 5, whereas all (NA+HA)Min-infected mice remained healthy. (NA+HA)Min was eventually cleared at 8–9 d p.i. (Fig. 4A). Generally, when mice were inoculated at different doses, the (NA+HA)Min titers were always 100- to 1,000-fold lower in lungs compared with WT PR8 titers on day 3 p.i. (Fig. 4B). Interestingly, a vaccination dose of 10 PFU, at which (NA+HA)Min barely replicated in the lungs of the animals, nevertheless provided complete protection against WT PR8 challenge (Figs. 3D and 4B). In addition, the attenuation of (NA+HA)Min in mice correlates with the attenuation of (NA+HA)Min in A549 cells (Fig. 1D). The phenotype of the (NA+HA)Min variant that we observed in MDCK cells and mice share similarity with an HA receptor binding site mutant virus (Y98F), in which the sialic acid binding ability of HA was reduced (24–26). In contrast, in our variants, amino acid sequence is 100% preserved, and the observed phenotypes are a direct result of the reduced protein expression, rather than of a change in protein function per se.
Fig. 4.
Virus titers in the lungs of infected mice. (A) Groups of three male Balb/C mice were intranasally infected with 104 PFU of WT PR8 or (NA+HA)Min. On days 1, 3, 5, 7, 9, and 11 p.i., the mice were euthanized, and their lungs were harvested and homogenized. Viral titers in the homogenates were determined by plaque assays on MDCK cells. †All WT PR8-infected mice were dead by day 5 p.i. ‡The virus titers in (NA+HA)Min-infected mice after day 9 p.i. were undetectable (<4 PFU). (B) Comparison of virus titers in lungs of three mice each infected with WT PR8 or (NA+HA)Min at a dose of 101–104 PFU. The lungs of the animals were harvested on day 3 p.i., and plaque assays were performed to determine virus titers. Error bars represent SD. *P < 0.05.
Cross-Protection and Long-Term Protection Induced by the (NA+HA)Min Variant.
The (NA+HA)Min variant of PR8 belongs to the influenza H1N1 subtype. Considering the significant ability of this variant to protect against homologous challenge, we were interested in exploring whether (NA+HA)Min could cross-protect animals against infections with a heterologous influenza virus strain, such as a mouse-adapted H3N2 strain (A/Aichi/2/1968) (27). Groups of five BALB/c mice were vaccinated with (NA+HA)Min virus at doses of 102–104 PFU and challenged at 28 d postvaccination with a 100 × LD50 dose of H3N2 virus (1.5 × 104 PFU). Here an (NA+HA)Min dose of 1,000 PFU was sufficient to protect mice from the heterologous lethal challenge, corresponding to a PD50 value of only 237 PFU (Fig. 5 A and B) . A similar result was obtained when the vaccinated (NA+HA)Min mice were challenged with a different strain of mouse- adapted H3N2, A/Victoria/3/75. Again, as little as 1,000 PFU of the H1N1 PR8-(NA+HA)Min variant protected all mice from lethal challenge with a 100 × LD50 dose (3.2 × 104 PFU) of A/Victoria/3/75. The PD50 of (NA+HA)Min protecting against A/Victoria/3/75 (H3N2) was only 147 PFU (Fig. 5 C and D). The foregoing results suggest that (NA+HA)Min of H1N1 PR8 can induce a robust cross-protective immune response in mice against H3N2 subtypes. Further experiments, including passive transfer in mice, may be conducted to determine whether the CD8+ T cells contribute to the heterosubtypic immunity. In addition, the same experiment might need to be repeated in ferrets, which more closely represent influenza infection in humans.
Fig. 5.
Cross-protection against H3N2 virus infections in (NA+HA)Min(H1N1)-vaccinated mice. (A and B) Groups of five BALB/c mice were vaccinated intranasally with (NA+HA)Min at different doses. On day 28 postvaccination, the mice were challenged with 100 × LD50 heterologous viruses A/Aichi/2/1968 (H3N2) virus (1.5 × 104 PFU). Survival rate and relative body weights were monitored for 14 d. The cross-protection PD50 against H3N2 Aichi virus was calculated to be 237 PFU. (C and D) Mice vaccinated intranasally with (NA+HA)Min virus were also challenged with 100 × LD50 A/Victoria/3/75 (H3N2) virus (3.2 × 104 PFU). Survival rate and relative body weights were monitored for 14 d. The cross-protection PD50 against H3N2 Victoria virus was calculated as 147 PFU based on the Reed–Muench method (23). Error bars represent SD.
We also asked whether (NA+HA)Min-vaccinated animals were protected against challenge after an extended period. Groups of five mice were vaccinated with different doses (101–105 PFU) of (NA+HA)Min and then challenged 7 mo later with 105 PFU of WT PR8. All vaccinated animals were completely protected, with no signs of disease (Fig. S2).
Robust Antibody Response Induced by the (NA+HA)Min Variant.
The host response to (NA+HA)Min inoculation suggested a strong host response, including adaptive immunity. We tested this by vaccinating groups of five BALB/c mice with different doses of (NA+HA)Min or WT PR8 and collecting sera on day 28 p.i. The antibody response was then determined by a hemagglutination inhibition (HAI) assay. The (NA+HA)Min virus was capable of inducing an equally high HAI titer as the WT virus, albeit at higher doses (Fig. 6 A and B). In parallel, we challenged the mice with a lethal dose of PR8 (105 PFU). An HAI titer of ≥40 in the serum is generally considered protective (28). This level was reached with 101 PFU of (NA+HA)Min, which protected vaccinated mice from challenge with 105 PFU of WT PR8 virus (Fig. 6B).
Fig. 6.
HAI assay with serum of vaccinated mice. Groups of five male BALB/c mice were vaccinated intranasally with different doses of WT PR8 virus (A) or the (NA+HA)Min variant (B). Note that all mice “vaccinated” with 102 PFU of WT virus died (Fig. 3F). Sera were collected on day 28 p.i., and serum antibody titers were determined by HAI assays as described in Materials and Methods. All mice were then challenged with 105 PFU of WT PR8, and their survival rates were monitored for an additional 14 d. Each dot represents a mouse. Gray symbols represent mice that did not survive after the WT challenge.
Concluding Remarks.
We have used a unique strategy, which we call “synthetic attenuated virus engineering” (SAVE) (12), to modify genes of influenza virus by computer-aided design and chemical synthesis. This procedure leads to viral variants with hundreds of nucleotide changes in ORFs without changing codon use or amino acid sequences. The sequence changes result from rearranging existing synonymous codons such that new codon pairs with suboptimal viral gene expression are formed (13).
We targeted the HA and NA genes, which encode the two glycoproteins most important in conferring immunity to influenza virus infection, thereby generating viral variants NAMin, HAMin (12), and (NA+HA)Min. The deliberate reduction of NA and HA expression may seem completely counterintuitive for the development of an influenza vaccine, yet it allowed us to evaluate the roles of HA and NA in influenza virus pathogenesis. Our first surprising finding was that the suppression of HA protein synthesis and significantly decreased levels of NA mRNA in (NA+HA)Min-infected cells did not significantly affect viral growth in MDCK cells. Our second surprising finding was that, in contrast to the efficient replication in MDCK cells, (NA+HA)Min not only was significantly attenuated in mice, but also conferred an unusually wide range of vaccine safety against lethal challenges (5 logs) from WT virus to these mice. Comparing LD50 values also led to unexpected observations of the very different effects of gene recoding: WT PR8, 32 PFU; HAMin, 1.7 × 103 PFU; NAMin, 2.4 × 105 PFU; (NA+HA)Min, ≥3.16 × 106 PFU. Remarkably, recoding solely the NA gene attenuated the WT PR8 by nearly 10,000-fold, 100-fold more than recoding solely the HA gene. It is possible that that the substantial attenuation phenotype of NAMin is the consequence of reduced spread of this variant in the respiratory tract and lungs of the host. When combined with the deoptimized HA gene, the production of suboptimal quantities of NA and HA may delay normal maturation of virus particles, further inhibiting viral spread. Although this is pure speculation, HA molecules in (NA+HA)Min-infected animals might be degraded, perhaps already during synthesis from the recoded mRNAs, leading to the presentation of HA fragments and novel species of neutralizing antibodies.
These considerations may explain the very small PD50 of only 2.4 PFU achieved with (NA+HA)Min, perhaps the lowest PD50 of any experimental vaccine in the mouse model. At the same time, (NA+HA)Min induced significant production of virus-specific antibodies. Moreover, (NA+HA)Min-vaccinated animals were significantly cross-protected against heterologous virus challenge with two H3N2 subtypes (A/Aichi/2/1968 and A/Victoria/3/75). Considering that the H3N2 Aichi strain and our original PR8 strain share ∼42% similarity of their HA and NA protein sequences, the dose required to achieve full cross-protection against the heterologous H3N2 viruses (PD50 = 147 and 237 PFU, respectively) seems low.
We envision that LAIVs, based on codon pair-deoptimization of HA and NA, can be derived each year anew in their entirety according to the strains annually recommended by the World Health Organization. Designing a codon pair-deoptimized virus can be done in a matter of minutes, and new seed viruses can be produced by de novo DNA synthesis in as little as 3 wk from the time the sequences of the upcoming season have been announced. Because our method does not depend on any specific virus backbone or any reassortant virus, such a vaccine candidate would be 100% identical in all viral proteins to the circulating target strain. Thus, the vaccine virus would keep pace with the evolving seasonal target virus in all eight gene segments, not only in HA and NA as is the case with all currently used influenza vaccines. This would ensure the best possible antigenic match for cellular immune epitopes, and also likely would enhance the cross-protective capacity of such a vaccine against HA-drifted viruses. In addition, a codon pair-deoptimized vaccine strain would not introduce any new gene segments into the seasonal human influenza virus pool that are not already circulating in the population, thus eliminating the possibility of novel reassortants between vaccine virus and circulating WT viruses. Of course, the response of national or international regulators to codon pair-deoptimized vaccines remains to be tested; nonetheless, we suggest that suppressing HA and NA expression in live virus variants presents a unique and counterintuitive strategy in influenza vaccine development.
Materials and Methods
Cells and Viruses.
MDCK (dog kidney epithelial), A549 (human lung epithelial carcinoma), and HEK293 T-cell lines were maintained in DMEM supplemented with 10% FBS at 37 °C. Influenza A/PR/8/34 (PR8) was cultured in MDCK cells. The genes of NAMin, HAMin, and (NA+HA)Min variants were codon pair-deoptimized. They were designed, synthesized, and characterized as described previously (12, 13).
Western Blot and Northern Blot Analyses.
MDCK cells were infected with WT PR8 or (NA+HA)Min viruses or mock-infected at an MOI of 5. Western blot analysis was performed as described previously (14). Mouse monolclonal antibodies against influenza M1 (FluAc), HA (C102), and NS1(23-1) and goat polyclonal antibody against PB1 (vK20) were purchased from Santa Cruz Biotechnology. Total cytoplasmic RNA was extracted with an RNeasy Kit (Qiagen). Viral RNAs were detected with a DIG Northern Starter Kit (Roche). All probes were designed as described in the figure legends.
LD50, PD50, Long-Term Protection, and Lung Titers.
After intranasal inoculation of anesthetized animals with 50 μL of inoculum, the LD50 and PD50 values and the titers of virus in the lungs were determined as reported previously (14). Long-term protection assays were performed in the same way as the PD50 assays, with the challenge time points at 7 mo p.i. All animal studies were conducted under approved institutional protocols and in accordance with the guidelines established by Stony Brook University’s Institutional Animal Care and Use Committee.
Cross-Protection Assay.
Groups of five male BALB/c mice were vaccinated intranasally with 50 μL containing (NA+HA)Min at different doses. On day 28 postvaccination, mice were challenged with A/Aichi/2/1968 (H3N2) or A/Victoria/3/75 (H3N2) at a dose of 100 × LD50, and their survival rate and relative body weight were monitored for 14 d.
HAI Assay.
Groups of five male BALB/c mice were infected intranasally with 50 μL containing WT PR8 virus or (NA+HA)Min at different doses. On day 28 p.i., sera were collected, and HAI assays were performed as described in the protocol of the World Health Organization’s Manual on Animal Influenza Diagnosis and Surveillance (29).
Supplementary Material
Acknowledgments
We thank Dr. Ioanna Skountzou for influenza virus strains A/Aichi/2/1968 (H3N2) and A/Victoria/3/75 (H3N2), Dr. Adolfo Garcia Sastre and Dr. Peter Palese for plasmids, and Aniko Paul and Jeronimo Cello for a critical reading of the manuscript. This work was supported by National Institutes of Health Grant R01 AI07521901A1 (to E.W.), R01GM098400 (to B.F.), and National Science Foundation Grant DBI-1060572 (to S.S.).
Footnotes
Conflict of interest statement: The authors declare a conflict of interest. Stony Brook University has a patent pending covering the strategy described in this paper; S.S., B.F., S.M., and E.W. are coinventors named in the patent.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307473110/-/DCSupplemental.
References
- 1.Thompson WW, Comanor L, Shay DK. Epidemiology of seasonal influenza: Use of surveillance data and statistical models to estimate the burden of disease. J Infect Dis. 2006;194(Suppl 2):S82–S91. doi: 10.1086/507558. [DOI] [PubMed] [Google Scholar]
- 2.Smith DJ, et al. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305(5682):371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 3.Palese P, Shaw ML. Orthomyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. 5th Ed. Vol 2. Philadelphia: Lippincott Williams & Wilkins; 2006. pp. 1647–1690. [Google Scholar]
- 4.Simonsen L, et al. Impact of influenza vaccination on seasonal mortality in the US elderly population. Arch Intern Med. 2005;165(3):265–272. doi: 10.1001/archinte.165.3.265. [DOI] [PubMed] [Google Scholar]
- 5.Osterholm MT, Kelley NS, Sommer A, Belongia EA. Efficacy and effectiveness of influenza vaccines: A systematic review and meta-analysis. Lancet Infect Dis. 2012;12(1):36–44. doi: 10.1016/S1473-3099(11)70295-X. [DOI] [PubMed] [Google Scholar]
- 6.Belshe RB, et al. CAIV-T Comparative Efficacy Study Group Live attenuated versus inactivated influenza vaccine in infants and young children. N Engl J Med. 2007;356(7):685–696. doi: 10.1056/NEJMoa065368. [DOI] [PubMed] [Google Scholar]
- 7.Hussain AI, Cordeiro M, Sevilla E, Liu J. Comparison of egg and high- yielding MDCK cell-derived live attenuated influenza virus for commercial production of trivalent influenza vaccine: In vitro cell susceptibility and influenza virus replication kinetics in permissive and semi-permissive cells. Vaccine. 2010;28(22):3848–3855. doi: 10.1016/j.vaccine.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Tobler S, Roayaei J, Eick A. Live attenuated or inactivated influenza vaccines and medical encounters for respiratory illnesses among US military personnel. JAMA. 2009;301(9):945–953. doi: 10.1001/jama.2009.265. [DOI] [PubMed] [Google Scholar]
- 9.Bouvier NM, Palese P. The biology of influenza viruses. Vaccine. 2008;26(Suppl 4):D49–D53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutman GA, Hatfield GW. Nonrandom utilization of codon pairs in Escherichia coli. Proc Natl Acad Sci USA. 1989;86(10):3699–3703. doi: 10.1073/pnas.86.10.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moura G, et al. Large-scale comparative codon-pair context analysis unveils general rules that fine-tune evolution of mRNA primary structure. PLoS ONE. 2007;2(9):e847. doi: 10.1371/journal.pone.0000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang FP, Li H. Codon-pair usage and genome evolution. Gene. 2009;433(1-2):8–15. doi: 10.1016/j.gene.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 13.Coleman JR, et al. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320(5884):1784–1787. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller S, et al. Live attenuated influenza virus vaccines by computer-aided rational design. Nat Biotechnol. 2010;28(7):723–726. doi: 10.1038/nbt.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cello J, Paul AV, Wimmer E. Chemical synthesis of poliovirus cDNA: Generation of infectious virus in the absence of natural template. Science. 2002;297(5583):1016–1018. doi: 10.1126/science.1072266. [DOI] [PubMed] [Google Scholar]
- 16.Sutejo R, et al. Activation of type I and III interferon signalling pathways occurs in lung epithelial cells infected with low-pathogenic avian influenza viruses. PLoS ONE. 2012;7(3):e33732. doi: 10.1371/journal.pone.0033732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dove BK, et al. A quantitative proteomic analysis of lung epithelial (A549) cells infected with 2009 pandemic influenza A virus using stable isotope labelling with amino acids in cell culture. Proteomics. 2012;12(9):1431–1436. doi: 10.1002/pmic.201100470. [DOI] [PubMed] [Google Scholar]
- 18.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440(7083):561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palese P, Tobita K, Ueda M, Compans RW. Characterization of temperature-sensitive influenza virus mutants defective in neuraminidase. Virology. 1974;61(2):397–410. doi: 10.1016/0042-6822(74)90276-1. [DOI] [PubMed] [Google Scholar]
- 20.Liu C, Eichelberger MC, Compans RW, Air GM. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J Virol. 1995;69(2):1099–1106. doi: 10.1128/jvi.69.2.1099-1106.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J Virol. 2004;78(22):12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner R, Matrosovich M, Klenk HD. Functional balance between haemagglutinin and neuraminidase in influenza virus infections. Rev Med Virol. 2002;12(3):159–166. doi: 10.1002/rmv.352. [DOI] [PubMed] [Google Scholar]
- 23.Reed L, Muench M. A simple method for estimating fifty percent endpoints. Am J Hyg. 1938;27(3):493–497. [Google Scholar]
- 24.Bradley KC, et al. Analysis of influenza virus hemagglutinin receptor binding mutants with limited receptor recognition properties and conditional replication characteristics. J Virol. 2011;85(23):12387–12398. doi: 10.1128/JVI.05570-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meisner J, et al. Infectivity studies of influenza virus hemagglutinin receptor binding site mutants in mice. J Virol. 2008;82(10):5079–5083. doi: 10.1128/JVI.01958-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martín J, et al. Studies of the binding properties of influenza hemagglutinin receptor-site mutants. Virology. 1998;241(1):101–111. doi: 10.1006/viro.1997.8958. [DOI] [PubMed] [Google Scholar]
- 27.Koutsonanos DG, et al. Transdermal influenza immunization with vaccine-coated microneedle arrays. PLoS ONE. 2009;4(3):e4773. doi: 10.1371/journal.pone.0004773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Jong JC, et al. Haemagglutination-inhibiting antibody to influenza virus. Dev Biol (Basel) 2003;115:63–73. [PubMed] [Google Scholar]
- 29.WHO 2002. WHO Manual on Animal Influenza Diagnosis and Surveillance. Available at http://www.who.int/csr/resources/publications/influenza/whocdscsrncs20025rev.pdf. Accessed April 20, 2013.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.