Abstract
The jasmonate family of phytohormones plays central roles in plant development and stress acclimation. However, the architecture of their signaling circuits remains largely unknown. Here we describe a jasmonate family binding protein, cyclophilin 20-3 (CYP20-3), which regulates stress-responsive cellular redox homeostasis. (+)-12-oxo-phytodienoic acid (OPDA) binding promotes CYP20-3 to form a complex with serine acetyltransferase 1, which triggers the formation of a hetero-oligomeric cysteine synthase complex with O-acetylserine(thiol)lyase B in chloroplasts. The cysteine synthase complex formation then activates sulfur assimilation that leads to increased levels of thiol metabolites and the buildup of cellular reduction potential. The enhanced redox capacity in turn coordinates the expression of a subset of OPDA-responsive genes. Thus, we conclude that CYP20-3 is a key effector protein that links OPDA signaling to amino acid biosynthesis and cellular redox homeostasis in stress responses.
Keywords: wounding, cysteine biosynthesis, GSH, TGA, plant plasticity
The phytohormone, (-)-jasmonic acid [JA; (1R,2R)-3-oxo-2-(2Z)-2-pentenyl-cyclopentaneacetic acid] and its precursors/derivatives, collectively known as jasmonates, are derived from a trienoic fatty acid (TFA) through the octadecanoid pathway, and widely distributed throughout the plant kingdom. Jasmonates modulate the expression of numerous genes and mediate responses to various forms of abiotic and biotic stresses, including wounding, insect attack, pathogen infection, and UV damage. They also play pivotal roles in reproduction and other plant developmental processes, such as fruit ripening, root growth, tuberization, senescence, and tendril coiling. These important but diverse activities of jasmonates must be reflected by their versatility as molecular modulators. However, our current knowledge regarding their modes of action is incomplete (1, 2).
A recent breakthrough unraveled the mechanism of CORONATINE INSENSITIVE 1 (COI1)/JASMONATE ZIM-domain (JAZ)-mediated JA signaling that leads to global changes in gene expression (3, 4). More specifically, when it has been produced, JA is conjugated to isoleucine to form (+)-7-iso-jasmonoyl-L-isoleucine (JA-Ile) that binds the COI1 protein, a part of an SCF (SKP1-Cullin-F-box) ubiquitin E3 ligase. This complex then interacts and ubiquitinates JAZ proteins acting as negative transcriptional regulators (TRs) of JA-responsive genes. JAZ protein degradation by 26S proteasomes then activates transcription factors (TFs) and allows gene expression (1, 2). These discoveries have underscored the role of JA-Ile as an important bioactive form of JA.
However, jasmonate signaling must involve a more complex network, as a number of JA-responsive genes respond independently of COI1 (5). Furthermore, a primary JA precursor, (+)-12-oxo-phytodienoic acid [OPDA; (1S,2S)-3-oxo-2-(2Z-pentenyl)-cyclopent-4-ene-1-octanoic acid], triggers autonomous pathways that regulate unique subsets of jasmonate-responsive genes, coordinated with and without the canonical JA pathway (6). OPDA signaling is presumed independent of COI1, as it is unable to bind the COI1/JAZ complex (3). In addition, a recent study elucidated a distinct role of a jasmonoyl-L-tryptophan conjugate, linking jasmonate with auxin signaling (7), further supporting the notion that distinct messages sent out by specific “jasmonates” coordinate essential molecular and cellular processes. Hence, additive or synergistic activities of jasmonates, and other reactive electrophilic species (RES; also derived from TFA) (8), are needed for complete resistance to necrotrophic pathogens (e.g., Alternaria brassicicola) in Arabidopsis, as we demonstrate in the present study (Fig. S1). Arabidopsis mutants disrupted in TFA synthesis (fad3/7/8) or the octadecanoid pathway (aos/dde2-2) were impaired in disease resistance to a greater extent than mutants inhibited in JA-Ile derivatization (jar1) and JA signaling (coi1-1). Moreover, WT-like resistance of opr3, which arrests conversion of OPDA to JA, demonstrated a critical role of OPDA signaling in defense responses against A. brassicicola in the absence of JA (9) (Fig. S1 B–D).
A recent study, however, reported that opr3 is not a null mutant, concluding that fungal resistance of opr3 likely results from JA and not OPDA (10). In contrast, our studies confirmed that opr3 is impaired in OPR3 expression upon A. brassicicola infection, supporting results by Stintzi et al. (9) (Fig. S1E). The cause for this contradiction is not understood, but it may result from different experimental conditions, experimental errors, and/or pathogen strains (A. brassicicola vs. Botrytis cinerea). Further investigations must clarify the nature of opr3 mutant. Nonetheless, a growing body of evidence strongly supports distinct roles of OPDA signaling, although little is known about mechanisms that mediate the downstream effects of OPDA (reviewed in refs. 11, 12). Several studies underlined the importance of Arabidopsis TGA TFs in OPDA signaling, as a large number of OPDA-responsive genes (ORGs) are differentially regulated in TGA mutant (tga2/5/6) compared with WT plants upon OPDA application (13, 14). Thus, a redox-dependent transmission of OPDA signaling has been hypothesized whereby OPDA-responsive redox changes may activate TGA TFs (11). Indeed, redox-imbalanced mutants [gr1 (glutathione reductase1) and cad1 encoding γ-GLUTAMYLCYSTEINE SYNTHETASE1 (γECS1)] showed the modified expression of TGA transcripts (15, 16). GR1 maintains a high cellular glutathione (GSH):glutathione disulfide (GSSG) ratio by reducing GSSG (the oxidized disulfide form of GSH), whereas γECS1 initiates a two-step GSH biosynthetic process by metabolizing cysteine (Cys) and glutamic acid to form GSH (17), suggesting a potential role of cellular GSH and redox potentials in OPDA signaling.
Cys, a precursor of GSH and thiols, is synthesized through a hetero-oligomeric Cys synthase complex (CSC), composed of serine acetyltransferase (SAT) and O-acetylserine(thiol)lyase (OAS-TL), in cytosol, plastid, and mitochondria. CSC is a part of metabolic flux control systems that regulate cellular sulfur uptake and reduction. SAT transfers an acetyl moiety of acetyl-CoA to serine, producing O-acetylserine (OAS), which in turn accepts sulfide by OAS-TL, releasing Cys. Sulfate uptake and reduction initiates CSC formation that activates SAT, but inactivates OAS-TL, whereas OAS production dissociates CSC and activates OAS-TL. The precise function of CSC is uncharacterized, but its regulatory model indicates an intrinsic role in sulfide sensing to maintain sulfur homeostasis (18, 19).
Collectively, available data suggest that several effectors and/or receptors turn on and modulate a signaling circuitry that mediates the diverse effects of multiple members of jasmonate family. In the present study, a jasmonate pathway effector was identified, in the form of a jasmonate binding protein (JBP), cyclophilin 20-3 (CYP20-3). We demonstrate that CYP20-3 relays an OPDA signal in plastid Cys biosynthesis and buildup of cellular reduction potential. The CYP20-3–dependent enhanced redox capacity in turn coordinates a subset of ORG expression and plant stress acclimation. Thus, the present study highlights the importance of CYP20-3 as a key, physically interacting, OPDA effector.
Results
Jasmonate Affinity Purification of CYP20-3.
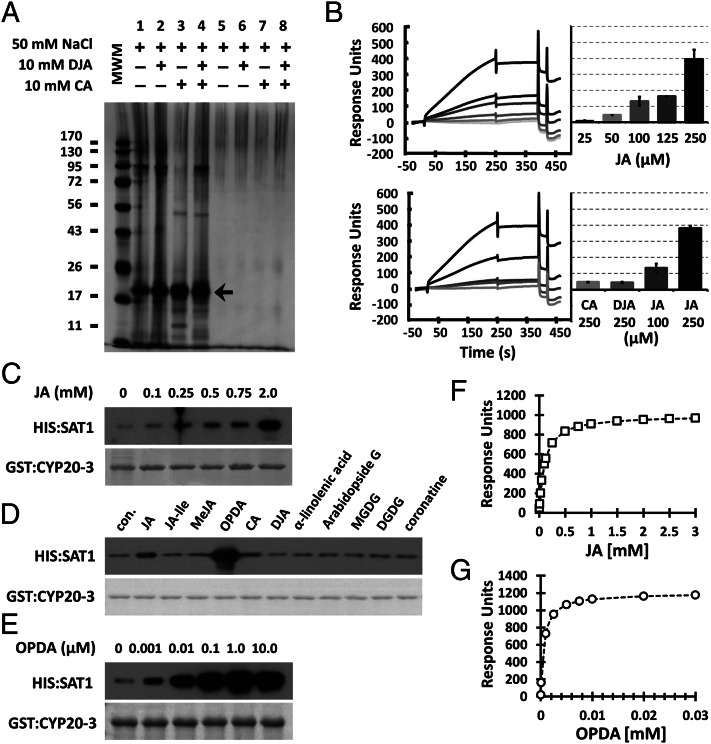
As an initial step to further delineate a jasmonate signaling circuitry, we searched for novel jasmonate effectors and/or receptors in extracts of Arabidopsis resisting infection to A. brassicicola through affinity chromatography with JA as ligand and subsequent MS analyses (Fig. 1A and Fig. S2 A and B). Repeated chromatographic separations predominantly enriched a protein of ∼20 kDa that was identified by MS as CYP20-3 (At3g62030; Fig. 1A and Fig. S2C). Surface plasmon resonance (SPR) analyses showed a concentration-dependent interaction between CYP20-3 and JA, assuring CYP20-3 as a JBP (Fig. 1B, Upper). JA tightly bound to CYP20-3 as indicated by the minor loss in SPR signal during washing (∼250–350 s), whereas the binding of CYP20-3 with biologically inactive JA analogues [i.e., (+)-cucurbic acid (CA) and (-)-didehydro-JA (DJA)] (20) was negligible (Fig. 1B, Lower).
Fig. 1.
Affinity identification of CYP20-3 as a JBP. (A) A representative batch affinity chromatogram. The pool of JA eluates, collected from flow-through affinity chromatography (e.g., fractions 16–19; Fig. S2), was concentrated and again subjected to JA (lanes 1–4) or mock (lanes 5–8) columns. The columns were washed by using loading buffer without and then with NaCl, CA, and/or DJA to remove nonspecifically bound proteins. Resin-retained proteins were fractionated by SDS/PAGE and silver-stained (arrow indicates CYP20-3, identified by subsequent MS analyses). MWM, molecular weight marker. (B) SPR analyses of concentration-dependent binding of CYP20-3 by JA (Upper) and CA, DJA, and JA (Lower). (C–E) In vitro pull-down assays for CYP20-3 and SAT1 in the presence of various concentrations of JA (C) and OPDA (E), or a broader effector screening with 500 μM of jasmonates and TFAs (D). GST:CYP20-3 fusion protein was used as a bait to pull down HIS:SAT1. (Lower) Coomassie blue-stained gels, indicating amount of bait proteins used in each pull-down assay. Parallel immunoblots of proteins that copurified with GST:CYP20-3 were probed with monoclonal anti-His antibody (Upper). con., control; DGDG, digalactosyldiacylglycerol; MGDG, monogalactosyldiacylglycerol. (F and G) Dissociation of CYP20-3 interactions with increasing JA (F; Kd = 140 µM) and OPDA (G; Kd = 196 nM) concentration as determined by SPR. The global fit of all data (n = 3) was integrated and plotted as millimolar ratio by using ANEMONA.XLT (46).
CYPs belong to a peptidyl-prolyl cis-trans isomerase (PPIase) family and were first characterized in mammals as a target of an immunosuppressive drug, cyclosporin A, preventing proinflammatory cytokine production (21). The Arabidopsis genome encodes 29 CYPs and CYP-like proteins, involved in diverse processes including transcriptional regulation and stress acclimation (22). CYP20-3 is the only isoform localized in the chloroplast stroma, and functions in plastid Cys biosynthesis by binding and possibly activating SAT1 (22, 23). The related protein CYP20-2 localizes in the thylakoid lumen (22), shares 50% amino acid sequence identity with CYP20-3 but lacks affinity to JA (< 10% of CYP20-3, Fig. S3), and thus can be excluded as a JBP candidate.
CYP20-3 Binds and Mediates OPDA Signal.
CYPs usually exert their function either as PPIases or interacting proteins (21). We found that JA binding shows little effect on the PPIase activity of CYP20-3 (Fig. S4). Thus, we examined if JA regulates the binding of CYP20-3 toward SAT1. To test this hypothesis, serial concentrations of JA were supplemented in in vitro CYP20-3–SAT1 (C–S) interaction assays (23). Immunoblot analysis detecting CYP20-3–copurified SAT1 illustrated that JA promotes the C–S interaction (Fig. 1C). We hence surveyed the potential effect of other jasmonates and TFAs on the C–S interaction (Fig. 1D). Surprisingly, OPDA fostered the interaction much stronger than JA, whereas other ligands failed to stimulate the interaction (Fig. 1 D and E). To scrutinize significance of this finding beyond Arabidopsis, the soybean [Glycine max L. (Gm)] counterparts GmCYP20-3 (NP001240987) and GmSAT1 (BM523465) were subjected to pull-down assays (Fig. S5). The GmC–S interaction was likewise promoted by OPDA, but not by other ligands including JA, indicating the preferential binding of OPDA to GmCYP20-3. The differential interaction was corroborated by SPR analyses; the dissociation constant Kd of CYP20-3 for OPDA was 196 nM and thus 700-fold higher than to JA (Kd of 140 µM; Fig. 1 F and G). Noticeably, JA-Ile and coronatine exhibited little, if any, effect on the C–S interactions, thereby suggesting that CYP20-3 conveys the COI1-independent activity of OPDA.
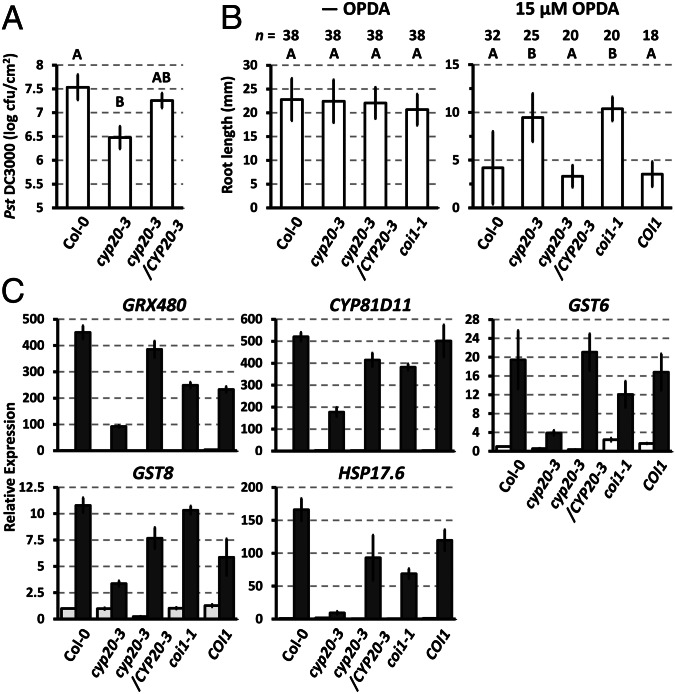
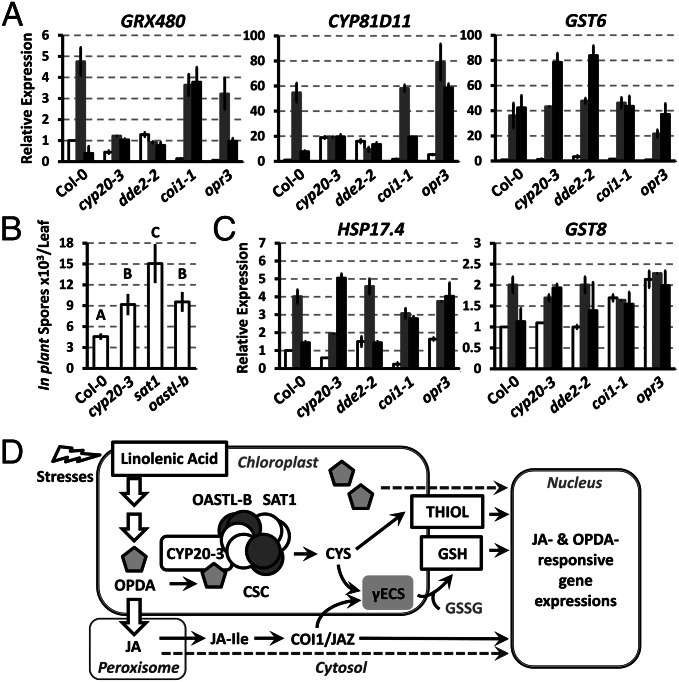
In agreement with the in vitro results, CYP20-3 was shown to be involved in OPDA-dependent signaling because its KO mutant (cyp20-3) (23) exhibited jasmonate/OPDA-insensitive phenotypes; as indicated by (i) enhanced resistance to infection by Pseudomonas syringae pv. tomato DC3000 (24), (ii) insensitivity to OPDA-mediated root growth inhibition, and (iii) attenuated expression of the ORGs GLUTAREDOXIN 480 (GRX480), CYTOCHROME P450 (CYP81D11), GLUTATHIONE S-TRANSFERASE 6 (GST6), GST8, and HEAT SHOCK PROTEIN 17.6 (HSP17.6) upon OPDA application (13) (Fig. 2 and Fig. S6A). In contrast, JA-insensitive phenotypes, i.e., alteration of JA-mediated root growth inhibition and JA-responsive gene expression, were not observed in cyp20-3 (Fig. S7). The significance of CYP20-3 in stress acclimation was apparent by the growth inhibition of cyp20-3 at higher light intensities (23), which largely antagonized the insensitivity of cyp20-3 to OPDA-mediated root growth inhibition (Fig. S6B).
Fig. 2.
CYP20-3 functions in the OPDA signaling pathway. (A) P. syringae (Pst DC3000) growth (shown as cfu/cm2 leaf area) in WT (Col-0), cyp20-3, and complementation (cyp20-3/CYP20-3) Arabidopsis plants at 4 d postinoculation (mean ± SD; n = 3). (B) Root length of WT (Col-0) and mutant [cyp20-3, cyp20-3/CYP20-3, coi1-1, and COI1 (COI1/coi1)] plants in the absence and presence of 15 µM OPDA at 5 d after germination (mean ± SD for n shown). (A and B) Different letters indicate statistically significant differences between genotypes (Tukey–Kramer honestly significant difference test on all pairs; α = 0.05). (C) Transcript quantification by quantitative RT-PCR of ORGs in OPDA-treated (black bars) and untreated (white bars) WT (Col-0) and mutant (cyp20-3, cyp20-3/CYP20-3, coi1-1, and opr3) plants. Values were normalized to the expression of POLYUBIQUITIN (means ± SD; n = 3).
OPDA–CYP20-3 Interaction Promotes Plastid CSC Formation.
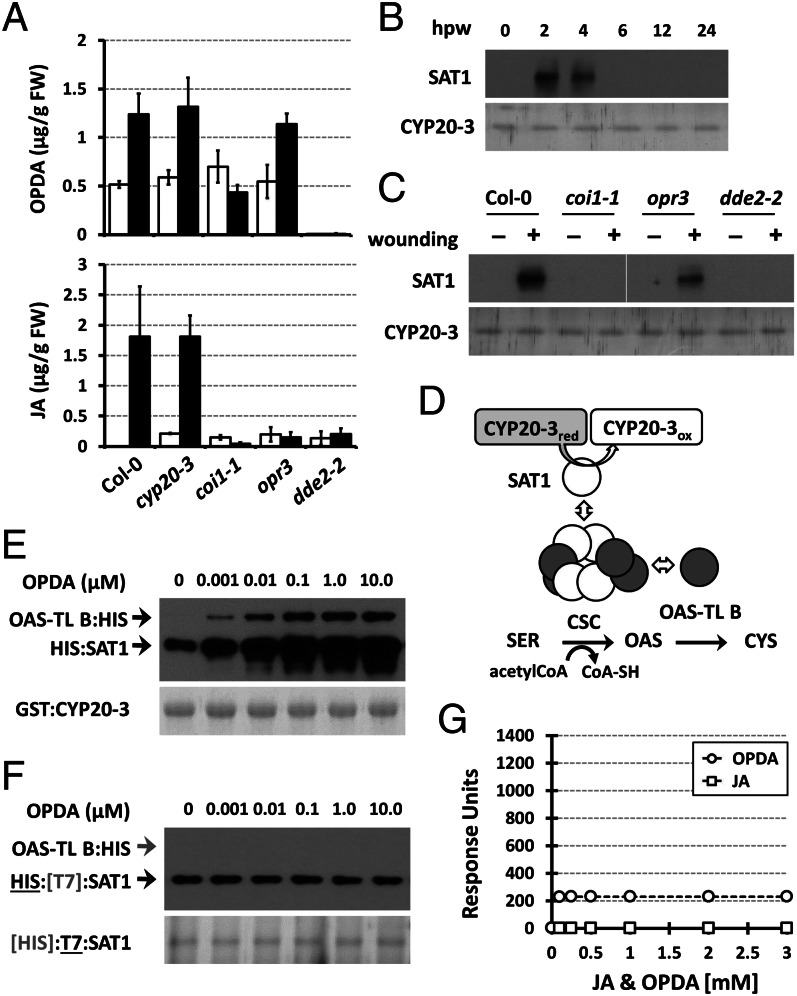
Our data suggest that CYP20-3 is a potential OPDA effector, especially considering that OPDA is produced in plastids where it has immediate access to CYP20-3 and SAT1 (1, 19, 22). In contrast, JA is not primarily present in plastids, as the enzymatic conversion from OPDA to JA takes place in peroxisomes (1). To substantiate our hypothesis, we combined ex vivo coimmunoprecipitation (co-IP) assays with wound-responsive jasmonate biosynthesis. Wounding induced the accumulation of OPDA with a peak at ∼1 to 3 h postwounding (hpw) (9, 25), subsequently JA, and ultimately the differential expression of various genes (1, 2). As predicted, the C–S interaction was promoted and/or stabilized in parallel with OPDA accumulation (Fig. 3 A–C). CYP20-3–coimmunoprecipitated SAT1 increased between 2 and 4 hpw in WT and opr3 (i.e., +OPDA/−JA), but not in aos/dde2-2 (i.e., −OPDA/−JA) and coi1-1. As previously reported, wound-induced JA accumulation is impaired in coi1-1 (26) (Fig. 3A). Similarly, coi1-1 lacked wound-inducible OPDA accumulation and an OPDA-mediated C–S interaction (Fig. 3 A and C). By contrast, the wound-induced production of OPDA but not JA in opr3 stimulated the binding of SAT1 by CYP20-3 (Fig. 3 A and C). In addition, basal levels of OPDA in unwounded WT, opr3, and coi1-1 paralleled a nondetectable C–S interaction (Fig. 3C), indicating that wound-induced OPDA accumulation stimulated the association of CYP20-3 and SAT1.
Fig. 3.
OPDA stimulates CYP20-3–SAT1 interaction and CSC formation. (A) Accumulation of OPDA and JA in wounded WT (Col-0) and mutant (cyp20-3, coi1-1, opr3, and dde2-2) plants. Jasmonates were extracted from leaves at 0 hpw (white bars) and 3 hpw (black bars; mean ± SD; n = 3). (B) Time-resolved in situ co-IP assays determining CYP20-3–SAT1 interaction in wounded WT (Col-0) plant. (C) In situ co-IP assays determining the CYP20-3-SAT1 interaction in WT (Col-0) and mutant (coi1-1, opr3, and dde2-2) plants at 3 hpw. (B and C) Total protein extracts were subjected to co-IP using anti-CYP20-3 antibody-coupled resin. (Lower) Silver-stained gels indicating amount of bait proteins (CYP20-3) used in each IP assay. Parallel immunoblots of proteins from co-IP with CYP20-3 were probed with polyclonal anti-SAT antibody (Upper). (D) Schematics of the plastid Cys biosynthetic pathway. (E and F) In vitro pull-down assays for determining the physical association of CYP20-3, SAT1, and OAS-TL B (E), and SAT and OAS-TL B (F), at various OPDA concentrations. GST:CYP20-3 fusion protein (E) or T7-tagged SAT1 (F) were used as a bait to pull down HIS:SAT1 and/or OAS-TL B:HIS proteins. (Lower) Coomassie blue-stained gels indicating amount of bait proteins used in each pull-down assay. Parallel immunoblots of proteins that copurified with the baits were probed with monoclonal anti-His antibody (Upper). (G) SPR analyses of SAT1 interactions with increasing concentrations of JA (□) and OPDA (○). The global fit of all data (n = 3) was integrated and plotted as concentration dependency by using ANEMONA.XLT (46).
We previously proposed that the interaction of CYP20-3 with SAT1 assists in the folding or tetramerization of SAT1 to form a CSC with OAS-TL B (23) (Fig. 3D). Indeed, a sequential pull-down assay demonstrated that OPDA stimulates CSC formation. OPDA–CYP20-3 binding assisted or stabilized the assembly of SAT1, which subsequently recruited OAS-TL B (Fig. 3E). Control experiments showed that OPDA (and JA) neither directly binds SAT1 nor promotes SAT1–OAS-TL B interaction (Fig. 3 G and F).
CYP20-3 Relays OPDA Signal in Wound-Responsive Regulation of Cellular Redox Homeostasis.
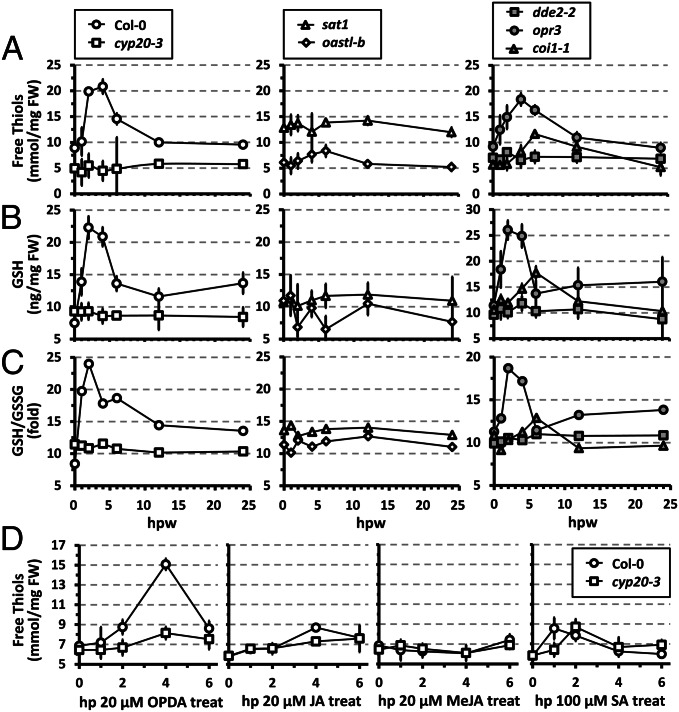
Our results explain the role of OPDA in stimulating Cys biosynthesis, a key step in sulfur assimilation and production of thiol-based metabolites (18). In line with this scenario, low molecular weight thiols increased rapidly following wounding in WT and opr3 (peaking at ∼2–4 hpw), but remained largely at basal levels in cyp20-3, sat1, oastl-b, and aos/dde2-2, or marginally increased in coi1-1 (Fig. 4A). Unexpectedly, wounded coi1-1 showed delayed, yet moderate, increases of thiols despite the lack of an OPDA-stimulated C–S interaction (Fig. 3C). This partial redundancy may rely on increased protein turnover or other regulatory mechanisms (e.g., GRs) (17). Nevertheless, similar kinetic changes in GSH, the most abundant nonprotein thiol in plants, were observed and dominated the GSH/GSSG ratio in each genotype (Fig. 4B and C). Taken together, our data indicate that wounding triggered a rapid adjustment of redox homeostasis, shifting the thiol redox potential to more negative values, which requires (i) OPDA (not JA) production, (ii) CYP20-3, and (iii) plastid Cys biosynthesis. On the contrary, the expression of CYP20-3, SAT1, and OASTL-B was constitutive regardless of wounding (Fig. S8), and the wound-induced up-regulation of cytosolic GR1 appeared to contribute little to the increased thiol and GSH concentrations (Fig. S9A). The results concur with the conclusion that CYP20-3 relays the OPDA signal in the wound-responsive regulation of cell redox homeostasis.
Fig. 4.
CYP20-3 relays the OPDA signal in wound-dependent up-regulation of sulfur assimilation and adjustment of redox homeostasis. (A–C) Levels of soluble reduced nonprotein thiols (A), GSH (B), and GSH/GSSG ratios (C) in wounded WT (Col-0) and mutant (cyp20-3, dde2-2, opr3, and coi1-1) plants (mean ± SD; n = 3). (D) The levels of thiols in WT (Col-0) and cyp20-3 plants at various times after exogenous application of OPDA, JA, methyl (+)-7-iso-jasmonate (MeJA), or SA (mean ± SD; n = 3).
As expected from these results, exogenous OPDA application triggered a strong increase of thiols in WT but not in cyp20-3, whereas JA and salicylic acid (SA) led to only modest increases (< 35% that of OPDA) and methyl (+)-7-iso-jasmonate was ineffective (Fig. 4D). Noticeably, the modest effect of JA required CYP20-3, whereas that of SA was CYP20-3–independent. The weaker JA–CYP20-3 interaction (Fig. 1 B–F) might contribute to the thiol production, whereas SA-dependent redox regulation may rely on alternative mechanisms, perhaps operating in the cytoplasm (2). Note, however, that OPDA-induced Cys biosynthesis may not be limited to plastids, as OAS is membrane-permeable and thus available as a substrate for cytosolic OAS-TLs (17, 18). However, little difference in the kinetic changes of thiol and GSH levels in wounded oastl-b (Fig. 4 A–C) indicates a major role of plastid CSC in OPDA signaling. In contrast, OPDA-induced cellular redox changes, enhanced reduction potential, were observed in plastids and cytoplasm (Fig. S9B), suggesting the diffusion of Cys, GSH, and/or thiols from plastids to cytoplasm.
Redox-Dependent Transmission of OPDA Signal Is Intrinsic in Plant Stress Acclimation.
Maintenance of cellular redox homeostasis is central in transcriptional regulation and plant stress acclimation (27). For instance, we observed the differential expression of ORGs (i.e., CYP81D11, GRX480, and GST6) in redox-imbalanced cyp20-3 and aos/dde2-2 in response to wounding (Fig. 5A). Moreover, both types of mutants showed enhanced susceptibility toward A. brassicicola compared with WT (Fig. 5B and Fig. S1D), underpinning the decisive role of redox homeostasis in stress and defense responses.
Fig. 5.
OPDA-dependent regulation of wound-responsive transcripts and defense response against A. brassicicola infection. (A and C) Time-resolved quantitative RT-PCR analyses of wound-responsive genes in wounded WT (Col-0) and mutant (cyp20-3, dde2-2, coi1-1, and opr3) plants. Total RNA were prepared from leaves at 0 (white bars), 3 (gray bars), and 6 (black bars) hpw. Values were normalized to the expression of POLYUBIQUITIN (means ± SD; n = 3). (B) Average number of in planta-formed spores per lesion ± SD. Measurements were taken at 5 d postinoculation, and each data point is the average of three pools of 16 inoculated leaves from four plants per genotype. Statistical analyses were carried out with Tukey–Kramer honestly significant difference test on all pairs (α = 0.05). (D) Proposed OPDA signaling pathway: OPDA binds CYP20-3, stimulating sulfur assimilation by promoting CSC formation that shifts the redox potential and controls the expression of a subset of ORGs, indicating the action of a COI-1–independent but OPDA-dependent signaling pathway. The increased GSH:GSSG ratio is initiated by γECS that is concomitantly induced by the activation of CSC (17) and by the COI1/JAZ-mediated JA signaling pathway (5).
Stress acclimation (e.g., upon wounding) requires complex cellular mechanisms, orchestrated by various signals and metabolic pathways (28). We found that a subset of ORGs (e.g., GST6, GST8, and HSP17.6) still accumulated in wounded cyp20-3 and aos/dde2-2 (Fig. 5C). The wound-responsive, but CYP20-3- and OPDA-independent, induction of those genes may involve alternative signaling pathways, perhaps centered upon RES and/or reactive oxygen species (ROS) (13, 16, 28, 29). Dysregulation of the redox state in wounded cyp20-3 and aos/dde2-2 may lead to increased ROS accumulations (e.g., H2O2), known to stimulate detoxification (GSTs) and general stress tolerance (HSPs) machineries through ROS-responsive NAC TFs (16, 29). Alternatively, wound-responsive breakage of TFA most likely produces RES in parallel to jasmonates, which are also capable of regulating TGA-dependent transcriptions, including GST6, GST8, and HSP17.6 (13). This signal redundancy must be an intrinsic function for environmental plasticity in plant survival.
Discussion
Plastids are a biochemical factory that facilitates diverse regulatory mechanisms, coordinated by bidirectional signaling from nucleus to plastids and from plastids to nucleus (i.e., retrograde signaling). Retrograde signaling involves elaborate regulatory pathways critical in mediating the rapid adjustment of nuclear gene expression in response to environmental constraints. During environmental stresses, plastids experience drastic metabolic and redox changes that trigger signaling cascades to modify nuclear transcription mediating acclimations (i.e., operational control) (30, 31). In the present study, OPDA is described as a unique player in retrograde signaling (Fig. 5D). Wound-induced OPDA activates CYP20-3/SAT1–dependent Cys biosynthesis that employs redox-dependent signal transmission in nuclear transcriptional regulation. The CYP20-3/SAT1 pathway appears to be critical in linking stress effects to redox homeostasis and acclimation. For instance, excess light and oxidative stress also promote the accumulation of OPDA on a time scale of hours with concomitant accumulation of reduced, nonprotein thiols (17, 32, 33). Recently, another metabolic pathway was identified that conveys stress-responsive retrograde signaling (34). The metabolic intermediate of the plastid isoprenoid biosynthetic pathway (i.e., methylerythritol cyclodiphosphate) displayed an unequivocal activity, regulating a subset of stress-responsive genes in nucleus. This and our data confirm the previously proposed hypothesis that metabolite signals, in addition to redox signals, decisively contribute to the operational control of nuclear gene expression (30, 31).
Interestingly, a number of ORGs contain TGA motifs in their promoter region (6, 9, 13, 14), suggesting that TGAs are a key activator in OPDA signaling. TGAs belong to a basic leucine zipper TF family that functions in various signaling pathways involving hormones, RES, and redox cues (13, 14, 16, 35). In the present study, cyp20-3 showed attenuated TGA gene expression upon OPDA and wound treatments (Figs. 3C and 5A), indicating that activity of TGAs is critical in OPDA signaling and requires changes in cellular redox state. In line with this scenario, OPDA signaling facilitates yet uncharacterized redox-controlled TRs that might target TGAs. GRX480 can immediately be considered as an OPDA-associated TR candidate, as its induction is CYP20-3- and OPDA-dependent (Figs. 3C and 5A), and it functions to bind and regulate TGAs in the nucleus (36).
However, GRX480 may be multifunctional, as it can also be induced by JA and SA (13, 35, 37). JA induction of GRX480 is rather modest (13, 35), but may explain its residual expression in OPDA-treated cyp20-3 (Fig. 3C). The up-regulation of GRX480 is though TGA-/COI1-independent, but MYC2-dependent (37). However, SA induction of GRX480 is TGA-/NPR1-dependent, and its subsequent formation of a ternary complex with TGA/NPR1 negatively regulates JA signaling (35). Such a redundancy and crosstalk of multiple signaling likely enables environmental plasticity in plants, perhaps conveying the activation of general stress responses. For instance, the induction of GSTs and HSPs is orchestrated by OPDA, JA, SA, ROS, and RES, or involves multiple cis elements in respective promoters (13, 14, 16, 29, 38). On the contrary, effects of a specific signal are mediated through multiple pathways. JA induction of GRX480 elucidates a unique jasmonate signaling, COI1-independent MYC2 regulatory pathway (37). MYC2 is a key transcription activator of JA-Ile/COI1 signaling, whose activity is suppressed by JAZ in a resting condition (1, 2). A similar pathway may explain the induction of COI1-dependent genes in OPDA-treated opr3, as OPDA is unable to bind COI1/JAZ complex (2, 3, 9). In comparison, OPDA induction of PHO;H10 is COI1-dependent, whereas JA-activated MAP kinase cascades (e.g., MPK1, MPK2, and BIK1) and GST25 is regulated in a COI1- and/or MYC2-independent manner (14, 39–41). Future studies are needed to resolve mechanisms underlying these alternative or perhaps redundant pathways. Indeed, we were able to detect additional JBPs in this study that may assist our understanding of novel jasmonate signaling in future studies (e.g., Fig. 1A).
CYP20-3 belongs to the thiol-dependent redox regulatory network in plastids, whose operation is governed by 2-Cys peroxiredoxin (2-CysPrx) (42). 2-CysPrx is a multifunctional regulator that adopts different conformational states in dependence on its redox state. Our fluorescence resonance energy transfer assays and coexpression analyses determined that CYP20-3 is able to form a stable complex with 2-CysPrx. These results lead us to hypothesize that, under normal conditions, the CYP20-3/2-CysPrx complex is maintained, whereas, during stress, increased levels of OPDA bind and release CYP20-3 that then recruits SAT1 to form CSC. Finally, OPDA/CYP20-3–dependent buildup of cellular reduction potential may reverse the conformational states of 2-CysPrx and/or CYP20-3, leading to the reformation of CYP20-3/2-CysPrx complex. In addition, another redox sensor/regulator, thioredoxin, has shown to physically interact with CYP20-3 or 2-CysPrx, which perhaps stimulates Cys biosynthesis and/or detoxification machineries in coordination with/without OPDA (23, 43). The finer aspects of dynamic relationships between OPDA, 2-CysPrx, thioredoxin, CYP20-3, CSC, and redox changes will have to be explored in future work.
Materials and Methods
Plant Growth Condition.
Genetic backgrounds of Arabidopsis thaliana WT and mutants were in Columbia (i.e., Col-0; cyp20-3, cyp20-3/CYP20-3, sat1, oastl-b, cyp20-2, fad3/7/8, aos/dde2-2, jar1, coi1-1, and jin1) or Wassilewskija (opr3). Plants were grown in a chamber with a 10-h day cycle (80–100 μE/m2/s) at 22 °C and 60% to 80% relative humidity.
The sat1 (SALK_120440) and cyp20-2 (SALK_024971) transfer (T)-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center (http://abrc.osu.edu). The homozygous KO plants were selected by PCR amplification with T-DNA- and gene-specific primers (Table S1), and confirmed by semiquantitative RT-PCR using gene-specific primers (Table S2 and Fig. S10).
Batch JA-Affinity Chromatography.
JA-eluted fractions (e.g., fractions 16–19 in Fig. S2) were pooled, concentrated, and dialyzed against loading buffer. The eluate was resubjected to JA affinity or mock resin, and incubated for 2 h at 4 °C with agitation. The resins were packed into empty columns (Bio-Rad) and washed with loading buffer, without and then with 50 mM NaCl, 10 mM DJA, and/or 10 mM CA. The column-retained proteins were analyzed by SDS/PAGE.
SPR.
Optical biosensor analyses were performed with BIAcore T100 and Reichert SR7000 spectrometers. For protein immobilization, an amine coupling kit (BIAcore) was used for CM5 chips (BIAcore) following manufacturer’s instructions, and chemicals purchased from Sigma was used for SC CMPD-5 chips (Xantec) following Reichert in-house protocol (www.reichert.com/life_sciences.cfm). The protein–ligand interactions were monitored by injecting various concentrations of jasmonates over the reference and/or protein-immobilized flow cells, with periodic buffer blank injections for double referencing [25 °C, flow rate of 20 µL/min (BIAcore) or 50 µL/min (Reichert) with PBS solution with Tween 20]. Binding responses in the sensorgrams were corrected for reference cell responses or blank injections.
In Vitro Protein–Protein Interaction Assay.
Coding sequences for full-length proteins were cloned into pGEX-4T-1 (GE Healthcare), pET28a, or pET21c (Novogen) to obtain N-terminal GST-fused CYP20-3, N-terminal 6×HIS/T7-tagged SAT1, or C-terminal 6×HIS-tagged OAS-TL B, respectively. Recombinant proteins were prepared from Escherichia coli coexpressing (Gm)CYP20-3 and (Gm)SAT1, or expressing an individual protein. The coexpressed proteins or individually purified proteins (1:1:1 molar ratio) were immobilized with/without ligands on affinity beads for 1 h at room temperature. GSH beads (Sigma) were used to immobilize GST:(Gm)CYP20-3, whereas T7 antibody beads (Novagen) were used to immobilize T7: SAT1. After washing and elution with GSH or citric acid, proteins were resolved by SDS/PAGE and probed with a monoclonal anti-His antibody (Invitrogen).
Ex Vivo Protein–Protein Interaction Assay.
Polyclonal antibody was raised in rabbits against AtCYP20-3 and subsequently purified (HiTrap Protein G HP; Amersham) and cleaned (Antibody Clean-up Kit; Pierce). The generation of anti–CYP20-3 antibody-immobilized beads, preparation of total protein lysates, and co-IP assays were carried out by using a Direct Magnetic Co-IP Kit (Pierce) in accordance with the manufacturer’s instructions. Protein was quantified by Bradford method and NanoDrop ND-1000 (Thermo Scientific). Equal amounts of eluates were fractionated by SDS/PAGE and visualized by silver staining (Pierce) and Western blot by using a polyclonal anti-SAT antibody (44).
Determination of Soluble (Nonprotein) Thiols, GSH, and GSSG.
Thiol levels present in leaf tissue samples were measured as previously described by Blaszczyk et al. (45), and levels of GSH and GSSG in leaf tissues were measured by using a GSH assay kit (BioVision) according to the manufacturer’s instructions.
Supplementary Material
Acknowledgments
We thank J. Browse, S. Gazzarrini, J. Glazebrook, C. Gatz, and A. Meyer for sharing T-DNA insertion KO mutants and transgenic lines: oastl-b, fad3/7/8, aos/dde2-2, jar1, coi1-1, jin1, opr3, GRX1-roGFP2, and TKTP-GRX1-roGFP2; and P. Staswick and D. E. Salt for providing (+)-7-iso-jasmonoyl-L-isoleucine and anti-serine acetyltransferase antibody. This work was partially supported by the Virginia Bioinformatics Institute, US Department of Agriculture Grant 2010-65108-20527 and Deutsche Forschungsgemeinschaft Grant Di346.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 9197.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218872110/-/DCSupplemental.
References
- 1.Acosta IF, Farmer EE. The Arabidopsis Book. 2010. Jasmonates. 8:e0129. Available at http://thearabidopsisbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM. Hormonal modulation of plant immunity. Annu Rev Cell Dev Biol. 2012;28:489–521. doi: 10.1146/annurev-cellbio-092910-154055. [DOI] [PubMed] [Google Scholar]
- 3.Thines B, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448(7154):661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 4.Chini A, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448(7154):666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 5.Devoto A, et al. Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol Biol. 2005;58(4):497–513. doi: 10.1007/s11103-005-7306-5. [DOI] [PubMed] [Google Scholar]
- 6.Taki N, et al. 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 2005;139(3):1268–1283. doi: 10.1104/pp.105.067058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Staswick PE. The tryptophan conjugates of jasmonic and indole-3-acetic acids are endogenous auxin inhibitors. Plant Physiol. 2009;150(3):1310–1321. doi: 10.1104/pp.109.138529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farmer EE, Davoine C. Reactive electrophile species. Curr Opin Plant Biol. 2007;10(4):380–386. doi: 10.1016/j.pbi.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 9.Stintzi A, Weber H, Reymond P, Browse J, Farmer EE. Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc Natl Acad Sci USA. 2001;98(22):12837–12842. doi: 10.1073/pnas.211311098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chehab EW, et al. Intronic T-DNA insertion renders Arabidopsis opr3 a conditional jasmonic acid-producing mutant. Plant Physiol. 2011;156(2):770–778. doi: 10.1104/pp.111.174169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bӧttcher C, Pollmann S. Plant oxylipins: Plant responses to 12-oxo-phytodienoic acid are governed by its specific structural and functional properties. FEBS J. 2009;17:4693–4704. doi: 10.1111/j.1742-4658.2009.07195.x. [DOI] [PubMed] [Google Scholar]
- 12.Dave A, Graham IA. Oxylipin signaling: A distinct role for the jasmonic acid precursor cis-(+)-12-oxo-phytodienoic acid (cis-OPDA) Front Plant Sci. 2012;3:42. doi: 10.3389/fpls.2012.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mueller S, et al. General detoxification and stress responses are mediated by oxidized lipids through TGA transcription factors in Arabidopsis. Plant Cell. 2008;20(3):768–785. doi: 10.1105/tpc.107.054809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stotz HU, Mueller S, Zoeller M, Mueller MJ, Berger S. TGA transcription factors and jasmonate-independent COI1 signalling regulate specific plant responses to reactive oxylipins. J Exp Bot. 2013;64(4):963–975. doi: 10.1093/jxb/ers389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ball L, et al. Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell. 2004;16(9):2448–2462. doi: 10.1105/tpc.104.022608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mhamdi A, et al. Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol. 2010;153(3):1144–1160. doi: 10.1104/pp.110.153767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noctor G, Queval G, Mhamdi A, Chaouch S, Foyer CH. The Arabidopsis Book. 2011. Glutathione. 9:e0142. Available at http://thearabidopsisbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi H, Kopriva S, Giordano M, Saito K, Hell R. Sulfur assimilation in photosynthetic organisms: Molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol. 2011;62:157–184. doi: 10.1146/annurev-arplant-042110-103921. [DOI] [PubMed] [Google Scholar]
- 19.Wirtz M, Hell R. Functional analysis of the cysteine synthase protein complex from plants: Structural, biochemical and regulatory properties. J Plant Physiol. 2006;163(3):273–286. doi: 10.1016/j.jplph.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 20.Miersch O, Kramell R, Parthier B, Wasternack C. Structure-activity relations of substituted, deleted or stereospecifically altered jasmonic acid in gene expression of barley leaves. Phytochemistry. 1999;50:353–361. [Google Scholar]
- 21.Wang P, Heitman J. The cyclophilins. Genome Biol. 2005;6(7):226. doi: 10.1186/gb-2005-6-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romano PGN, Horton P, Gray JE. The Arabidopsis cyclophilin gene family. Plant Physiol. 2004;134(4):1268–1282. doi: 10.1104/pp.103.022160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dominguez-Solis JR, et al. A cyclophilin links redox and light signals to cysteine biosynthesis and stress responses in chloroplasts. Proc Natl Acad Sci USA. 2008;105(42):16386–16391. doi: 10.1073/pnas.0808204105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katagiri F, Thilmony R, He SY. The Arabidopsis Book. 2002. The Arabidopsis Thaliana-Pseudomonas Syringae Interaction. 1:e0039. Available at http://thearabidopsisbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koo AJK, Gao X, Jones AD, Howe GA. A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J. 2009;59(6):974–986. doi: 10.1111/j.1365-313X.2009.03924.x. [DOI] [PubMed] [Google Scholar]
- 26.Chung HS, et al. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008;146(3):952–964. doi: 10.1104/pp.107.115691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foyer CH, Noctor G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell. 2005;17(7):1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wasternack C, et al. The wound response in tomato—role of jasmonic acid. J Plant Physiol. 2006;163(3):297–306. doi: 10.1016/j.jplph.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 29.Wu A, et al. JUNGBRUNNEN1, a reactive oxygen species-responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell. 2012;24(2):482–506. doi: 10.1105/tpc.111.090894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pogson BJ, Woo NS, Förster B, Small ID. Plastid signalling to the nucleus and beyond. Trends Plant Sci. 2008;13(11):602–609. doi: 10.1016/j.tplants.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 31.Pesaresi P, Schneider A, Kleine T, Leister D. Interorganellar communication. Curr Opin Plant Biol. 2007;10(6):600–606. doi: 10.1016/j.pbi.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 32.Kazan K, Manners JM. The interplay between light and jasmonate signalling during defence and development. J Exp Bot. 2011;62(12):4087–4100. doi: 10.1093/jxb/err142. [DOI] [PubMed] [Google Scholar]
- 33.Riemann M, et al. Impaired induction of the jasmonate pathway in the rice mutant hebiba. Plant Physiol. 2003;133(4):1820–1830. doi: 10.1104/pp.103.027490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao Y, et al. Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell. 2012;149(7):1525–1535. doi: 10.1016/j.cell.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 35.Ndamukong I, et al. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 2007;50(1):128–139. doi: 10.1111/j.1365-313X.2007.03039.x. [DOI] [PubMed] [Google Scholar]
- 36.Li S, et al. Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. Plant Cell. 2009;21(2):429–441. doi: 10.1105/tpc.108.064477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Köster J, et al. Xenobiotic- and jasmonic acid-inducible signal transduction pathways have become interdependent at the Arabidopsis CYP81D11 promoter. Plant Physiol. 2012;159(1):391–402. doi: 10.1104/pp.112.194274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clarke SM, et al. Jasmonates act with salicylic acid to confer basal thermotolerance in Arabidopsis thaliana. New Phytol. 2009;182(1):175–187. doi: 10.1111/j.1469-8137.2008.02735.x. [DOI] [PubMed] [Google Scholar]
- 39.Ribot C, Zimmerli C, Farmer EE, Reymond P, Poirier Y. Induction of the Arabidopsis PHO1;H10 gene by 12-oxo-phytodienoic acid but not jasmonic acid via a CORONATINE INSENSITIVE1-dependent pathway. Plant Physiol. 2008;147(2):696–706. doi: 10.1104/pp.108.119321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ortiz-Masia D, Perez-Amador MA, Carbonell J, Marcote MJ. Diverse stress signals activate the C1 subgroup MAP kinases of Arabidopsis. FEBS Lett. 2007;581(9):1834–1840. doi: 10.1016/j.febslet.2007.03.075. [DOI] [PubMed] [Google Scholar]
- 41.Veronese P, et al. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell. 2006;18(1):257–273. doi: 10.1105/tpc.105.035576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muthuramalingam M, et al. Multiple redox and non-redox interactions define 2-Cys peroxiredoxin as a regulatory hub in the chloroplast. Mol Plant. 2009;2(6):1273–1288. doi: 10.1093/mp/ssp089. [DOI] [PubMed] [Google Scholar]
- 43.Meyer Y, Buchanan BB, Vignols F, Reichheld JP. Thioredoxins and glutaredoxins: Unifying elements in redox biology. Annu Rev Genet. 2009;43:335–367. doi: 10.1146/annurev-genet-102108-134201. [DOI] [PubMed] [Google Scholar]
- 44.Na G, Salt DE. Differential regulation of serine acetyltransferase is involved in nickel hyperaccumulation in Thlaspi goesingense. J Biol Chem. 2011;286(47):40423–40432. doi: 10.1074/jbc.M111.247411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blaszczyk A, Sirko L, Hawkesford MJ, Sirko A. Biochemical analysis of transgenic tobacco lines producing bacterial serine acetyltransferase. Plant Sci. 2002;162:589–597. [Google Scholar]
- 46.Hernández A, Ruiz MT. An EXCEL template for calculation of enzyme kinetic parameters by non-linear regression. Bioinformatics. 1998;14(2):227–228. doi: 10.1093/bioinformatics/14.2.227. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.