Abstract
The Hawaiian Islands provide the venue of one of nature’s grand experiments in evolution. Here, we present morphological, behavioral, genetic, and geologic data from a young subterranean insect lineage in lava tube caves on Hawai‘i Island. The Oliarus polyphemus species complex has the potential to become a model for studying rapid speciation by stochastic events. All species in this lineage live in extremely similar environments but show strong differentiation in behavioral and morphometric characters, which are random with respect to cave age and geographic distribution. Our observation that phenotypic variability within populations decreases with increasing cave age challenges traditional views on founder effects. Furthermore, these cave populations are natural replicates that can be used to test the contradictory hypotheses. Moreover, Hawaiian cave planthoppers exhibit one of the highest speciation rates among animals and, thus, radically shift our perception on the evolutionary potential of obligate cavernicoles.
Keywords: density-dependent selection, dynamic adaptive landscape, nonadaptive speciation, sexual behavior, vibrational communication
The role of extrinsic factors such as environmental changes in driving genetic change in populations is largely undisputed. Genetic drift (i.e., random changes in gene frequency attributable to stochastic allele assortment) also affects all populations, but its role in yielding significant differences between populations in interplay with selective forces is controversially discussed. Genetic drift is most effective in small populations, the best-known examples being drastic population bottlenecks or founding individuals (e.g., during the colonization of islands). These observations led to the development of the much-debated founder-effect concept by Mayr (1). This concept was further developed by including population structure and sexual selection (2, 3) but has remained contentious (4–6) (for reviews, see refs. 7 and 8). Here, we revisit the founder-effect concepts using the blind planthopper Oliarus polyphemus in the Hawaiian lava tube caves as a model system. The Hawaiian cave planthopper system provides excellent opportunities to test models of stochastic effects in evolution in a natural setting, because it is simple enough for distinguishing the influence of many of the major factors involved and encompasses natural populations undergoing repeated events (replicates of “natural experiments”) under similar conditions where many of the relevant biotic and abiotic factors can be assessed.
Grounds and Habits of the Hawaiian Cave Planthoppers
The Hawaiian archipelago, the most remote group of high islands in the world, hosts a highly diverse endemic fauna. Much of this diversity is the result of radiations following rare colonization events on the islands (9). The Hawaiian chain was formed by outpourings of lava from a volcanic “hot spot” (10), and volcanoes are still active on its youngest and largest island, Hawai‘i. This active volcanism causes rapid landscape dynamics [e.g., 90% of the entire surface area of Kīlauea Volcano has been replaced within the past 1,500 y (11)]. The nearly continuous flow of lava creates a unique subterranean environment consisting of interconnected systems of air-filled voids of varying sizes up to lava tube caves that can extend up to several dozen kilometers. These subterranean voids host diverse root communities (12–14), with food webs largely sustained by living roots of the pioneer plant Metrosideros polymorpha (Myrtaceae) (SI Appendix, Fig. S1). Roots are ephemeral resources, because their abundance decreases with increasing cave age through ecological succession on the surface (SI Appendix, Text S2).
Species of Oliarus are the only obligatory cave-dwelling primary consumers in this ecosystem (15). The Hawaiian Oliarus (Nesoliarus) clade is a monophyletic endemic radiation (16) comprising about 85 known species. The latter include seven exclusively cave-dwelling species that have been described to date from three islands (17). Three cave taxa are endemic to Hawai‘i Island, but only O. polyphemus is widely distributed, inhabiting lava tubes on all major volcanoes except Kohala. The inhabited caves occur from sea level to about 2,000 m and range in age from less than 50 to several thousand years (18) (Fig. 1 and SI Appendix, Fig. S3). O. polyphemus exhibits extreme character reduction (eyes, wings, pigmentation) associated with its troglobitic habit (SI Appendix, Fig. S2).
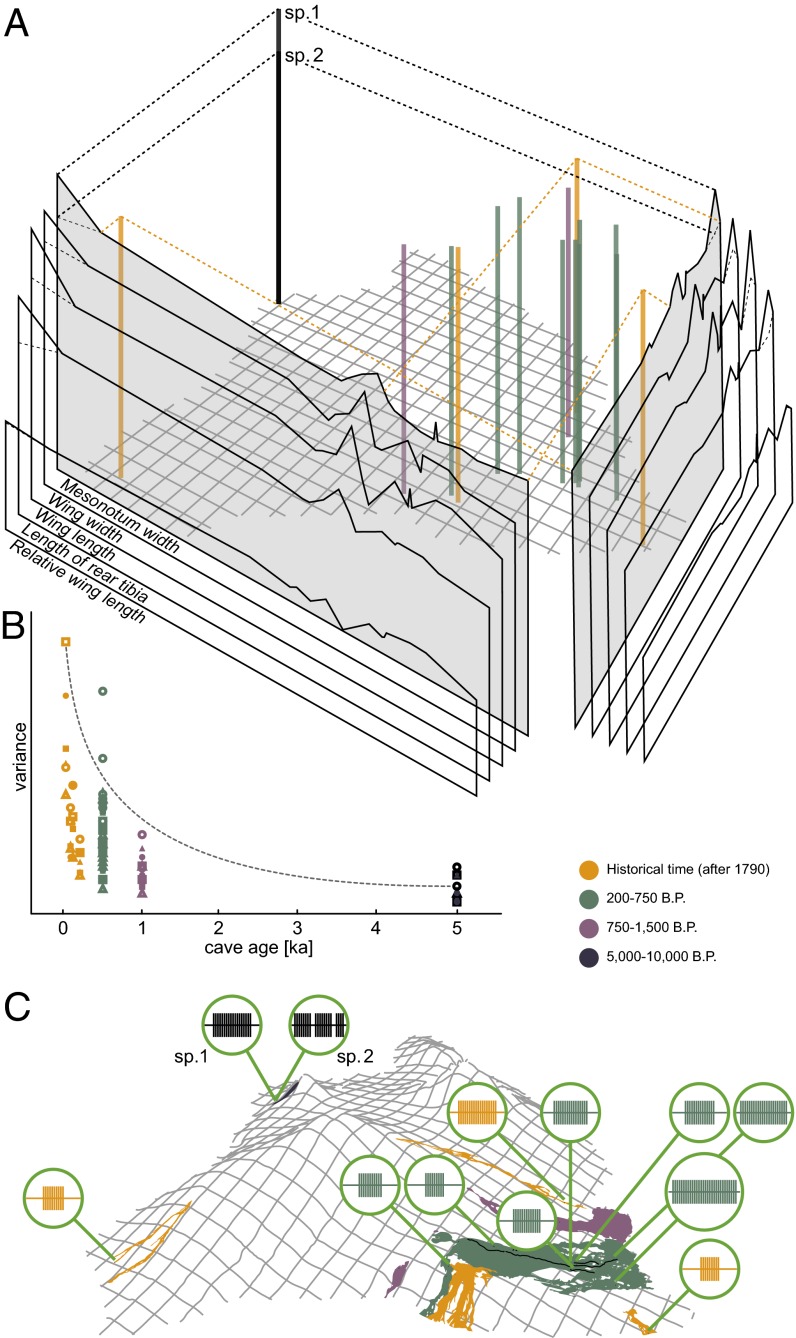
Fig. 1.
Phenotypic variation in the O. polyphemus complex. (A) Morphometric variation in five characters (population mean, relative scale) plotted against geography (compare C). Columns shown for mesonotum width only; for all other characters, only the resulting 2D projections are shown. (B) Scatterplot of variance in seven morphometric characters (denoted by symbols) against cave age. (C) Call-pattern variation (schematic representation is proportional to mean call length and pulse number) mapped on topography of Hawai‘i Island. Colors indicate cave age (see the legend in the figure).
Given the extreme degree of troglomorphy in O. polyphemus, dispersal is only possible by subterranean migration, which is constrained by the patchy distribution of resources, both at the intralava tube (root patches within lava tubes) and interlava tube (and lava flow) levels. A certain level of migration is necessary to maintain populations despite the risk of extinction through ecological succession or catastrophic events (see SI Appendix, Text S2 for a discussion of modes of subterranean migration). The high level of phenotypic and genetic differentiation found between geographically proximate, young caves suggests largely isolated populations and low migration rate through rare and accidental dispersal. This essentially implies a series of many founder events in the establishment of new populations (SI Appendix, Fig. S8 and Text S2). If the migration rate is low, each new cave population would be descended from a single or few founding events from neighboring older established populations. Because even young caves have established populations (SI Appendix, Table S5), the age of the founding event probably approximates the age of the lava flow. The minimum speed of dispersal has been estimated at >10 m/y (SI Appendix, Text S2), which is compatible with the assumption of underground dispersal across the study area within the last 10,000 y.
Planthoppers worldwide use low-frequency substrate-borne vibration for communication (19–21). The signals are crucial for species-specific recognition (22) and have led to the discovery of “cryptic acoustic species,” which are morphologically indistinguishable (23, 24). In the case of O. polyphemus, vibrations are transmitted along the roots on which the blind animals feed and are their only means of communication. Differences in male and female courtship calls of O. polyphemus from lava tubes on three different volcanoes indicate the existence of cryptic species (18, 25).
Here, we use morphological, acoustic, genetic, and geologic data to investigate patterns of differentiation in populations of O. polyphemus from 20 caves of varying age (SI Appendix, Text S1, Fig. S3, and Tables S1 and S2).
Results
Phenotypic Differentiation.
Significant intercave differences in call patterns were observed in all 10 parameters measured for 12 populations (Fig. 1C and SI Appendix, Fig. S5 and Table S7). The significant differentiation found even between caves in close proximity indicates an interruption of gene flow between these caves. In “Pink Pistillaria,” two completely different call patterns were found (SI Appendix, Fig. S6), which, along with morphometric differences, indicate the coexistence of two sympatric species in the oldest (5,000–8,000 y) cave system known to harbor cave planthoppers (SI Appendix, Table S5). Moreover, in one of the two Pink Pistillaria populations and in the “Kaumana” population, variation of some call parameters did not overlap with these parameters in all other populations studied (SI Appendix, Fig. S5). The population from “McKenzie Park” even showed a unique song structure with a regularly alternating duet, again suggesting the existence of a distinct species. Remarkably, McKenzie Park and Kaumana are among the youngest caves [i.e., formed after 1790 (SI Appendix, Fig. S3 and Table S5)]. Similarly, significant morphometric differentiation between 18 cave populations was found in all 14 parameters measured, albeit to a lesser degree, because no gaps in the ranges of character variation were observed (SI Appendix, Table S7). A discriminant analysis including both song and morphometric parameters revealed a 100% assignment for 15 populations in at least one sex for at least one character complex (SI Appendix, Table S8). The differences between populations do not follow a pattern of clinal variation in either call patterns or morphology (Fig. 1A). In addition, no correlation between the degree of morphological differentiation and cave age was found. Interestingly, the only correlation found is a negative one of phenotypic character variability to cave age; this correlation is significant or highly significant for 9 of 14 morphological characters (Fig. 1B and SI Appendix, Table S11). None of these morphologic or ethologic differences appears to be adaptations to differences in habitat.
Genetic Differentiation.
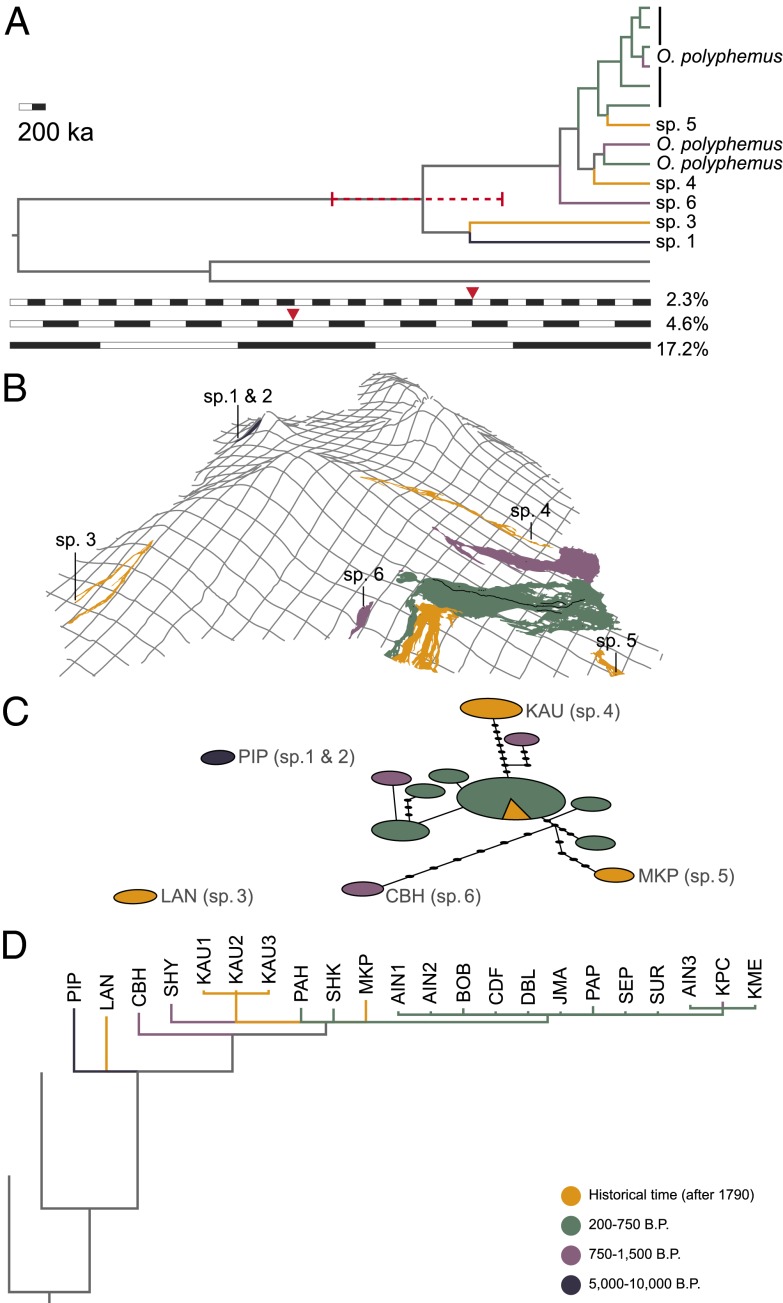
An mtDNA-based molecular phylogeny supports the monophyly [i.e., a single initial cave colonization by the ancestor of O. polyphemus (Fig. 2D and SI Appendix, Fig. S7)]. The basal splits involve populations from the western and southernmost caves Pink Pistillaria, “Lanikai,” and “Calabash” and are consistent with the assumption of dispersal from the older Hualālai Volcano to the southern flows of Mauna Loa. All other populations in the southeastern part of the island, including those on Kīlauea, the youngest volcano, form a monophyletic group. Although the branching sequence within this clade cannot be resolved, some populations within this group are genetically quite distinct (Fig. 2C and SI Appendix, Table S13). The largest genetic distance (p-distance) between two haplotypes in the O. polyphemus complex is 4.7%. In contrast to the acoustic and morphological data, genetic distance does correlate with geography (Fig. 2C) but, again, not with cave age.
Fig. 2.
Genetic variation in the O. polyphemus complex. (A) Bayesian Inference (BEAST) phylogram; the three scales represent the three different rates used. Red triangles indicate the origin of Hawai‘i Island ca. 1 Ma for each rate. The two unlabeled bottom lineages have been used as the outgroup (see Material and Methods for details). The red dashed bar indicates the 95% confidence interval for the age of the O. polyphemus complex. (B) Distribution of cavernicolous Oliarus species on Hawai‘i Island. Unlabeled flows are inhabited by O. polyphemus s.str. (C) Haplotype network. Circle size is relative to number of sequences per haplotype. (D) Bayesian Inference (MrBayes) phylogram. Three-letter codes in C and D are cave abbreviations (SI Appendix, Table S6).
The application of “standard,” frequently applied substitution rates for cytochrome c oxidase subunit I (COI) derived from other insects (2.3% per Ma) (26) and cave arthropods (4.6% per Ma) (27) to the O. polyphemus complex yields divergence time estimates based on molecular clock analyses for the basal intracave split ranging from 0.64 to 1.29 Ma (Fig. 2A and SI Appendix, Figs. S9 and S10). These dates seem reasonable if recent estimates for the age of Hawai‘i Island of ca. 1 Ma are accepted prima facie (11) (SI Appendix, Fig. S11). However, it is not very likely that the O. polyphemus complex is as old as the basal rocks of Hawai‘i Island. The extremely complex geologic history of the island, especially the catastrophic collapses of large segments of each volcano in turn, suggests that the current cave Oliarus populations must be much younger than the maximum age of the island and each volcano. Divergence time estimates gained from a much faster COI rate of 17.2% per Ma (28) would, for example, be compatible with assuming that the maximum age of the species complex is constrained by the transition from shield to post shield volcanics of Hualālai ca. 130–100 ka ago (11) (Fig. 2A and SI Appendix, Fig. S12). The volcanic dynamics of the postshield phase might even constrain the maximum age further to 25 ka (see SI Appendix, Text S3 and Text S4 for additional information). If the dispersal of O. polyphemus indeed involved a series of thousands or tens of thousands of founder events, this would lead to the amplified accumulation of mutations in a neutral marker and might provide a plausible explanation for the high genetic distances observed. This interpretation is corroborated by comparative studies on Hawaiian Drosophila lineages experiencing founder events (29).
Discussion
Rapid Speciation.
Irrespective of the factors leading to the apparently high substitution rate in O. polyphemus, some of the fastest speciation processes found to date occur in this clade of subterranean planthoppers. The highest speciation “rate” reported for invertebrates to date is 4.17 species per million years (Sp/Ma) for the Laupala cricket species of Hawai‘i Island, assuming six species and an island age of 430 ka (30). This is more than an order of magnitude higher than the one usually assumed for arthropods and only surpassed by that of African cichlids (6) or, potentially, Hawaiian drosophilids (31). However, the island age has been revised to ca. 1 Ma (11). Assuming that the O. polyphemus clade comprises seven species (see SI Appendix, Text S5 for a discussion of species numbers), a slightly higher rate of speciation as for Laupala is obtained under any estimate of island age. The assumption of clade age corresponding to island age is the most conservative estimate possible. If the transition to postshield volcanics of the Hualālai (11, 32) is used as a somewhat more realistic estimate of the maximum age of the clade, a roughly 10-fold higher rate emerges. Assuming, in addition, that dispersal and cave colonization originated from the Hualālai, and using the three species of the Mauna Loa/Kīlauea clade and a maximum age of 10 ka, the resulting rate estimate is again 10 times higher. This estimate, which exceeds the highest yet-recorded speciation rate in any taxon—75.6 Sp/Ma in Lake Victoria cichlids (33)—seems not overly extreme given the specific conditions of the Hawaiian cave system.
Founder Effects and Adaptively Neutral Change.
The O. polyphemus species complex has a very fast substitution rate and possibly the highest diversification rate in animals. The responsible mechanisms are still largely unknown, but our data suggest that stochastic events, along with changing selective forces, may play an important role in this system. The lack of any correlation of phenotypic evolution with distance and habitat differences precludes a major role of geography and adaptation in diversification. In addition, the predominant role of random individual dispersal (i.e., where single or very few individuals found new populations) results in parallel and frequent sequential genetic bottlenecks. These naturally occurring “replicates” can be used to test the contradictory hypotheses of the different founder-effect concepts.
The observed genetic changes associated with founder events led to the original founder-effect concept, which refers to the changes brought about by genetic drift on the reduced and arbitrary sample of a population’s total genetic variation carried by the few individuals that establish a new population. In its classical formulation by Mayr (1), these changes principally involve a decrease in genetic variation and the increased fixation of rare and common alleles. Subsequently, selection in the altered genetic environment causes the breakup and reorganization of coadapted gene complexes, leading to a hypothesized “genetic revolution.” Mayr’s founder-effect concept has been criticized for requiring unrealistic conditions to work (4), such as an exceedingly small number of founding individuals surviving for several generations. Moreover, it is barely possible to distinguish the impact of population bottlenecks in small founder populations from the effects of isolation, environmental differences, and genetic drift in moderately sized populations.
In recognition of these problems, Carson (2, 5, 34) developed the “founder-flush” concept that proposes a mechanism for the successful shift of the balanced genotype in founder populations. Under Carson’s model, a strong increase in variability occurs during the process of rapid population growth during which the founding population expands to occupy the new habitat (the founder flush) after a founder event and the associated bottleneck. This increase in variation is the result of relaxed selection resulting from reduced intraspecific competition for resources and, especially, of relaxed sexual selection on both sexes resulting from low availability of mating partners (see also ref. 35). With further population growth and accompanying increase in population density, strong selection is resumed, and the population collapses. The envisioned population collapse may not be a decrease in the census population size but, rather, a collapse of the effective population size after strong sexual selection has resumed and random mating is replaced once more by assortative mating (see SI Appendix, Fig. S13, time b → d). The breakup of some coadapted gene complexes in the founder-flush phase and random processes can cause the state of some characters in the now-stabilized population to differ significantly from those of the parental population (5). Carson’s founder-flush concept was subsequently complemented with Templeton’s genetic-transilience theory, which also invokes an increase of selectable genetic variation after a founder event (3, 8). Carson’s and Templeton’s concepts are compatible and even synergistic with one another, but they are both incompatible with Mayr’s genetic revolution theory (5, 8).
Repeated attempts to test Carson’s and Templeton’s concepts in the laboratory yielded ambiguous results (36–38), although a statistical reanalysis of the individual experimental results found strong support for the predictions made by this founder concept (8). However, these findings are difficult to interpret with respect to the role of founder effects in evolution because this can only be tested using a natural population with an undisturbed mating system. O. polyphemus offers just these conditions, and the system is simple enough to distinguish between the influence of most factors involved. The populations of O. polyphemus essentially constitute naturally occurring replicates of an evolutionary experiment at different time stages, permitting a space-for-time approach to test the predictions of the founder-effect models of Mayr (1) and Carson and Templeton (2, 5, 34), which differ in a crucial aspect. Mayr predicts that the initial founder event is followed by an immediate further decrease in genetic variability from which the population will only recover slowly. In contrast, Carson proposed that a founder event is immediately followed by the founder flush, leading to an instant rise in variability. The variance of morphological characters, which is used here as a proxy of genetic variability, is highest in the youngest caves and decreases with increasing age of the caves and thus presumably the age of the populations of the O. polyphemus complex (Fig. 1B). This pattern would be expected under the founder-flush model but does not fit the prediction of Mayr. Furthermore, it corroborates the counterintuitive prediction of Carson’s and Templeton’s theories that founder events can actually increase the additive phenotypic variance when dealing with epistatic systems, as confirmed by diverse experimental systems (e.g., ref. 39; for review, see ref. 8).
However, as succinctly pointed out by Mayr, “the real problem of speciation is not how to produce difference but rather to escape from the cohesion of the gene complex” (ref. 40, p. 518). According to Carson’s model (5), coadapted gene complexes can be broken up in the flush phase, leading to new combinations. This model can be translated into Sewall Wright’s heuristic analytical tool of fitness landscapes (41–43) to integrate concepts of fitness maximization and adaptive change. At the core of this approach is the interpretation of speciation as a shift of a population from one adaptive peak to the next. Assuming a static landscape surface this would pose the classical problem for the founder-flush models, how and why the founders leave an adaptive peak and cross the adaptive valley (Fig. 3A). Some reviews even unjustifiably subsumed the concepts of Mayr, Carson, and Templeton under the term “peak shift models” (see, for example, ref. 6), ignoring differing conceptual approaches to the problem that led us to a new thinking about fitness landscapes.
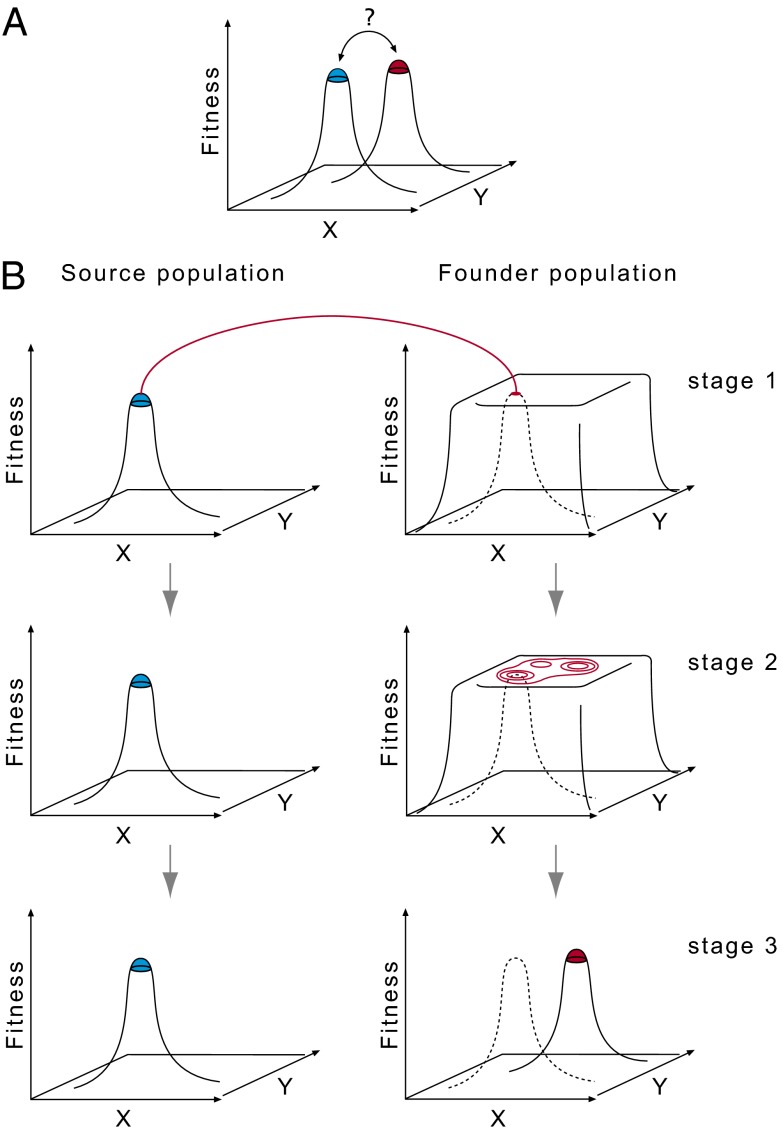
Fig. 3.
Peak move in a fitness landscape. Schematic representation of the Wrightian peak shift problem (A) and the envisioned Carsonian peak move in a fitness landscape (B) (see Discussion for further explanations). The fitness landscape is a diagrammatic representation of the field of gene combinations in two dimensions (axes x and y represent allele frequencies), graded with respect to adaptive value under a particular set of conditions (60).
In sexually reproducing species under sexual selection, the structure of the landscape can only be estimated from a population perspective or is defined by population structure, respectively. There is no “objectively” recognizable landscape that is independent from the population nor does a landscape for a species or an individual exist. Thus, the adaptive landscape can change even in a stable environment if the population structure changes (bottleneck, founder flush), so the landscape is in continuous flux through extrinsic and intrinsic (population) factors.
A founder event would accordingly involve the establishment of a new landscape (Fig. 3B, stage 1). The relaxation of sexual selection during the flush phase allows the temporary existence of a fitness plateau instead of a fitness peak for the characters involved in sexual selection; in other words, the plateau is the local relaxation in attractors establishing a section in the multidimensional fitness space, which can be freely occupied by the population during the flush phase (Fig. 3B, stage 2). When sexual selection sets in again, accompanied by a breakdown of effective population size (i.e., effective breeding size), the plateau collapses into a peak (stage 2 → 3), which will, with some probability, occupy a different place from the one in the source population (Fig. 3B, stage 3). This can be regarded as a “peak move” rather than as a peak shift. The movements of the population on the plateau are almost entirely random, and this generates an undirected peak movement in a series of founder events. This would explain why there is no observable cline or trend in the changes of the O. polyphemus populations (Fig. 1). Similarities between populations can also be attributable to random peak movements (i.e., may have originated in parallel).
Our observation that phenotypic variability decreases with increasing cave age is consistent with the founder-flush model (2, 5) but challenges traditional views on founder effects (1). The interpretation of the founder-flush concept as a “peak-move model” in the framework of the adaptive landscape (Fig. 3) allows an understanding of adaptively neutral [i.e., “nonadaptive” sensu (2)] evolutionary change. The O. polyphemus species complex now offers an opportunity to develop a compelling quantitative concept further toward numerical simulations using data from natural populations to model the interplay of stochastic effects and density-dependent selection in evolution.
Methods
Cave Exploration, Sampling, Mapping, and Age Determination.
Since the discovery of the cave ecosystems in July 1971 (12)—in the course of the International Biological Program, Hawaii Subprogramme—the caves were systematically explored through the Hawaii Biological Survey (14). To protect the sensitive ecosystems and cultural sites, the cave entrances have not been officially mapped, and the respective data are treated as confidential. Two caves are open to the public as designated tourist caves and can be found on maps: Thurston Lava Tube and Kaumana Cave. O. polyphemus occurs in both. Overall, populations of O. polyphemus are known from more than 30 caves, of which 21 were included in this study (SI Appendix, Tables S1 and S5). The position of the cave entrances was determined with a global positioning system (GPS) device (Garmin GPS 12XL; 12 channels). The Digital Terrain Model (DTM) (SI Appendix, Fig. S4) was calculated from data provided by the US Geological Survey (USGS): http://hawaii.wr.usgs.gov/oahu/data.html. All data were calculated in the North American Datum of 1983 (NAD83), universal transverse Mercator grid zone 4 coordinate system. The compressed Shape file providing an equidistance of 100 feet was processed using LISA, to calculate DTM from contour lines (see www.lisa-geosoftware.de/prod_1e.htm for more information). The fundamental 2D and 3D terrain models were overlaid with the lava flows mapped by USGS. In all cases, it was possible to assign cave positions unambiguously to a dated lava flow (see Figs. 1 and 2 and SI Appendix, Fig. S3). The course of the ca. 60-km-long Kazumura master tube was mapped using data from (44), and the course of Carson’s Cave could be charted for about 3.7 km using measurements by A.W.
Morphology (Morphometry).
For the morphometric analyses, individuals from 22 populations were studied (SI Appendix, Table S1). Nine parameters (SI Appendix, Table S3) were measured using a measuring ocular with an Olympus SZH 10 at 50-fold magnification (accuracy ± 10 µm). In the analyses, these measurements were complemented by five indices computed from seven of the measured parameters (see also ref. 25).
Acoustic Recording and Analyses.
For the sound recordings, adults and fifth-instar nymphs from 15 caves (SI Appendix, Table S2) were taken to the laboratory and kept under controlled conditions closely resembling those of their natural habitat (complete darkness; constant temperature, about 18 °C). Adults were kept individually on roots of Metrosideros or fresh sprouts of soybeans, as a substitute; the nymphs were separated following final molting (for details, see refs. 45 and 18).
For recording of vibrational signals, a male and female (if available) from the same population were placed together onto the substrate. The natural substrate (living Metrosideros roots) was substituted by fresh Metrosideros leaves. Light exposition of the recording area was considerably reduced; however, enough light was retained to observe the animals and record their behavior. The vibrational signals were received with a magnetodynamic induction converter system [“MD-system” sensu (46)] and amplified ∼1,000 times in the process. The signals were recorded with a Sony TCD-D8 digital audio tape (DAT) recorder (on TDK DA-RXG DAT tapes; sampling rate, 48 kHz).
For time-pattern measurements, signals were digitized using Mac Lab/4s (ADInstruments) running on a Power Macintosh 7600/132 (Apple) with a sampling rate of 44 kHz. Measurements were taken using Chart Version 3.5.4/s with an accuracy of ± 0.15 ms. Ten time-pattern parameters of single calls (composed of more or less homogenous pulse trains; SI Appendix, Fig. S6) were taken (SI Appendix, Table S4). After omission of the first and last three pulses of the call, the parameters 18–24 were included in the analysis as parameters 25–31 to assess intraindividual variability (SI Appendix, Table S4 and ref. 25).
Statistical Analyses.
Statistical analyses were performed with SPSS Version 17.0 for Windows. A sufficient set of measured parameters for statistical analyses was available for 18 (morphometry) and 12 (acoustics) populations, respectively (SI Appendix, Tables S1 and S2). Discriminant analyses were conducted using all measured morphological and acoustic parameters for 18 populations (SI Appendix, Tables S8–S10). Confidence for correlation between character variability and population age was tested using Kendall’s τ (47) and correlation strength using Spearman’s ρ (48, 49). The statistical treatment of the DNA sequence data are described below.
Molecular Genetics and Sequence Analyses.
Specimens from 18 caves were used for the genetic analyses (SI Appendix, Table S1). DNA was purified from whole nymphs (fourth- and fifth-instar larvae) with a Qiagen DNeasy Tissue kits. A 658-bp COI fragment was amplified and sequenced (primers LCO1490 and HCO2198) (50). PCR was performed in 25-µL volumes containing double-distilled H2O, 1× Taq buffer, 1.5 mM MgCl2, 200 µM each dNTP, 1–2.5 U of Taq polymerase, and ca. 100 nM DNA, with an initial denaturation step of 3 min at 94 °C; cycling conditions of 35 cycles of 1 min each at 94 °C, 40–45 °C, and 72 °C; and a final elongation step of 5 min. PCR products were purified with QiaQuick PCR purification kits and cycle-sequenced with Big Dye Terminator Chemistry Version 1.1 (Applied Biosystems). Sequences were assembled and corrected using CodonCode Aligner Version 3.7.1. Sequences have been deposited in the ENA (see SI Appendix, Table S12 for accession numbers). Substitution model was estimated with jModeltest Version 0.1.1 (24 models; Akaike information criteria: GTR+G) (51). Haplotypes were identified using DAMBE (software package for extensive Data Analysis in Molecular Biology and Evolution) Version 5.1.1 (52), uncorrected genetic p-distances were calculated using MEGA (Molecular Evolutionary Genetics Analysis) Version 4.1 (53).
Phylogenetic Analyses.
Phylogenetic analyses were performed using maximum parsimony (MP) as implemented in PAUP* Version 4.0b010 for Windows [heuristic search with 10 random addition cycles, tree bisection and reconnection (TBR)] (54); maximum likelihood (ML) using TREEFINDER Version June 2008 (search depth: 2; bootstrap replicates: 1,000) (55); and Bayesian inference (BI) using MrBayes Version 3.1.2 (ngen: 1,000,000; samplefreq: 20; burnin: 35,001) (56). Trees were rooted using two epigean species of Oliarus from Hawai‘i Island. A minimum spanning COI network was generated using TCS Version 1.21 (57) with a connection limit (parsimony criterion) of 96%.
Molecular Clock Analysis.
Relaxed lognormal molecular clock analyses were performed using BEAST (Bayesian Evolutionary Analysis Sampling Trees) Version 1.6.2 (HKY+I; Yule/birth–death process; ngen: 10,000,000; log: 200; burnin: 35,001) (58); clock rates: 2.3%/Ma [mitochondrial insect rate (26)]; 4.6%/Ma [cave amphipods; COI and cytochrome c oxidase subunit II (COII) (27)]; 17.2%/Ma [Coleoptera; COI (28)], and in-group calibration [1.0 ± 0.0001 Ma (11), resulting in a mean rate of 3.19%/Ma]. Bayes factor analysis was conducted in Tracer Version 1.5 (1,000 bootstrap replicates) (59) to test support for a Yules vs. birth–death process. Bayes factor analyses resulted in low positive values and, thus, slightly favored a birth–death process (Bayes factors: 2.3%/Ma, 0.542; 4.6%/Ma, 0.343; 17.2%/Ma, 0.514; calibration, 0.444; however, node ages were almost identical).
Supplementary Material
Acknowledgments
We thank all of the individuals who have supported the study on O. polyphemus in Hawai‘i; the staff of Hawai‘i Volcanoes National Park, US Geological Survey, and Department of Land and Natural Resources for the permits under which these studies were conducted; and all private landowners who generously granted access to cave entrances on their properties, regardless of the risks involved. We also thank numerous colleagues from different disciplines, especially the late naturalists Hampton L. Carson, Ernst Mayr, and Günter Tembrock for inspiring discussions, and Kari Roesch Goodman for helpful comments on the manuscript. We thank two anonymous reviewers for their constructive comments, which helped us to improve the manuscript. This study was supported by the Graduate School “Evolutionary Transformations and Extinction Events” [Deutsche Forschungsgemeinschaft (DFG) Research Training Group 503 scholarship to A.W.], DFG Grants HO 1004/3-1 and HO 1004/7-1 (to H.H.), a grant from the Hawaii Bishop Research Institute (to. F.G.H.), and National Science Foundation Grants GB 23075, GB 75-23106, DEB 79-04760, and BSR 85-15183 (to F.G.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the European Nucleotide Archive (ENA), www.ebi.ac.uk/ena (accession nos. HF674815–HF674838).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1301657110/-/DCSupplemental.
References
- 1.Mayr E. In: Evolution as a Process. Huxley J, Hardy AC, Ford EB, editors. London: Allen & Unwin; 1954. pp. 157–180. [Google Scholar]
- 2.Carson HL. In: Population Biology and Evolution. Lewontin RC, editor. New York: Syracuse Univ Press; 1968. pp. 123–137. [Google Scholar]
- 3.Templeton AR. The theory of speciation via the founder principle. Genetics. 1980;94(4):1011–1038. doi: 10.1093/genetics/94.4.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barton NH, Charlesworth B. Genetic revolutions, founder effects, and speciation. Annu Rev Ecol Syst. 1984;15:133–164. [Google Scholar]
- 5.Carson HL, Templeton AR. Genetic revolutions in relation to speciation phenomena: The founding of new populations. Annu Rev Ecol Syst. 1984;15:97–131. [Google Scholar]
- 6.Coyne JA, Orr HA. Speciation. Sunderland, MA.: Sinauer; 2004. [Google Scholar]
- 7.Provine WB. In: Genetics, Speciation, and the Founder Principle. Val Giddings L, Kaneshiro KY, Andreson WW, editors. New York, Oxford: Oxford Univ Press; 1989. pp. 43–76. [Google Scholar]
- 8.Templeton AR. The reality and importance of founder speciation in evolution. Bioessays. 2008;30(5):470–479. doi: 10.1002/bies.20745. [DOI] [PubMed] [Google Scholar]
- 9.Carson HL. In: The Origin and Evolution of Pacific Island Biotas, New Guinea to Eastern Polynesia: Patterns and Processes. Keast A, Miller SE, editors. Amsterdam: SPB Academic Publishing; 1996. pp. 7–17. [Google Scholar]
- 10.Carson HL, Clague DA. In: Hawaiian Biogeography: Evolution on a Hot Spot Archipelago. Wagner WL, Funk VA, editors. Washington, DC: Smithsonian Institute Press; 1995. pp. 14–29. [Google Scholar]
- 11.Sherrod DR, Sinton JM, Watkins SE, Brunt KM, Survey USG. In: Geological Map of the State of Hawai’i: U.S. Geological Survey Open-File Report 2007-1089. 1st Ed. Myers MD, editor. Reston, VA: US Department of the Interior, US Geological Survey; 2007. [Google Scholar]
- 12.Howarth FG. Cavernicoles in lava tubes on the island of hawaii. Science. 1972;175(4019):325–326. doi: 10.1126/science.175.4019.325. [DOI] [PubMed] [Google Scholar]
- 13.Stone FD, Howarth FG, Hoch H, Asche M. In: Encyclopedia of Caves. Culver DC, White WB, editors. Amsterdam: Elsevier Academic Press; 2005. pp. 477–484. [Google Scholar]
- 14.Howarth FG, James SA, Preston DJ, Imada CT. Identification of roots in lava tube caves using molecular techniques: Implications for conservation of cave arthropod faunas. J Insect Conserv. 2007;11(3):251–261. [Google Scholar]
- 15.Howarth FG. The cavernicolous fauna of Hawaiian lava tubes, 1. Introduction. Pacific Insects. 1973;15(1):139–151. [Google Scholar]
- 16.Asche M. A review of the systematics of Hawaiian planthoppers (Hemiptera: Fulgoroidea) Pac Sci. 1997;51(4):366–376. [Google Scholar]
- 17.Hoch H, Howarth FG. Multiple cave invasion by the species of the cixiid planthopper Oliarus in Hawaii. Zool J Linn Soc. 1999;127(4):453–475. [Google Scholar]
- 18.Hoch H, Howarth FG. Evolutionary dynamics of behavioral divergence among populations of the Hawaiian cave-dwelling planthopper Oliarus polyphemus (Homoptera: Fulgoroidea: Cixiidae) Pac Sci. 1993;47(4):303–318. [Google Scholar]
- 19.Ossiannilsson F. Insect drummers. A study on the morphology and function of the sound-producing organ of Swedish Homoptera Auchenorrhyncha with notes on their sound-production. Opuscula Entomologica Supplements. 1949;10:1–145. [Google Scholar]
- 20.Strübing H. Lautäußerung – der entscheidende Faktor für das Zusammenfinden der Geschlechter bei Kleinzikaden (Homoptera – Auchenorrhyncha) Zoologische Beiträge NF. 1958;4(1):15–21. German. [Google Scholar]
- 21.Michelsen A, Fink F, Gogala M, Traue D. Plants as transmission channels for insect vibrational songs. Behav Ecol Sociobiol. 1982;11(4):269–281. [Google Scholar]
- 22.Claridge MF. Acoustic recognition signals: Barriers to hybridisation in Homoptera Auchenorrhyncha. Can J Zool. 1990;68(8):1741–1746. [Google Scholar]
- 23.Claridge MF, Reynolds WJ. Male courtship songs and sibling species in the Oncopsis flavicollis species group (Hemiptera: Cicadellidae) Journal of Entomology Series B Taxonomy and Systematics. 1973;42(1):29–39. [Google Scholar]
- 24.Strübing H. Die Bedeutung des Kommunikationssignals für die Diagnose von Euscelis-Arten (Homoptera Cicadina) Zool Jahrb, Abt Allg Zool Physiol Tiere. 1983;87(2–3):343–351. [Google Scholar]
- 25.Wessel A, Hoch H. Remane’s statistic species criterion applied to Hawaiian cave planthoppers (Hemiptera: Auchenorrhyncha: Fulgoromorpha: Cixiidae) Reichenbachia. 1999;33(1):27–35. [Google Scholar]
- 26.Bowers N, Stauffer JR, Kocher TD. Intra- and interspecific mitochondrial DNA sequence variation within two species of rock-dwelling cichlids (Teleostei: Cichlidae) from Lake Malawi, Africa. Mol Phylogenet Evol. 1994;3(1):75–82. doi: 10.1006/mpev.1994.1009. [DOI] [PubMed] [Google Scholar]
- 27.Villacorta C, Jaume D, Oromí P, Juan C. Under the volcano: Phylogeography and evolution of the cave-dwelling Palmorchestia hypogaea (Amphipoda, Crustacea) at La Palma (Canary Islands) BMC Biol. 2008;6:7. doi: 10.1186/1741-7007-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pons J, Ribera I, Bertranpetit J, Balke M. Nucleotide substitution rates for the full set of mitochondrial protein-coding genes in Coleoptera. Mol Phylogenet Evol. 2010;56(2):796–807. doi: 10.1016/j.ympev.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 29.DeSalle R, Templeton AR. Founder effects and the rate of mitochondrial DNA evolution in Hawaiian Drosophila. Evolution. 1988;42(5):1076–1084. doi: 10.1111/j.1558-5646.1988.tb02525.x. [DOI] [PubMed] [Google Scholar]
- 30.Mendelson TC, Shaw KL. Sexual behaviour: Rapid speciation in an arthropod. Nature. 2005;433(7024):375–376. doi: 10.1038/433375a. [DOI] [PubMed] [Google Scholar]
- 31.Lapoint RT, Gidaya A, O’Grady PM. Phylogenetic relationships in the spoon tarsus subgroup of Hawaiian Drosophila: Conflict and concordance between gene trees. Mol Phylogenet Evol. 2011;58(3):492–501. doi: 10.1016/j.ympev.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 32.Moore JG, Clague DA. Volcano growth and evolution of the island of Hawaii. Geol Soc Am Bull. 1992;104(11):1471–1484. [Google Scholar]
- 33.Genner MJ, et al. Age of cichlids: New dates for ancient lake fish radiations. Mol Biol Evol. 2007;24(5):1269–1282. doi: 10.1093/molbev/msm050. [DOI] [PubMed] [Google Scholar]
- 34.Carson HL. The genetics of speciation at the diploid level. Am Nat. 1975;109:83–92. [Google Scholar]
- 35.Kaneshiro KY. In: Genetics, Speciation, and the Founder Principle. Val Giddings L, Kaneshiro KY, Andreson WW, editors. New York, Oxford: Oxford Univ Press; 1989. pp. 279–296. [Google Scholar]
- 36.Carson HL. Mate choice theory and the mode of selection in sexual populations. Proc Natl Acad Sci USA. 2003;100(11):6584–6587. doi: 10.1073/pnas.0732174100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ritchie MG. Sexual selection and speciation. Annu Rev Ecol Evol Syst. 2007;38:79–102. [Google Scholar]
- 38.Simões P, et al. How repeatable is adaptive evolution? The role of geographical origin and founder effects in laboratory adaptation. Evolution. 2008;62(8):1817–1829. doi: 10.1111/j.1558-5646.2008.00423.x. [DOI] [PubMed] [Google Scholar]
- 39.Carson HL, Wisotzkey RG. Increase in genetic variance following a population bottleneck. Am Nat. 1989;134(4):668–673. [Google Scholar]
- 40.Mayr E. Animal Species and Evolution. Cambridge, Mass.: Belknap Press of Harvard University Press; 1963. [Google Scholar]
- 41.Gavrilets S. Fitness Landscapes and the Origin of Species. Princeton: Princeton Univ Press; 2004. [Google Scholar]
- 42.Wright S. Evolution in Mendelian populations. Genetics. 1931;16(2):97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright S. Surfaces of selective value revisited. Am Nat. 1988;131(1):115–123. [Google Scholar]
- 44.Allred K, Allred C. Development and morphology of Kazumura Cave, Hawaii. J Caves Karst Stud. 1997;59(2):67–80. [Google Scholar]
- 45.Howarth FG. An inexpensive constant temperature chamber for field and laboratory use. Environ Entomol. 1979;8(2):236–237. [Google Scholar]
- 46.Strübing H, Rollenhagen T. Ein neues Aufnehmersystem für Vibrationssignale und seine Anwendung auf Beispiele aus der Familie Delphacidae (Homoptera-Cicadina) Zool Jahrb, Abt Allg Zool Physiol Tiere. 1988;92(2):245–268. German. [Google Scholar]
- 47.Marascuilo LA, McSweeney M. Nonparametric and Distribution-Free Methods for the Social Sciences. Monterey, California: Brooks/Cole; 1977. [Google Scholar]
- 48.Marascuilo LA, Serlin RC. Statistical Methods for the Social and Behavioral Sciences. New York: Freeman; 1988. [Google Scholar]
- 49.Siegel S, Castellan NJ., Jr . Nonparametric Statistics for the Behavioral Sciences. 2nd Ed. New York: McGraw-Hill; 1988. [Google Scholar]
- 50.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3(5):294–299. [PubMed] [Google Scholar]
- 51.Posada D. jModelTest: Phylogenetic model averaging. Mol Biol Evol. 2008;25(7):1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 52.Xia X, Xie Z. DAMBE: Software package for data analysis in molecular biology and evolution. J Hered. 2001;92(4):371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- 53.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 54. Swofford DL (2002) PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer Associates, Sunderland, MA), Version 4.0.
- 55.Jobb G, von Haeseler A, Strimmer K. TREEFINDER: A powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 56.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 57.Clement M, Posada D, Crandall KA. TCS: A computer program to estimate gene genealogies. Mol Ecol. 2000;9(10):1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 58.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rambaut A, Drummond AJ (2007) Tracer.
- 60. Wright S (1932) Proceedings of the Sixth International Congress of Genetics, Vol. I, Ithaca, 1932, ed Jones DF (Brooklyn Botanic Garden, Menasha, WI), pp 356–366.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.