Abstract
Regulation of an intracellular acidic environment plays a pivotal role in biological processes and functions. However, spatiotemporal analysis of the acidification in complex tissues of living subjects persists as an important challenge. We developed a photo-inactivatable bioluminescent indicator, based on a combination of luciferase-fragment complementation and a photoreaction of a light, oxygen, and voltage domain from Avena sativa Phototropin1 (LOV2), to visualize temporally dynamic acidification in living tissue samples. Bioluminescence of the indicator diminished upon light irradiation and it recovered gradually in the dark state thereafter. The recovery rate was remarkably sensitive to pH changes but unsusceptible to fluctuation of luciferin or ATP concentrations. Bioluminescence imaging, taken as an index of the recovery rates, enabled long-time recording of acidification in apoptotic and autophagous processes in a cell population and an ischemic condition in living mice. This technology using the indicator is widely applicable to sense organelle-specific acidic changes in target biological tissues.
Individual subcellular compartments in live cells stringently produce an acidic condition for the regulation of protein activities, metabolism, and signal transduction. Proton gradients, for example, build up across the membrane of liposomes during endocytosis. In macroautophagy processes, old proteins and organelles are sequestered into an autophagosome, which is converted to an acidic condition after fusing with lysosome. Oxidative stress during tissue ischemia and reperfusion induces reactive oxygen species and a low pH condition, which engenders apoptosis and necrosis. Accordingly, the ability to visualize temporal acidification is extremely important for the study of the underlying processes taking place in living cells and tissues (1, 2).
Among the possible methods to monitor the acidic environment, optical methods present advantages of sensitivity and higher spatial resolution. Fluorescent probes using small organic dyes are now widely used to monitor pH changes in single living cells (2, 3). However, the wavelengths of many fluorescent probes are short, and their background signal is high. Additionally, it is difficult to locate the probes in a specific intracellular compartment or organelle. Recently, low-pH-activatable red to near-infrared fluorescent probes have been developed and applied to imaging of viable cancer cells (4, 5). The probes present the advantage of a higher signal-to-background ratio and strong fluorescence in acidic conditions. However, the challenge remains of analyzing acidification in intracellular microenvironments in living cells and tissues. In contrast, genetically encoded fluorescent probes can be targeted into a specific intracellular organelle; the probes for pH are now applied for single-cell analysis (6–8). The fluorescence protein itself is, however, sensitive to pH, and intensity-based imaging techniques remain difficult to apply for in vivo imaging because of bleaching of chromophores by excitation light and low penetration of the emitting light.
Recently, bioluminescence imaging using luciferase has been used extensively for in vivo analysis of many biological functions (9, 10). It is highly advantageous for the luciferase to emit its photons at red to near-infrared wavelengths, at which tissue attenuation of emitted photons is minimized. The imaging is a nonradioactive modality, in contrast to other imaging modalities such as positron emission tomography, single-photon emission computed tomography, and computed tomography. Because of such advantages of bioluminescence imaging, luciferase-based probes have been developed over the last decade for visualizing intracellular events such as gene expression (11–13), calcium ions (14), protein translocation (15), protein–protein interactions (16–18), and enzyme activities (19).
Here we describe a genetically encoded bioluminescent indicator for imaging temporal acidification in living cells and mice based on luciferase-fragment complementation with a photoreactive protein region, a light, oxygen, and voltage domain from Avena sativa Phototropin1 (LOV2). The LOV2 domain is connected with luciferase fragments to produce photo-inactivatable luciferase as a pH indicator. The indicator visualizes acidification of intracellular organelles and enables the potential to precisely monitore pH changes in living mice.
Results
Development of Photo-Inactivatable Luciferase.
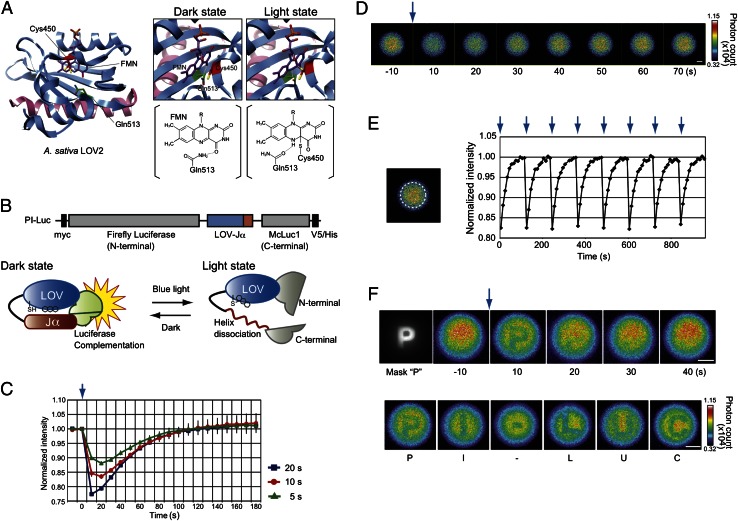
The usefulness of the LOV2 domain has been reported for controlling specific enzyme activities using external light (20–22). FTIR and NMR studies have revealed that the LOV2 domain comprises the LOV core domain region, which is bound to flavin mononucleotide (FMN), and a carboxyl-terminal helical extension (Jα) (23, 24) (Fig. 1A). In the dark, the Jα-helix interacts with the LOV core domain by hydrogen bonds between Gln513 of the LOV domain and atom O4 of the FMN ring (25) (dark state, Fig. 1A). Blue light irradiation induces the formation of a covalent bond between Cys450 and the flavin chromophore (26). The covalent bond leads temporarily to formation of a new hydrogen bond between Gln513 and the FMN ring and dissociation of the Jα-helix from the LOV core domain (light state). When turning off the light, the LOV2 domain returns to the dark state by base catalysis with base abstraction of FMN and reprotonation of Cys450. It has been investigated extensively that the reaction time is strongly dependent on pH (27). Based on this information, we connected the complete LOV2 domain sequence (A. sativa Phototropin1, 404–546 amino acids) with an N-terminal fragment of firefly luciferase (28) (Photinus pyralis, 1–415 amino acids) and a mutant of C-terminal fragment (F420I, G421A, E453S; 395–542 amino acids) of click beetle luciferase (Caribbean Pyrophorus plagiophthalamus) named McLuc1, which generates bioluminescence with a higher signal-to-background ratio upon complementation (17) (Fig. 1B). The LOV core domain interacts with Jα in the dark, causing complementation of the luciferase fragments to emit luminescence. Photon absorption of FMN would engender release of Jα-helix from the LOV core domain and dissociation between the luciferase fragments. When the light is turned off, the LOV core domain would again interact with Jα-helix to recover the bioluminescence.
Fig. 1.
Photoreaction of the pH indicator (PI-Luc) in mammalian cells. (A) Three-dimensional structures of LOV2 domain and light-induced signal transduction pathway. The LOV core domain and Jα are colored blue and pink. FMN (purple), amino acid residues of Cys450 (red), and Gln513 (green) are presented in the structures. Formation changes among FMN, Cys450, and Gln513 with light are shown on right (dark state and light state). The hydrogen bond is indicated with a yellow dashed line. R, ribityl side chain of FMN. (B) Schematic construct and a photoreaction model of PI-Luc. Black squares in the construct show epitope Myc and V5/His tags. (C) Time-course measurements of bioluminescence with different periods of irradiation (20, 10, or 5 s). Bioluminescence intensities of PI-Luc expressed in HEK293 cells were measured after blue light irradiation. The bioluminescence intensities were normalized against initial values of bioluminescence intensities (n = 3). Error bars, SD. The blue arrow indicates the irradiation time point. (D) Bioluminescence images of cell population in a cultured dish. The blue arrow indicates a light irradiation point. HEK293 cells harboring PI-Luc were cultured on a dish and sequential images of bioluminescence were taken every 10 s using a bioluminescence microscope. (Scale bar, 1 mm.) (E) Reproducibility of bioluminescence recovery after light irradiation. Average bioluminescence intensities within the region of broken line circle on the left image were calculated. The normalized values are shown as a function of time. Each arrow indicates the light irradiation point. (F) Bioluminescence images of cell population in a cultured dish upon letter-shaped irradiation of light. The cell population harboring PI-Luc was stimulated with “P” shaped light (Mask “P”), and the bioluminescence images were taken sequentially every 10 s (Upper). The cell population was stimulated successively with light in the following letter shapes P, I, -, L, U, and C. (Scale bar, 1 mm.)
To characterize the LOV–Jα sandwiched between the luciferase fragments, we generated a HEK293 cell line that stably expressed the fusion protein. The cells were irradiated with blue light and bioluminescence was measured thereafter. Different periods of irradiation with blue light revealed that the bioluminescence intensities decreased immediately up to 77% for the maximal level (Fig. 1C). The bioluminescence was then recovered gradually. It reached the initial level after turning off the irradiation. Next, we tested mutants of the LOV2 domain connected with the luciferase fragments. A fusion protein with a LOV2 domain mutant (Q513L), which induces the defect of LOV–Jα conformational change (25), caused no change in the bioluminescence intensity after blue light irradiation (Figs. S1 and S2). Another LOV2 domain mutant (V416I, L496I) or mutant (G528E, N538E) is known to make the conformational change more slowly than the wild type (29, 30). These mutants connected with the luciferase fragments showed longer recovery time in the dark state, confirming that the temporal changes in the bioluminescence originated from the structural change of LOV2 domain. We designate the unique fusion protein hereinafter as “photo-inactivatable luciferase” (PI-Luc).
Repeatability of PI-Luc was examined in the HEK293 cells using a bioluminescence microscope. The cells including the PI-Luc distributed uniformly on a dish. Images of bioluminescence recovery were taken sequentially after blue light irradiation (Fig. 1D). The light inactivation and recovery of the bioluminescence were repeated without decay of the intensities (Fig. 1E and Movie S1). By sequentially inserting six letter-inscribed masks conjugated to a focal plane in the microscope, we were able to inactivate the PI-Luc that was irradiated with the blue light through the shape of each letter (Fig. 1F and Movie S2), demonstrating that the area of PI-Luc inactivation is controllable under bioluminescence microscopy.
Responses of PI-Luc to Acidic Conditions.
To evaluate the rate of bioluminescence recovery of PI-Luc quantitatively, we reasoned that the recovery reaction should include two steps: LOV–Jα interaction initially, followed by luciferase-fragment complementation. The time-dependent bioluminescence in the recovery process was fitted using either a single or double exponential curve (Fig. S3). Both fitting curves were equally matched with the data, indicating that the recovery time is dominated by the LOV–Jα interaction and that the interaction is much slower than the luciferase complementation. To simplify the calculation of the curve-fitting process, the single exponential curve was used for evaluating the bioluminescence recovery time. The half-recovery time, which is calculated from curve fitting with a single exponential curve, is referred to as RT (Fig. S3).
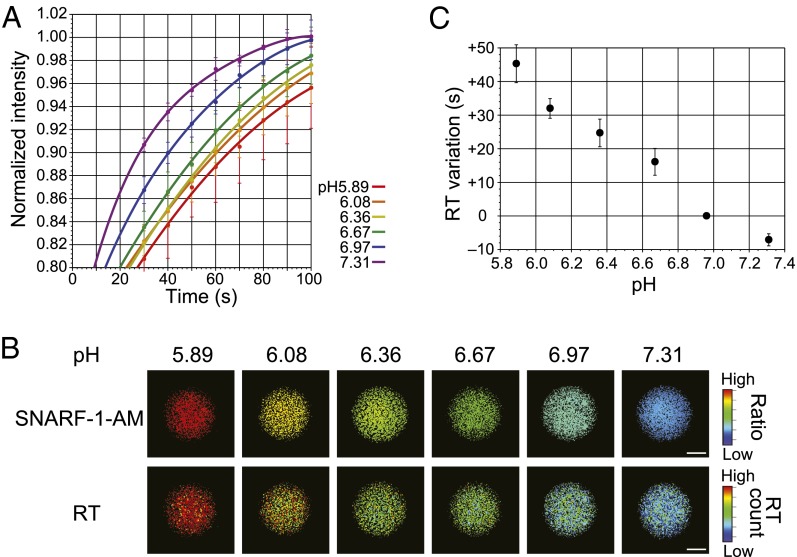
Next, we investigated the RT values under different environmental conditions. To calculate the RT values for different pH, the stable cell line expressing the PI-Luc was incubated in a pH-adjusting buffer containing the proton ionophore nigericin. Then the sequential bioluminescence images of the cells after light irradiation were acquired repeatedly. The decreased bioluminescence intensities were recovered with different speeds in each pH condition (Fig. 2A), although the bioluminescence spectra showed no remarkable change in different pH values (Fig. S4). Furthermore, the maximal wavelength of bioluminescence did not shift during the recovery process of LOV2–Jα conformational change (Fig. S5). The RT values of PI-Luc at cell population were calculated in each pixel of the images and converted into pseudocolor variation (called RT images) (Fig. 2B). To examine the pH values in the cells, we used a ratiometric fluorescence indicator, 5-(and-6)-carboxy seminaphthorhodafluor-1-acetoxymethylester (SNARF-1-AM), which has emission spectra depending on pH (580 nm/640 nm). The results of pH calibration by SNARF-1-AM showed that the RT values increased concomitantly with decreasing pH values from 7.3 to 5.9 (Fig. 2C). To confirm the effects of reactive oxygen species for LOV2–Jα conformational change, the RT values were calculated with hydrogen peroxide (Fig. S6A). There was little change in the RT values from cell lysates below the concentration of 1.0 mM hydrogen peroxide. Importantly, no effect was also found on RT values for different concentrations of d-luciferin or ATP, although absolute bioluminescence of PI-Luc varied upon changing their concentrations (Fig. S6 B and C). These results demonstrate that the RT value is a good index of acidic environment without subjecting to d-luciferin, ATP, or a low concentration of hydrogen peroxide.
Fig. 2.
Calibration of the recovery time for different pH conditions. (A) Time-dependent bioluminescence of PI-Luc in living cells after light irradiation. Bioluminescence intensities were measured repeatedly from the cells (n = 3). Error bars, SD. Fitting curves with the single exponential are shown. (B) Bioluminescence recovery time (RT) imaging of PI-Luc in living cells. HEK293 cells harboring PI-Luc were incubated in a culture medium containing the proton ionophore nigericin with different pH values. SNARF-1-AM was used as a fluorescence pH indicator in living cells. (Upper) The ratio image of SNARF-1-AM fluorescence (640 nm/ 580 nm) generated by 480 nm excitation light. (Lower) RT images calculated from sequential bioluminescence recovery images. (Scale bar, 1 mm.) (C) Calibration of the RT variation for different pH values. The average RT values were calculated using data from B. Error bars, SD.
Imaging Intracellular Acidification in Living Cells and Mice.
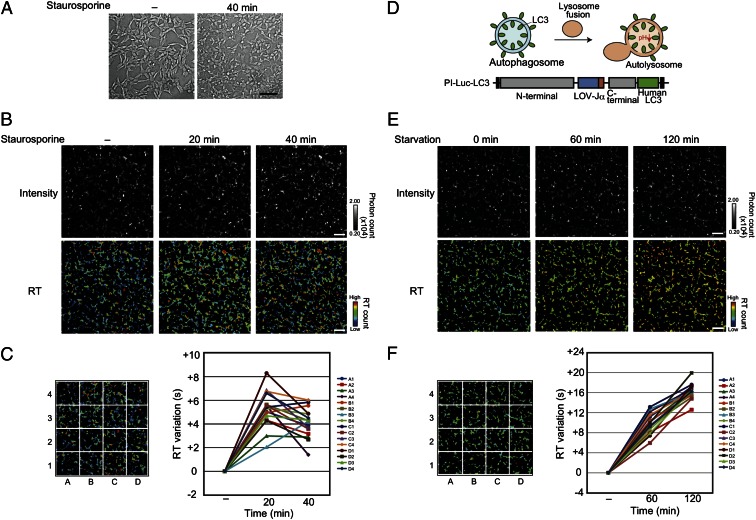
An acidic condition often occurs with cell death phenomena such as apoptosis and autophagy (31). Here, we applied the PI-Luc to examine the acidification in apoptotic cells using the RT values. HEK293 cells expressing the PI-Luc in cytosol were used for taking the sequential bioluminescence recovery images with an EM-CCD camera. Apoptosis was induced by staurosporine stimulation. Advancing apoptosis was confirmed by the observation of morphological changes in the cells (Fig. 3A). After staurosporine administration, the RT values in each cell fluctuated and some cell regions showed an increase in the RT values within 20 min (Fig. 3B). The RT values averagely decreased over the next 20 min but were still higher than the values without staurosporine (Fig. 3C). In contrast, no remarkable change existed in the absolute bioluminescence intensities in the cells, indicating that the RT values using PI-Luc showed a slight pH change in cytosol.
Fig. 3.
RT imaging to monitor the acidification for live cells. (A) Transmission (differential interference contrast) images of an HEK293 cell harboring PI-Luc. The cells were stimulated with 1 µM staurosporine for 40 min. (Scale bar, 100 µm.) (B) RT imaging of living cells upon apoptosis induction. The cells were treated with 1 µM staurosporine and incubated. (Upper) Intensity shows the absolute bioluminescence image. (Lower) RT images calculated from sequential bioluminescence recovery images. The RT values are shown as a pseudocolor bar. The incubated time are shown above the picture. (Scale bar, 100 µm.) (C) Relative analysis of RT variation upon staurosporine stimulation. Average RT values from sixteen partitioned areas were calculated using data (B). Each average RT variation was indicated in the graph, assuming that the RT value for the starting time was zero (n = 3). Error bars, SD. (D) Formation of autolysosome with pH changing and schematic construct of PI-Luc–LC3. Black squares in the construct show epitope Myc and V5/His tags. (E) RT imaging from PI-Luc–LC3 expressed HEK293 cells. (Upper) Intensity shows absolute bioluminescence images. (Lower) RT images calculated from sequential bioluminescence recovery images incubated in observation medium with 0.5% FBS. The incubated time are shown above the picture. (Scale bar, 100 µm.) (F) Variation of RT after serum starvation. Average RT values from sixteen partitioned areas were calculated using data from E. Each average RT variation is shown, assuming that the average RT value for the starting time was zero (n = 3). Error bars, SD.
In macroautophagy, the degraded targets are enveloped by membrane structure, which forms an autophagosome (Fig. 3D). It fuses with lysosomes, and the internal pH becomes acidic (autolysosome). To visualize the acidification in the lysosome fusion process, PI-Luc was connected with microtube-associated protein1 light chain 3 (LC3), which is located on the autophagosome membrane. Intercellular localization of PI-Luc–LC3 showed dot-like structures under a serum starvation condition (Fig. S7). Most of the dot-like structures were colocalized with lysosome. In contrast, PI-Luc–LC3 was dispersed uniformly in cytoplasm in the presence of 10% (vol/vol) FBS, demonstrating that PI-Luc–LC3 was incorporated in autolysosomes during serum starvation for 2 h. We next investigated the RT changes of the cells with the bioluminescence microscope. The images obtained by the RT values showed that the values in each cell gradually increased during serum starvation for 2 h (Fig. 3 E and F). Considering these results together, we concluded that PI-Luc enabled visualization of acidification in cytoplasm and the target organelles.
A low-oxygen condition upon ischemia treatment is known to induce a decrease in intracellular pH. Its successive reperfusion gives rise to oxygen stress (32–34). To demonstrate the applicability of PI-Luc for such hypoxic effects, the influence of oxygen concentration on RT values was examined for HEK293 cells expressing PI-Luc in cytosol. The low oxygen level only slightly affected the absolute intensity of PI-Luc (Fig. S8A). In contrast, RT images showed drastic changes: Low oxygen induced an increase in the RT values for 20 min. Then, an increase in the RT values continued for 10 min even if the oxygen concentration reverted to the normal level (Fig. S8B). Successive injection of oxygen caused lowering of the RT values near the initial level. When a low-oxygen condition was generated again, the RT values increased more rapidly than those for the first treatment, demonstrating that PI-Luc responded to the stress condition inside the cells.
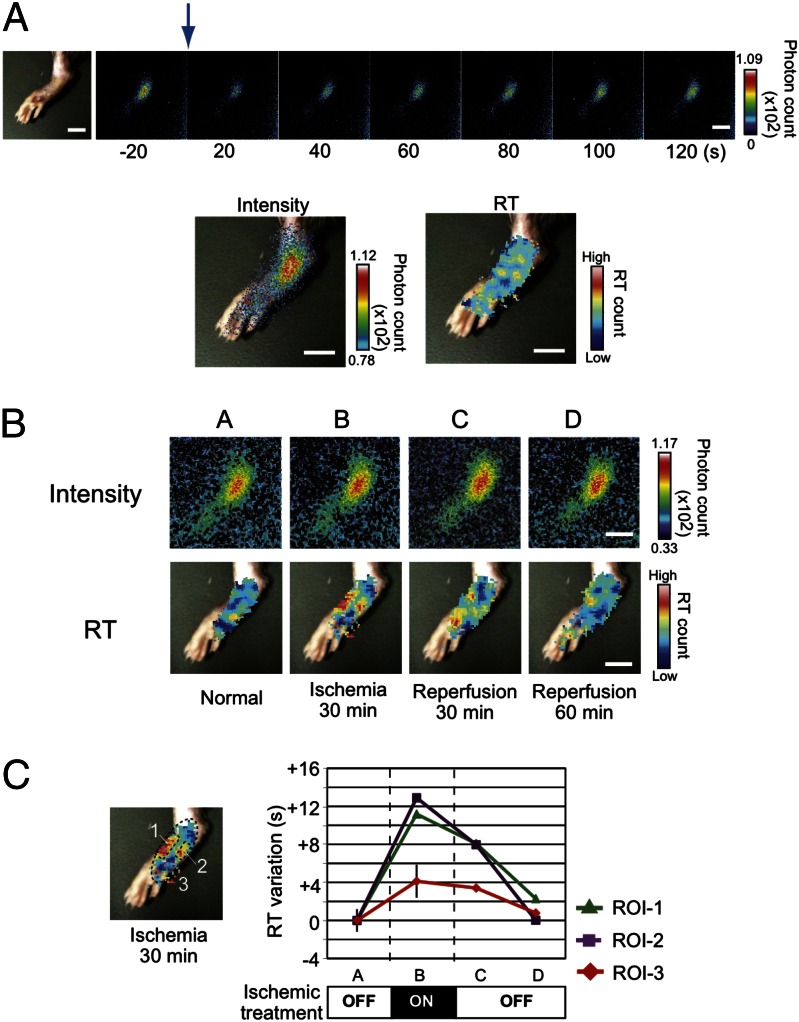
To monitor the effects of an ischemic condition on the intracellular pH environment in tissue of living mice directly, we expressed PI-Luc in the footpad because it is easy to control an ischemic condition by operating blood vessels in the base of the limb. The adenovirus coding PI-Luc was infected s.c. in the footpad and its expression was confirmed using bioluminescence imaging (Fig. 4A). The PI-Luc intensity was concentric and sufficient for taking images of bioluminescence recovery. The bioluminescence diminished immediately upon blue light irradiation from the outer skin, then it recovered gradually. From these data, we were able to calculate the RT images that displayed low RT values under normal blood flow. Next, we made an ischemic condition by clipping vessels of the base of the foot for 30 min. The RT images were obtained from the mouse footpad (Fig. 4B). Spots showing high RT values were generated at both sides of the footpad. The RT values recovered to the normal level in 60 min (Fig. 4C). The high-RT-value spots increased concomitantly with increasing time of ischemia and expanded to the entire footpad (Fig. S9). When the time of ischemia was extended for 180 min, the recovery speed of the RT values was found to be much slower and the values in some specific areas did not recover to the initial level. Long-term ischemia is known to induce severe cell damage. The observed irreversible increase in the RT values indicates deleterious effects of low-oxygen stress or apoptotic signals. Consequently, PI-Luc can be used to monitor such oxidative damage in living tissues.
Fig. 4.
RT imaging of a mouse footpad treated with ischemia. (A) Bright-field and bioluminescence imaging from living mouse footpad. A mouse footpad harboring PI-Luc was irradiated with blue light. The blue arrow indicates a light irradiation point. Bioluminescence images were taken every 20 s. The bioluminescence image and the RT image were overlaid with the bright-field image. (Scale bar, 10 mm.) (B) Bioluminescence and RT images from the mouse footpad with ischemic and reperfusion treatments. The mouse footpad was treated with vessel clipping and bioluminescence images were obtained. Each RT image was calculated from continuous images of bioluminescence. The RT images were overlaid with the bright-field one. (Scale bar, 10 mm.) (C) Variation of RT values upon ischemic treatment. Average RT values were evaluated using data from B. The areas of ROI-1–ROI-3 correspond to numbers 1–3 in the left image. Average RT variations are shown, assuming that the RT value for the starting time was zero. Variation of ROI-3 was an average from different mouse footpads (n = 3). Error bars, SD.
Discussion
We developed a photo-inactivated luciferase, PI-Luc, using luciferase-fragment complementation and a photoreactive protein, LOV2 domain. We introduced an original index of bioluminescence recovery time, RT, which enabled visualization of acidification in living cells and organelles. The PI-Luc technology revealed the applicability of image variation in the acidic environments of apoptosis and autophagy processes in cells and in an ischemic condition with a mouse model. A main advantage of the RT imaging is that there is no effect of either d-luciferin or ATP on the absolute RT values. A few reports have described the pH dependence of luciferase on bioluminescence intensities or spectral changes (35, 36), which can detect acidic environments in living cells. However, it was not applicable for long-time imaging or analysis in biological tissues because the luciferin–luciferase reaction is strongly dependent on the concentrations of d-luciferin and ATP, which fluctuate inside the cells. In fact, the substrate of d-luciferin is injected i.p. or i.v. and the bioluminescence images of living subjects are taken immediately. The signal intensity reaches the maximum level typically at 10–15 min, then it decreases immediately because of metabolism and clearance. These aspects often hamper quantitative imaging and analysis of bioluminescence inside cells when luciferase is used as a reporter. Analogously to the present study, fluorescence measurements using FRET-based probes are affected by the power of excitation light and bleaching of fluorescent molecules. However, fluorescence life-time imaging is independent of the fluorescence intensity, which enables detection of intracellular signaling with higher accuracy (37, 38). Consequently, in terms of the potential for acidic environmental monitoring, the RT imaging demonstrated in this study is expected to surpass other luminescence-based techniques for living organelles and tissue samples.
Another advantage of the present probe is that the probe enables visualization of an acidic environment in a specific intracellular microenvironment. The PI-Luc is genetically encoded. For that reason, it can be located at any specific organelle or compartment in living cells. We demonstrated an autolysosome process using PI-Luc–LC3, which was targeted to an autophagosome membrane. Previous reports have described a process of macroautophagy being visualized using an autophagic marker such as red fluorescent protein–GFP tandem-tagged LC3 and Keima-tagged LC3 (39). The probes were able to track both the formation of autophagosomes and acidic conditions with different-colored signals. They came to be used in a general approach for studying dynamic macroautophagy in quantitative terms. However, it is difficult to apply the probes to living animals because of the ratiometric analysis. The present method using PI-Luc–LC3 can compensate for the disadvantage of the fluorescence approach: The pH changes are assessed with the index of RT values, which are free from changes of bioluminescence intensity and substrate concentrations of the luciferase. The PI-Luc can be applied easily to a mouse model if the probe can be targeted and expressed to a specific organelle. Some compartments such as endocytic and secretory organelles also have a dynamic fluctuation with acidification upon extracellular stimuli (1, 40). These environments are suitable for managing cargo proteins and activation of some hydrolases for degradation. The present technique presents great potential for application in such organelles of living cells and biological tissues.
We demonstrated imaging of acidification caused by ischemia in the mouse footpad using PI-Luc. A clear difference between the present method and the previous one is the use of a unique index of the recovery time, the RT values. Generally, a key advantage of bioluminescence imaging is application of in vivo imaging to preclinical mouse models. However, repeated injection of the substrate in the mouse is needed for temporal analysis because clearance of the substrate occurs very rapidly. Repeated administration of the substrate can also cause severe damage to the injected organ, potentially limiting the number of images that can be acquired over time. The present method using PI-Luc can resolve these issues. The RT value is independent of the intensity. Consequently, there is no need to inject the substrate repeatedly until the bioluminescence of the PI-Luc diminishes to an undetectable level. Therefore, this RT imaging technique will facilitate the further design of luciferase-based probes applied for in vivo tissue imaging of living mice and rats.
Materials and Methods
Bioluminescence Analysis.
Measurement of PI-Luc bioluminescence from living cells with different periods of light irradiation was performed using Kronos (Atto) at 37 °C with a blue light-emitting diode (LED) handy light (480 nm, 2 mW, Mini Maglite 2AAA; Mag Instrument). Bioluminescence of a fusion protein with LOV2 domain mutant-expressed cells was also measured using Kronos. Effects of hydrogen peroxide on RT values were analyzed using PI-Luc–expressed cells lysed with Bright-Glo Luciferase Assay buffer (Promega). To examine the effects of d-luciferin and ATP on bioluminescence, the cells were digested with lysis buffer (Passive Lysis Buffer; Promega) and respective concentrations of d-luciferin and ATP were adjusted. The bioluminescence of these cell lysates was measured using a luminometer (MiniLumat LB9506; Berthold Technologies) at 37 °C. Light irradiation for the cell lysates was performed using an inverted fluorescence microscope (IX-70; Olympus Corp.) without an objective lens for 20 s. The light was generated from a mercury lamp through the band-pass excitation filter (BP470–495 nm, 2 mW). The bioluminescence recovery time (RT) of PI-Luc was calculated using DeltaGraph software (Japan Poladigital). Bioluminescence spectra of PI-Luc were measured from PI-Luc–expressed cell lysates adjusted for pH (AB-1850 LumiFL-spectrocapture; Atto) at room temperature. Cell lysates were irradiated for 20 s using the microscope.
Bioluminescence Measurement with Microscope.
Observation of bioluminescence and light stimulation of living cells were performed using an upright fluorescence microscope (BX-61WI; Olympus) with a 10× dipping objective lens (0.40 N.A.). The stage was incubated at 37 °C using a stage incubator (Tokai Hit). That microscope was set in a dark room. Bioluminescence images were acquired using a cooled EM-CCD camera (ImagEM; Hamamatsu Photonics). The images in masking assay and pH calibration were taken through a lens attachment (U-TV0.25×C; Olympus). For observation of cell apoptosis, the differential interference contrast and bioluminescence images of PI-Luc–expressed cells were obtained. In pH calibration, SNARF-1-AM in cells was excited by light through the band-pass excitation filter (BP470–495 nm). The fluorescence emissions at two wavelengths were observed through two independent band-pass excitation filters (BP570–590 nm or BP620–660 nm. Light stimulation for PI-Luc–expressed cells were performed with irradiation light generated from a metal–halide light source (KTX-60MT; Kenko Tokina) through the band-pass excitation filter (BP470–495 nm) and objective lens (0.15 mW). First, the cells were irradiated for 10 s. Subsequently the continuous bioluminescence images were acquired by a cooled EM-CCD camera with 10-s exposure for each. The light stimulation and image acquisition were programmed using Metamorph software (Molecular Devices). For masking assay, the optical path was partially blocked by a polyethylene terephthalate sheet with alphabet letters printed in black ink. The RT image was calculated using Igor Pro software (Hulinks).
In Vivo Bioluminescence Imaging System.
For whole-dish and in vivo mouse bioluminescence, images were taken using a semicustom bioluminescence imaging system in a dark room at 37 °C. All images were taken using a cooled CCD camera (Versarray 1300B; Princeton Instruments) with 20-s exposure. For whole-dish cell population imaging, the irradiation light source was a blue LED transilluminator (470 nm, 2.5 mW, LEDB-SBOXH; Optocode), and each cell dish was irradiated from the bottom for 20 s each. In the hypoxic effects assay for a cell population, a low-oxygen condition was established in the dark room by nitrogen filling. Autofluorescence images were taken using a monochrome CCD camera (Atto) with a UV transilluminator.
Irradiation of a mouse was done using a blue LED light source (UHP-Mic-LED-460; Prizmatix) via optical fiber (460 nm, 70 mW) for 40 s. The adenovirus-infected mouse (male C57BL/6, 9 wk old, 23–28 g body weight) was injected with d-luciferin (120 mg/kg of body weight) and PBS s.c. Mouse footpad ischemia was performed with clipping of the vessel by a Sugita clip (Mizuho). All animal experiments were conducted in accordance with a protocol approved by School of Science, the University of Tokyo.
Image processing was conducted using imaging software (SlideBook 4.1; Intelligent Imaging Innovation). The RT image was calculated using Igor Pro software.
Plasmid construction, selection of a stable cell line, sample preparation, immunostaining protocols, and adenovirus experiments are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by the Japan Society for the Promotion of Science, Japan Science and Technology Corporation, and the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by grants from Sasakawa Scientific Research Grant (to H.M.) and the Mitsubishi Foundation (to T.O.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304056110/-/DCSupplemental.
References
- 1.Casey JR, Grinstein S, Orlowski J. Sensors and regulators of intracellular pH. Nat Rev Mol Cell Biol. 2010;11(1):50–61. doi: 10.1038/nrm2820. [DOI] [PubMed] [Google Scholar]
- 2.Han J, Burgess K. Fluorescent indicators for intracellular pH. Chem Rev. 2010;110(5):2709–2728. doi: 10.1021/cr900249z. [DOI] [PubMed] [Google Scholar]
- 3.Thivierge C, Han J, Jenkins RM, Burgess K. Fluorescent proton sensors based on energy transfer. J Org Chem. 2011;76(13):5219–5228. doi: 10.1021/jo2005654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urano Y, et al. Selective molecular imaging of viable cancer cells with pH-activatable fluorescence probes. Nat Med. 2009;15(1):104–109. doi: 10.1038/nm.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee H, et al. Near-infrared pH-activatable fluorescent probes for imaging primary and metastatic breast tumors. Bioconjug Chem. 2011;22(4):777–784. doi: 10.1021/bc100584d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abad MF, Di Benedetto G, Magalhães PJ, Filippin L, Pozzan T. Mitochondrial pH monitored by a new engineered green fluorescent protein mutant. J Biol Chem. 2004;279(12):11521–11529. doi: 10.1074/jbc.M306766200. [DOI] [PubMed] [Google Scholar]
- 7.Bizzarri R, Serresi M, Luin S, Beltram F. Green fluorescent protein based pH indicators for in vivo use: a review. Anal Bioanal Chem. 2009;393(4):1107–1122. doi: 10.1007/s00216-008-2515-9. [DOI] [PubMed] [Google Scholar]
- 8.Katayama H, Kogure T, Mizushima N, Yoshimori T, Miyawaki A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol. 2011;18(8):1042–1052. doi: 10.1016/j.chembiol.2011.05.013. [DOI] [PubMed] [Google Scholar]
- 9.Niu G, Chen X. Molecular imaging with activatable reporter systems. Theranostics. 2012;2(4):413–423. doi: 10.7150/thno.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozawa T, Yoshimura H, Kim SB. Advances in fluorescence and bioluminescence imaging. Anal Chem. 2013;85(2):590–609. doi: 10.1021/ac3031724. [DOI] [PubMed] [Google Scholar]
- 11.Massoud TF, Gambhir SS. Molecular imaging in living subjects: Seeing fundamental biological processes in a new light. Genes Dev. 2003;17(5):545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 12.Tannous BA, Kim DE, Fernandez JL, Weissleder R, Breakefield XO. Codon-optimized Gaussia luciferase cDNA for mammalian gene expression in culture and in vivo. Mol Ther. 2005;11(3):435–443. doi: 10.1016/j.ymthe.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 13.Dothager RS, et al. Advances in bioluminescence imaging of live animal models. Curr Opin Biotechnol. 2009;20(1):45–53. doi: 10.1016/j.copbio.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito K, et al. Luminescent proteins for high-speed single-cell and whole-body imaging. Nat Commun. 2012;3:1262. doi: 10.1038/ncomms2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SB, Ozawa T, Watanabe S, Umezawa Y. High-throughput sensing and noninvasive imaging of protein nuclear transport by using reconstitution of split Renilla luciferase. Proc Natl Acad Sci USA. 2004;101(32):11542–11547. doi: 10.1073/pnas.0401722101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paulmurugan R, Umezawa Y, Gambhir SS. Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc Natl Acad Sci USA. 2002;99(24):15608–15613. doi: 10.1073/pnas.242594299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hida N, et al. High-sensitivity real-time imaging of dual protein-protein interactions in living subjects using multicolor luciferases. PLoS ONE. 2009;4(6):e5868. doi: 10.1371/journal.pone.0005868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dragulescu-Andrasi A, Chan CT, De A, Massoud TF, Gambhir SS. Bioluminescence resonance energy transfer (BRET) imaging of protein-protein interactions within deep tissues of living subjects. Proc Natl Acad Sci USA. 2011;108(29):12060–12065. doi: 10.1073/pnas.1100923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanno A, Yamanaka Y, Hirano H, Umezawa Y, Ozawa T. Cyclic luciferase for real-time sensing of caspase-3 activities in living mammals. Angew Chem Int Ed Engl. 2007;46(40):7595–7599. doi: 10.1002/anie.200700538. [DOI] [PubMed] [Google Scholar]
- 20.Strickland D, Moffat K, Sosnick TR. Light-activated DNA binding in a designed allosteric protein. Proc Natl Acad Sci USA. 2008;105(31):10709–10714. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu YI, et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461(7260):104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strickland D, et al. TULIPs: Tunable, light-controlled interacting protein tags for cell biology. Nat Methods. 2012;9(4):379–384. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swartz TE, Wenzel PJ, Corchnoy SB, Briggs WR, Bogomolni RA. Vibration spectroscopy reveals light-induced chromophore and protein structural changes in the LOV2 domain of the plant blue-light receptor phototropin 1. Biochemistry. 2002;41(23):7183–7189. doi: 10.1021/bi025861u. [DOI] [PubMed] [Google Scholar]
- 24.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301(5639):1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 25.Nash AI, Ko WH, Harper SM, Gardner KH. A conserved glutamine plays a central role in LOV domain signal transmission and its duration. Biochemistry. 2008;47(52):13842–13849. doi: 10.1021/bi801430e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crosson S, Moffat K. Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc Natl Acad Sci USA. 2001;98(6):2995–3000. doi: 10.1073/pnas.051520298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexandre MT, Arents JC, van Grondelle R, Hellingwerf KJ, Kennis JT. A base-catalyzed mechanism for dark state recovery in the Avena sativa phototropin-1 LOV2 domain. Biochemistry. 2007;46(11):3129–3137. doi: 10.1021/bi062074e. [DOI] [PubMed] [Google Scholar]
- 28.Paulmurugan R, Gambhir SS. Firefly luciferase enzyme fragment complementation for imaging in cells and living animals. Anal Chem. 2005;77(5):1295–1302. doi: 10.1021/ac0484777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zoltowski BD, Vaccaro B, Crane BR. Mechanism-based tuning of a LOV domain photoreceptor. Nat Chem Biol. 2009;5(11):827–834. doi: 10.1038/nchembio.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strickland D, et al. Rationally improving LOV domain-based photoswitches. Nat Methods. 2010;7(8):623–626. doi: 10.1038/nmeth.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waibel M, et al. Mitochondria are not required for death receptor-mediated cytosolic acidification during apoptosis. Apoptosis. 2007;12(3):623–630. doi: 10.1007/s10495-006-0006-z. [DOI] [PubMed] [Google Scholar]
- 32.Granger DN, Rutili G, McCord JM. Superoxide radicals in feline intestinal ischemia. Gastroenterology. 1981;81(1):22–29. [PubMed] [Google Scholar]
- 33.Chambers DE, et al. Xanthine oxidase as a source of free radical damage in myocardial ischemia. J Mol Cell Cardiol. 1985;17(2):145–152. doi: 10.1016/s0022-2828(85)80017-1. [DOI] [PubMed] [Google Scholar]
- 34.McCord JM. Oxygen-derived free radicals in postischemic tissue injury. N Engl J Med. 1985;312(3):159–163. doi: 10.1056/NEJM198501173120305. [DOI] [PubMed] [Google Scholar]
- 35.Ando Y, et al. Firefly bioluminescence quantum yield and colour change by pH-sensitive green emission. Nat Photonics. 2008;2:44–47. [Google Scholar]
- 36.Viviani VR, Silva Neto AJ, Arnoldi FG, Barbosa JA, Ohmiya Y. The influence of the loop between residues 223-235 in beetle luciferase bioluminescence spectra: A solvent gate for the active site of pH-sensitive luciferases. Photochem Photobiol. 2008;84(1):138–144. doi: 10.1111/j.1751-1097.2007.00209.x. [DOI] [PubMed] [Google Scholar]
- 37.Nakabayashi T, Wang HP, Kinjo M, Ohta N. Application of fluorescence lifetime imaging of enhanced green fluorescent protein to intracellular pH measurements. Photochem Photobiol Sci. 2008;7(6):668–670. doi: 10.1039/b800391b. [DOI] [PubMed] [Google Scholar]
- 38.Klarenbeek JB, Goedhart J, Hink MA, Gadella TW, Jalink K. A mTurquoise-based cAMP sensor for both FLIM and ratiometric read-out has improved dynamic range. PLoS ONE. 2011;6(4):e19170. doi: 10.1371/journal.pone.0019170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Demaurex N. pH Homeostasis of cellular organelles. News Physiol Sci. 2002;17:1–5. doi: 10.1152/physiologyonline.2002.17.1.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.