Abstract
Hepatitis C virus (HCV) is a major cause of chronic liver disease, with an estimated 170 million people infected worldwide. Low yields, poor stability, and inefficient binding to conventional EM grids have posed significant challenges to the purification and structural analysis of HCV. In this report, we generated an infectious HCV genome with an affinity tag fused to the E2 envelope glycoprotein. Using affinity grids, previously described to isolate proteins and macromolecular complexes for single-particle EM, we were able to purify enveloped particles directly from cell culture media. This approach allowed for rapid in situ purification of virions and increased particle density that were instrumental for cryo-EM and cryoelectron tomography (cryo-ET). Moreover, it enabled ultrastructural analysis of virions produced by primary human hepatocytes. HCV appears to be the most structurally irregular member of the Flaviviridae family. Particles are spherical, with spike-like projections, and heterogeneous in size ranging from 40 to 100 nm in diameter. Exosomes, although isolated from unfractionated culture media, were absent in highly infectious, purified virus preparations. Cryo-ET studies provided low-resolution 3D structural information of highly infectious virions. In addition to apolipoprotein (apo)E, HCV particles also incorporate apoB and apoA-I. In general, host apolipoproteins were more readily accessible to antibody labeling than HCV glycoproteins, suggesting either lower abundance or masking by host proteins.
Keywords: enveloped virus, hepacivirus, lipoviral particle, virus structure, virus assembly
Hepatitis C virus (HCV) is an important human pathogen that infects the liver and establishes chronic infection in the majority of cases, leading to cirrhosis and hepatocellular carcinoma (HCC) over the course of many years. More than 170 million people, ∼3% of the world’s population, have been infected with HCV. Each year, 4–5% of patients with HCV-induced cirrhosis develop HCC, making HCV infection the leading indicator for liver transplantation in many areas of the world (1). Surgery, however, does not provide a cure because the donor organ universally becomes reinfected. A prophylactic vaccine is not available and despite the recent addition of HCV-specific protease inhibitors to the pegylated (peg)-IFN and ribavirin regimen, which has increased the cure rate, better therapies are still needed to solve the emergence of resistant variants, severe side effects and suboptimal response rates in cirrhotic patients (2).
HCV is a single-stranded, positive-sense RNA virus in the family Flaviviridae. The HCV genome is ∼9.6 kb in length and encodes a long polyprotein of more than 3000 amino acids that is proteolytically processed to generate 10 mature viral proteins. Viral structural proteins are encoded by the first third of the polyprotein and include core or capsid protein (C) and the envelope glycoproteins E1 and E2. p7 (a viroporin) and nonstructural proteins, encoded by the C-terminal two-thirds of the polyprotein, play various roles in virus assembly and RNA replication (3). The putative HCV particle consists of a nucleocapsid containing the viral genome, enveloped by an endoplasmic reticulum (ER)-derived lipid bilayer where E1 and E2 are assembled as heterodimers. The structure of the infectious unit of HCV, however, is likely more complex because several lines of evidence suggest that HCV circulates in the bloodstream as a hybrid lipoviral particle (LVP) (4). The majority of viral RNA in human-infected plasma coelutes with very low-density lipoproteins (VLDLs), whose size ranges from 30 to 80 nm. Moreover, the association of HCV to host lipoproteins may explain the atypically low buoyant density (<1.10 g/mL) for the most highly infectious material. HCV particles display a broad density profile, ranging from 1.03 to 1.2 5g/mL. Higher density fractions are less infectious and can often be precipitated by anti-human IgG, suggesting the presence of immune complexes. In contrast, HCV RNA in low-density fractions can be captured by antibodies against the protein components of VLDLs and LDLs, and often exhibits higher specific infectivity for chimpanzee and cell culture infection (5). In this vein, HCV entry is strikingly linked to lipoproteins and their receptors (6). HDL enhances HCV infectivity, possibly by exploiting the lipid transfer function of scavenger receptor class B type I (SR-BI), a receptor for both HDL and HCV. Moreover, the LDL receptor was shown to play a role in the internalization of HCV. Given the link between lipoproteins and HCV infectivity and the potential implications for vaccine design or other therapeutic options, determining the structure and composition of the infectious HCV particle remains a priority.
Despite substantial progress in producing viral particles in cell culture and several biochemical and morphological studies, the structure of the HCV virion has remained elusive. To date, no crystal structure of the HCV glycoproteins or 3D reconstruction of the virion exists. Classical approaches that proved successful for other members of the Flaviviridae (e.g., dengue virus, West Nile virus) have thus far failed to yield sufficient quantities of well-preserved, structurally intact HCV particles (7, 8). Here, we developed alternative strategies for purifying enveloped HCV virions produced in cell culture and by primary human hepatocytes, obtaining low-resolution 3D details of their ultrastructure. These results have implications for understanding HCV assembly, its interactions with the host cell, and the possible basis for escape from neutralization.
Results
Capture of HCV via Antibodies Targeting Envelope Glycoproteins.
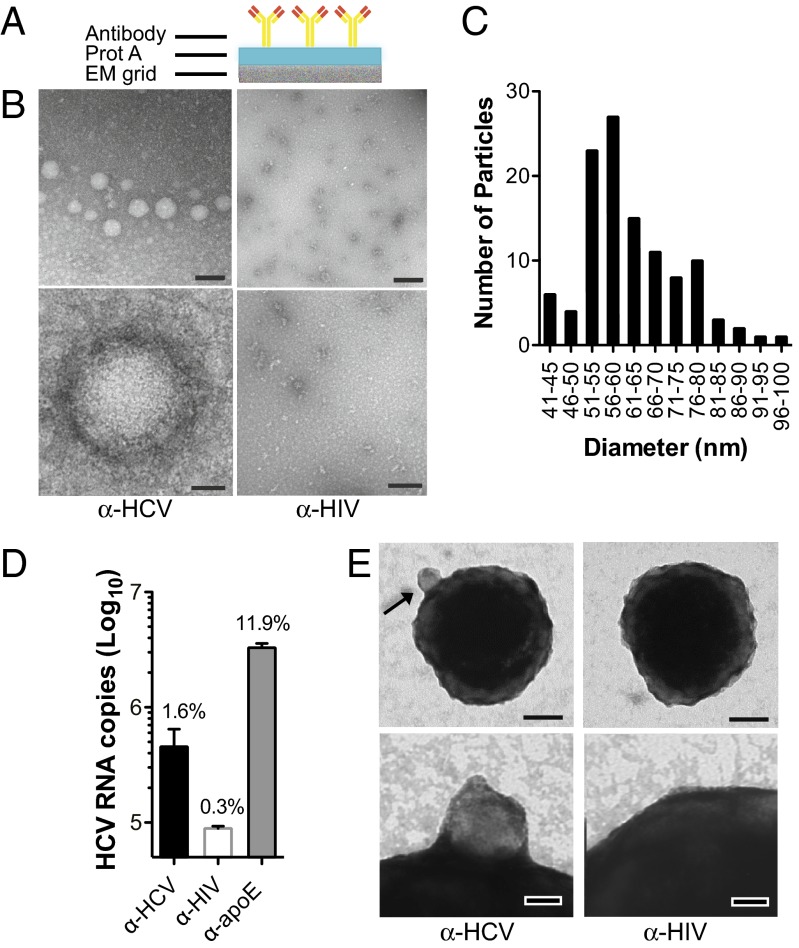
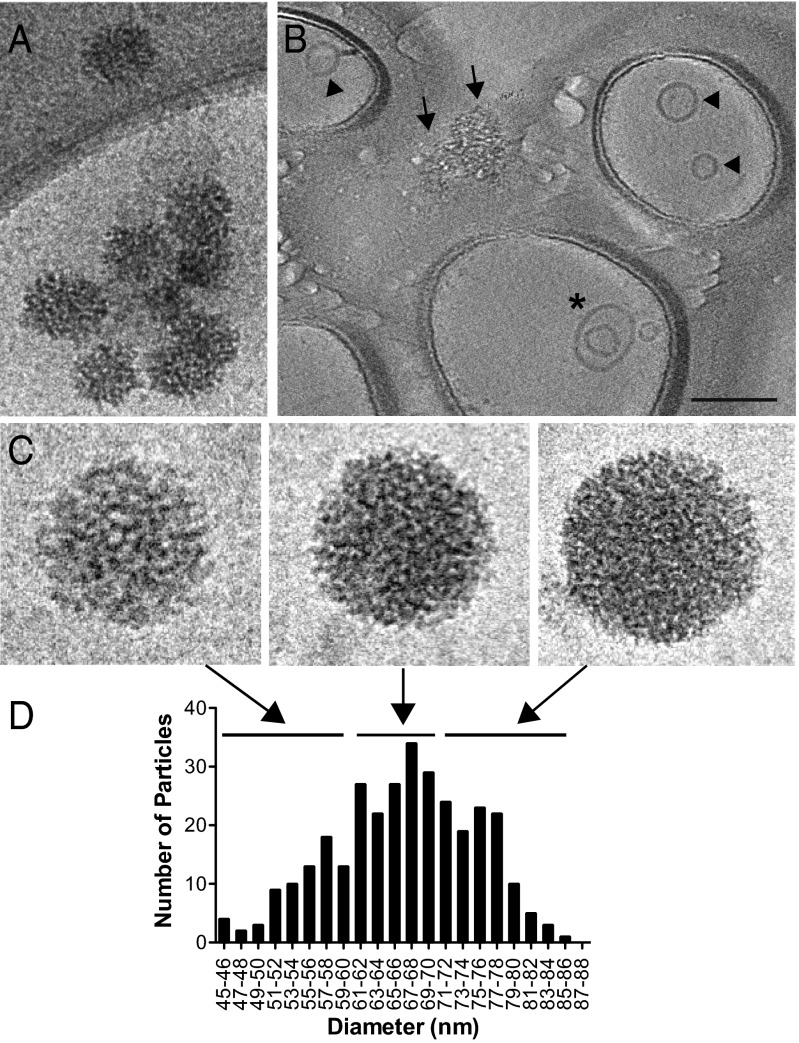
To capture and characterize extracellular virions, we used protein A–coated EM grids and α-HCV (AR4A) or α-HIV (B6) envelope antibodies as a negative control (Fig. 1A). Particles were affinity-purified on EM grids exclusively by AR4A, which recognizes an E1/E2 conformational epitope, but not by the B6 monoclonal antibody. This indicates that both HCV glycoproteins are displayed on the surface of released HCV particles. Particles were roughly spherical and well preserved, but with heterogeneous and not always symmetrical structures (Fig. 1B) and diameters ranging from 40 to 100 nm (Fig. 1C). Their distribution across the grid was not homogeneous and overall particle density was low. This observation was confirmed in a quantitative virus capture assay in which protein G–coated magnetic beads were conjugated to AR4A, B6, or an apolipoprotein (apo)E-reactive antibody and the amount of precipitated HCV RNA measured by RT-quantitative PCR. ApoE was chosen for HCV RNA capture because it is both required for the production of cell culture–derived HCV (HCVcc) and is a component of HCVcc particles (9, 10). Although AR4A only captured about 2% of the input viral genomes, 10-fold more viral RNA was precipitated using an apoE-targeting antibody (Fig. 1D). Similarly, a small fraction of the beads coupled to AR4A, but not B6, displayed bound particles by transmission EM (TEM), indicating that, albeit with low efficiency, intact virions were specifically captured using envelope-reactive antibodies (Fig. 1E).
Fig. 1.
Characterization of HCV virions captured via glycoproteins-specific antibody. (A) Schematic of protein (prot) A EM grids. (B) Representative images of negatively stained HCV virions captured using protein A EM grids coated with α-HCV or α-HIV antibodies. (Scale bar: Upper, 100 nm; Lower, 20 nm.) (C) Size histogram of HCV particles (n = 111; mean = 62 nm, SD = 11 nm). (D) HCV RNA capture assay using protein G beads coupled to α-HCV, α-HIV, or α-apoE antibody and incubated with equal amounts of virus-containing media (2 × 107 viral genome copies). RNA was extracted from each pull-down and HCV genome copy numbers were determined by RT-quantitative PCR. Means and SD from three independent experiments are shown. The percent of input HCV RNA captured by each antibody is indicated. (E) Images of negatively stained HCV virions adsorbed to protein G beads coupled to α-HCV or α-HIV antibody (Scale bars: Upper, 100 nm; Lower, 20 nm.) A bead-bound HCV particle is indicated (arrow).
Generation of E2-Tagged HCV Particles.
The number of virions captured using envelope-specific antibodies was suboptimal for further ultrastructural analyses. We hypothesized that the low yield might be due to poor accessibility of the viral glycoproteins. To increase particle density on the EM grid, we generated a J6/JFH-1 derivative with a 39-aa-long, tandem affinity tag fused to the N terminus of E2 (tag-HCV) immediately upstream of the hypervariable region 1. This region was shown to tolerate insertions, is a target of neutralizing antibodies and therefore exposed on infectious virus (6). The first two residues of E2 were duplicated to ensure proper cleavage at the E1/tagE2 junction followed by six histidine residues (6xHis) and two copies of a streptavidin tag II (StrepII) tag (Fig. S1A). Immunoblot analysis of lysates prepared from cells electroporated with tag-HCV demonstrated correct processing, with both tags detectable (Fig. S1B). Importantly, the insertion did not markedly impair infectious virus production (Fig. S1C). Incubation of virus-containing media with beads recognizing either tag captured both E2 and capsid protein, as expected for isolation of assembled particles (Fig. S1D).
Ultrastructural Characterization of tag-HCV with Affinity Grids.
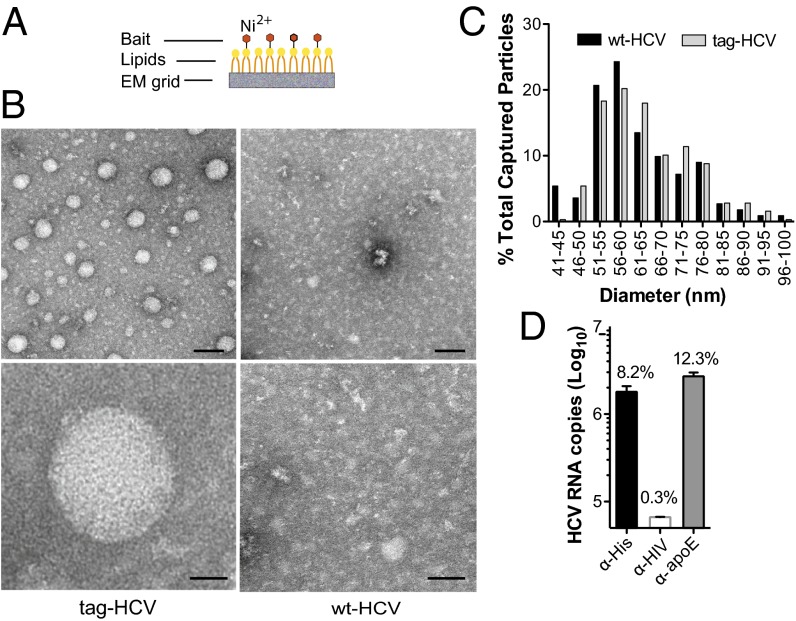
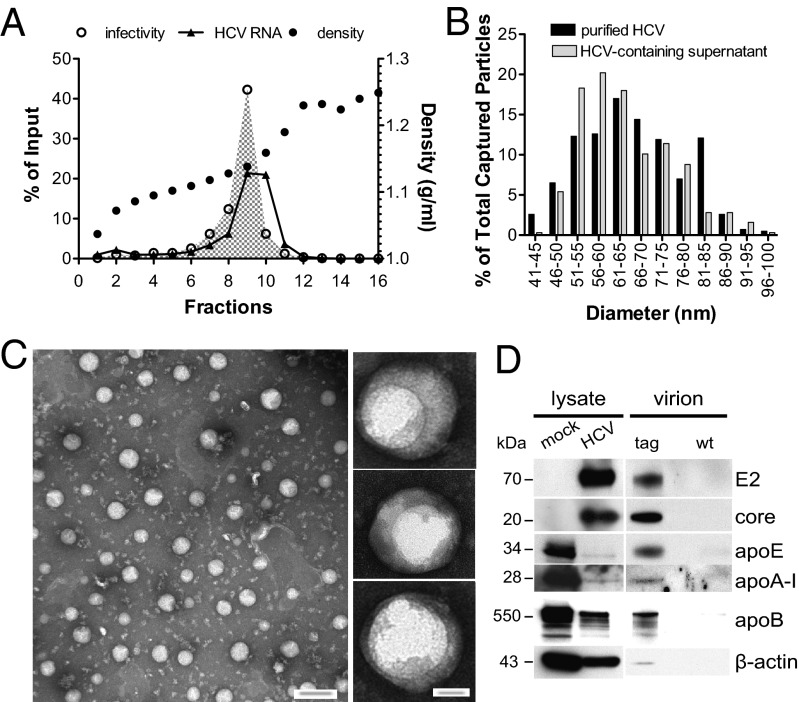
To purify tag-HCV, we developed custom-made EM affinity grids coated with a monolayer of lipids functionalized with a nickel-nitrilotriacetic acid group (Ni-NTA) to interact with the 6xHis-tag (Fig. 2A). High specificity of capture was achieved during the binding step by using low concentrations of imidazole (20 mM), as demonstrated by the lack of WT-HCV particles on the grids (Fig. 2B). The captured tag-HCV particles had a broad size range (n = 317; mean = 64 nm, SD = 11 nm) similar to that observed with glycoprotein-specific monoclonal antibodies and WT-HCV (Fig. 2C) but particle density on the grid improved significantly (compare Fig. 2B with Fig. 1B). Similarly, beads coupled to α-His antibody precipitated about eightfold more HCV RNA than AR4A (Fig. 2D vs. Fig. 1D) and bead-bound virions were easily visualized by TEM (Fig. S2).
Fig. 2.
Ultrastructural analysis of tag-HCV virions with affinity grids. (A) Schematic of the affinity grid, an EM grid coated with a Ni-NTA lipid monolayer. (B) Representative images of negatively stained HCV virions captured using 2% (vol/vol) Ni-NTA affinity grids. (Scale bars: Upper, 100 nm; Lower, 20 nm.) (C) Comparative analysis of HCV size distribution with different capture methods. The diameter of WT-HCV particles captured on protein A grids via α-E1/E2 antibody (black bars) and tag-HCV bound to affinity grids through the His-Ni interaction (gray bars) was measured and the number of virions in each size group is expressed as percent of total captured particles. (D) The copy number of tag-HCV genomes precipitated by His-, HIV-, or apoE-specific antibodies coupled to protein G beads was measured by RT-quantitative PCR. Means and SDs from three independent experiments are shown and the percent of input HCV RNA captured by each antibody is indicated.
HCV Virions Are Assembled as Lipoviral Particles.
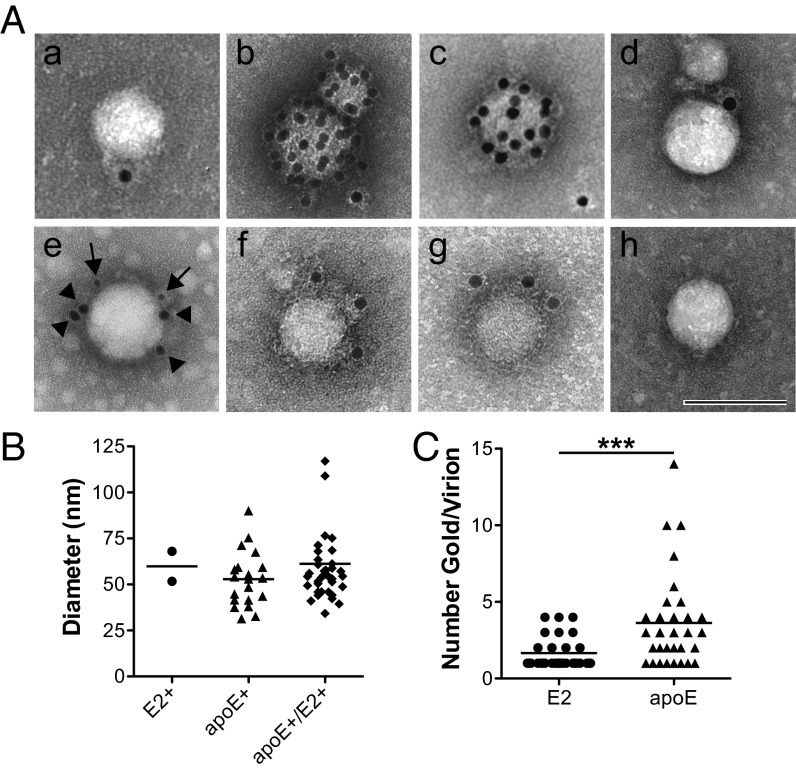
ApoE-specific antibodies were superior at capturing viral RNA than α-E1/E2 or His- and Strep-tag antibodies (Figs. 1D and 2D). Immunolabeling paralleled these results with particles staining more efficiently for apoE than E2 (Fig. 3A). Immunogold labeling revealed additional apolipoproteins associated with cell culture–produced HCVcc: apoA-I and apoB. Double-immunolabeling experiments for apoE and E2 showed that more than 95% of the stained particles were apoE+, whereas a low percentage (∼3%) stained only for E2 (Fig. 3A). Single- and double-stained virus particles had similar size ranges (Fig. 3B). However, significantly more apoE- than E2-reacting gold particles was observed per virion in the apoE+/E2+ population (Fig. 3C), suggesting that this viral envelope epitope may be less accessible and possibly masked. Taken together, our results are consistent with the LVP model for extracellular HCV.
Fig. 3.
Viral and host proteins exposed on the envelope of HCV virions. (A) Representative images of tag-HCV particles purified on 2% (vol/vol) Ni-NTA affinity grids and immunolabeled for the (a) viral glycoprotein E2, (b) apoE, (c) apoA-I, (d) apoB, and then (e) double-stained for E2 (arrows, 8 nm gold) and apoE (arrowheads, 18 nm gold), (f) StrepII tag, (g) 6xHis tag, (h) or incubated with secondary antibody only. (Scale bar: 100 nm.) (B) Size distribution of tag-HCV particles immunoreactive for E2 (n = 2, mean = 60 nm, SD = 11 nm), apoE (n = 20, mean = 53 nm, SD = 15 nm), or double-positive for E2 and apoE (n = 38, mean = 61 nm, SD = 22 nm). (C) Number of E2- and apoE-reactive gold particles per virion in apoE+/E2+ group (***P = 0.0003).
HCV Produced by Primary Human Hepatocytes.
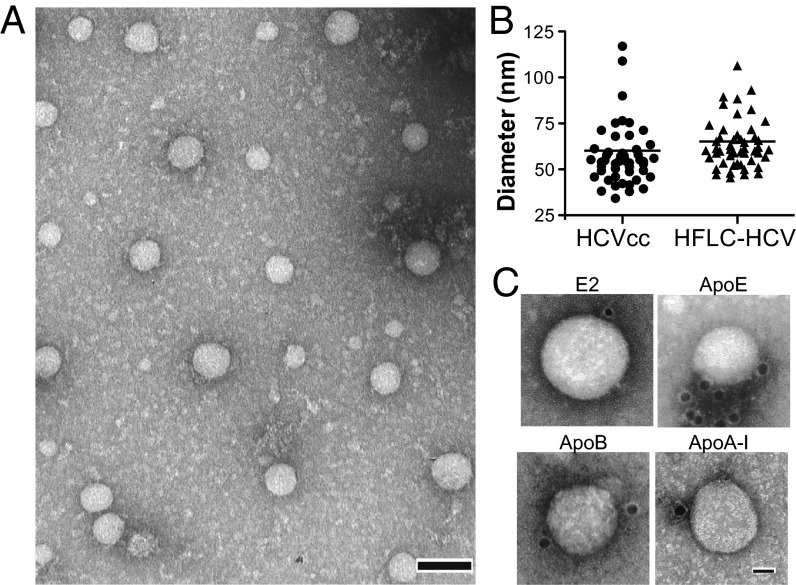
Given that hepatoma cells are unable to produce authentic VLDLs (4), we were interested in characterizing HCV particles grown in more physiologic cultures, human fetal liver cells (HFLCs), which are polarized and may better recapitulate in vivo lipoprotein and virus assembly. These cells are permissive for HCV but infection is short-lived with low virus yields and little evidence of spread. However, inhibition of innate immune responses enhances permissiveness, spread, and virus yield (11). To reduce possible background resulting from input cell culture–produced virus, HCV infection was initiated by RNA transfection in the presence of a tank binding kinase 1 (TBK1) inhibitor, BX795 (Fig. S3A). Both WT- and tag-HCV titers in HFLC approached those of HCVcc in Huh-7.5.1 cells by the third week posttransfection (Fig. S3B). Nuclear translocation of an HCV protease-targeted tagRFP-nls-IPS reporter was used to visualize the number of cells successfully infected over time (12). At peak virus titers (day 26), the majority of HFLC transfected with WT- and tag-RNA was HCV+ (Fig. S3C). Despite limited starting material, well-preserved, HFLC-produced virions (HFLC-HCV) could be captured using both protein A–coated EM grids with AR4A (Fig. S3D) or affinity grids (Fig. 4A), with the latter method being more efficient, as previously observed for HCVcc. When the same supernatant was applied to regular EM grids, no particles were observed, suggesting that the affinity purification in situ was crucial. As for HCVcc, no particles were detected with α-HIV antibody or when WT-HCV was applied to affinity grids. The size range of apoE+ particles captured from Huh-7.5.1 and HFLC culture media was similar (60.17 ± 21.03 nm and 65.13 ± 16.56 nm respectively; P = 0.193) (Fig. 4B). Particles immunoreactive for apoA-I and apoB were detected, albeit to a lesser extent than apoE (Fig. 4C). These results suggest that virions derived from HFLC, like HCVcc, are assembled as LVPs incorporating host-derived apolipoproteins.
Fig. 4.
Ultrastructure of HCV purified from primary human hepatocytes cultures. TEM images of virions from HFLC supernatant harvested at day 26 posttransfection and concentrated 10-fold. (A) Tag-HCV particles purified on 2% (vol/vol) Ni-NTA affinity grids. (Scale bar: 100 nm.) (B) Size distribution of apoE+ particles produced in Huh-7.5.1 (HCVcc) or HFLC (HFLC-HCV), captured on affinity grids. (C) TEM images of HFLC-tag-HCV showing immunolabeling with the indicated antibodies. (Scale bar: 20 nm.)
Cryo-EM of HCV.
To attempt 3D reconstruction of the HCV virion, structurally preserved enveloped HCVcc particles were purified directly from cell culture media using affinity grids with 20% (vol/vol) Ni-NTA and frozen in their native state (Fig. 5 A–C and Fig. S4). A total of 318 particle images were isolated and processed with RobEM software. Particle sizes ranged from 45 to 86 nm in diameter, with a mean diameter of 68 nm (Fig. 5C). A total of 150 particles were randomly selected to generate a random model using Auto3DEM. Random model generation yielded a model of 58 Å resolution as determined by 0.5 Fourier shell correlation (FSC). The resulting model appeared as a smooth sphere, as expected for a heterogeneous set of particles, where any identifying features would be averaged out. This model was used for iterative orientation search on the entire dataset using Auto3DEM. No result better than 58 Å was achieved. Because of the disparate particle size range, the dataset was sorted into three size classes (50–60, 61–70, and >71 nm) and the procedure was repeated on each size class with similar results. Efforts to use reconstructions of the alphavirus Ross River virus (22 Å) or the flavivirus dengue virus (30 Å) as initial models also yielded featureless spheres. Finally, the two smaller size classes were put through a common lines reconstruction method using the EMICOFV program. Orientation for particles was hand selected and the resulting reconstruction was yet another smooth sphere. Single-particle reconstruction methods failed because of extreme pleomorphism of the particles. Exosome-like structures 50–100 nm in diameter were occasionally visible, some of which were multivesicular with two or more distinguishable bilayers (Fig. 5B and Movie S1). Exosome-like particles were observed on affinity grids only when tag-HCV samples were applied, suggesting that they contain accessible HCV E2.
Fig. 5.
Cryo-EM analysis of HCVcc virions. (A) Cluster of HCV virions from Huh-7.5.1 cultures purified on 20% (vol/vol) Ni-NTA affinity grids. (B) Area of the grid showing HCV virions on the carbon film (arrows), exosomes (arrowheads), and multivesicular structures (*) inside the holes. (Scale bar: 100 nm.) A reconstructed tomogram of this field is available in Movie S1. (C) Low-dose images of HCV particles at 78,000× magnification, representative of three size classes (50–60, 61–70, and >71 nm). (D) Size distribution of HCV particles (n = 318; mean = 64 nm, SD = 11 nm).
Ultrastructural Characterization of Purified HCV.
With the goal of gaining insights into the ultrastructure of the infectious HCV particle, we generated highly purified HCVcc preparations. Infectious particles were concentrated by binding and elution from a heparin column followed by buoyant density fractionation on a 10–40% (wt/vol) iodixanol gradient. Particles with a density of ∼1.13 g/mL contained more than 40% total infectivity, 20% of the HCV RNA, and thus a higher specific infectivity compared with the input (range 1:10–1:50 vs. 1:1,000–1:5,000; Fig. 6A). TEM analysis of peak infectious fractions captured on affinity grids revealed well-preserved, spherical particles with a broad size range (Fig. 6 B and C). The average diameter and overall appearance of purified particles was similar to unpurified HCVcc (67 ± 12 nm vs. 64 ± 11 nm). However, virions 81–85 nm in size were enriched in purified samples (12.1% vs. 2.8% of total bound particles; Fig. 6B) and different structures were occasionally observed (Fig. 6C, Right). Given the 2D nature of TEM images, it was difficult to determine whether these structures represented discrete particles with internal features or separate particles that overlapped. Interestingly, despite being present in cell culture supernatant, exosomes were not found in purified samples of high specific infectivity. Immunoblot analyses of purified virions (tag-HCV) captured with His-tag Dynabeads revealed apoE, apoA-I, and apoB100, confirming the immunolabeling results (Fig. 6D).
Fig. 6.
Biophysical and ultrastructural characterization of purified HCV. (A) High-titer, heparin-purified tag-HCV virions were fractionated according to density on a 10–40% (wt/vol) iodixanol gradient. For each fraction, viral RNA, infectivity, and density were determined and expressed as percent of input. (B) Size distribution of highly infectious tag-HCV particles in fraction 9 (1.13 g/mL) of the buoyant density gradient (black bars; n = 430; mean = 67 nm, SD = 12 nm) is compared with unpurified HCV-containing supernatant (gray bars; n = 317; mean = 64 nm, SD = 11 nm). Data are expressed as percent of total captured particles. (C) Left: Low-magnification TEM image of tag-HCV virions from fraction 9 captured on 2% (vol/vol) Ni-NTA affinity grids. (Scale bar: 100 nm.) Right: Close up views of structures observed in purified preparations with high specific infectivity (Scale bar: 20 nm.) (D) Immunoblot for the indicated proteins in mock- or HCV RNA–electroporated Huh-7.5.1 cells, tag-, and WT-HCV extracellular virions that were heparin- and iodixanol-purified, precipitated with Dynabeads and eluted with 0.9 M imidazole.
Cryo-ET of Purified HCV.
In further attempts to obtain additional 3D structural information of purified HCV, we collected tomographic tilt series of vitrified, unfixed particles with high specific infectivity purified from iodixanol gradients. The quality of the tomograms was affected by the limited tilt range we were able to collect (∼−60° to +30°). The missing wedge made the tomograms streaky and limited the z-resolution of the reconstructions. Unfortunately, attempts at generating affinity grids using wider mesh grids resulted in insufficient binding of virus particles to enable cryoelectron tomography (cryo-ET) studies. Two reconstructed tomograms illustrating the general morphology of HCV particles are available in Movies S2 and S3. The majority of virions were round but not symmetrical with the outer rim heavily covered by globular proteins distributed unevenly around the viral envelope. Examples of particles with apparent patches of double-layer membrane, spikes, and internal structures are shown in Fig. 7 A–D. Striations in the viral envelope, indicated by transversal densities are visible in Fig. 7E and might represent transmembrane proteins (Movie S4).
Fig. 7.
Cryo-ET of purified HCVcc virions. (A–D) Two virtual slices through some of the reconstructed purified HCV particles with spikes (arrows), viral envelope (arrowheads), and internal structures (*) indicated. (E and F) Two sections through the 3D tomogram of an HCV particle showing striations in the envelope (arrows) and viral membrane (arrowheads).
Discussion
TEM has been instrumental to the discovery and characterization of many clinically relevant viruses, some from infected cell cultures, as in the case of adeno-, entero-, and reo-viruses, and others from unpurified clinical samples (i.e., hepatitis A and B) (13). HCV represents an unusual case in that it was first identified by molecular cloning and has proven very difficult to visualize from infected sera and tissues (14). Despite the development of systems to propagate infectious virus in cell culture (15), low titers and poor stability remained important challenges to the structural analysis of HCV. Attempts, by us and others, to scale up the number of purified particles applied to EM grids by concentration or precipitation were not successful because of aggregation and serum contaminants, whereas virus stocks produced in serum-free medium yielded fewer infectious particles.
To gain insights into the ultrastructure of fully assembled virions, we first used EM grids coated with AR4A, but only a very small percentage of HCV RNA was precipitated and very few particles were visualized by TEM. AR4A potently neutralizes the virus used in this study and displays one of the highest inhibitory effects among a panel of α-HCV antibodies screened (16, 17), arguing against insufficient binding properties (Fig. 1).
To increase the efficiency of particle capture, as an alternative approach we constructed a genome with a long, hydrophilic tandem affinity tag on E2 that was not impaired in infectious virus production (Fig. S1) and purified it directly on custom-made EM affinity grids interacting with the tags. The affinity grid, first described by Kelly et al. (18), enabled fast isolation, preservation, and enrichment of HCV virions that were instrumental to ultrastructural studies without the need for prior biochemical purification. Indeed, no particles were visualized if the same cell culture–derived media were applied to regular EM grids, indicating that the affinity purification in situ was a key step. Provided that tags can be inserted on accessible regions of the viral surface, the method described here can be applied to ultrastructural studies of other virions that are unstable, produced in low amounts, or not easily bound to conventional grids. The 6xHis tag allowed for better purification of HCV particles than the StrepII tag, possibly because of its higher accessibility (Fig. S1D); nevertheless, the presence of the StrepII tag was confirmed by immunolabeling with peptide-specific antibody (Fig. 3A, f).
We were interested in characterizing HCV particles grown in primary hepatocytes (HFLCs) that represent more physiologic cultures. We previously showed that paramyxovirus V protein expression promotes productive HCV infection in HFLCs by antagonizing IFN induction and signaling following infection with HCVcc (11). Here, we report a further improvement of virus production in HFLCs using HCV RNA transfection rather than HCVcc infection. Productive infection was facilitated by a chemical inhibitor of TBK-1, a downstream signaling molecule of several pathogen-associated pattern recognition receptors including retinoic acid-inducible gene-I (RIG-I), a primary sensor for HCV. Increasing HCV titers were observed over time, indicative of virus spread, and high numbers of infectious particles [up to 105 50% tissue culture infectious dose per milliliter (TCID50/mL) of unconcentrated supernatant] were obtained without the potential for contamination by hepatoma-derived HCVcc inocula. The combination of optimized titers and affinity grid enrichment enabled the characterization of primary human hepatocyte-derived HCV virions (HFLC-HCV; Fig. 4).
Both HCVcc and HFLC-HCV purified with affinity grids appeared well preserved (as suggested by the lack of positive staining), spherical, and very heterogeneous in size (40–100 nm in diameter), with spike projections but no distinguishable arrays. Thus, HCV does not seem to have structural similarities with other members of the Flaviviridae, such as dengue or West Nile virus, which display a relatively smooth surface in their mature stage, an icosahedral symmetry and a diameter of 50 nm (7, 8). Not only the size, but also the density of HCV particles was reported to be very broad both in vivo and in cell culture systems and to vary depending on the host cell in which the virus is assembled. This suggests that the host contributes to define the makeup of the virus in ways that might affect its structure and composition (5). Initial studies with human and chimpanzee plasma favored the hypothesis of association of HCV virions with the VLDL fractions (19, 20). Since then, a growing body of literature has emphasized how the maturation and release of HCV particles are tightly linked to the VLDL biosynthetic pathway, with the strongest evidence being that apoE is essential for virus assembly and found to be a component of HCVcc particles (9, 10).
In this study, we confirmed that apoE is incorporated in the majority of captured HCVcc and demonstrated that this is true for primary-derived HCV virions as well. In agreement with viral RNA immunocapture data (Fig. 1), double-immunolabeling experiments showed that significantly more copies of apoE than E2 were observed per particle (Fig. 3). These data indicate that a host-derived protein is better exposed on the outer rim of the viral envelope than the viral glycoproteins, which would be consistent with apoE playing a role in the initial attachment of HCV to the host cell. Interestingly, larger particles did not display stronger staining for apoE or E2. This may suggest that the sites accessible to antibody binding on the virion were saturated because of steric hindrance. Alternatively, a larger size may reflect higher lipid content or the presence of additional host proteins.
Other apolipoproteins were suggested to associate with serum- or cell culture-derived HCV (4). In this study, we show incorporation of apoA-I and apoB-100 into both HCVcc- and primary hepatocyte–derived virions. ApoA-I, the major structural protein of HDL, interacts with SR-BI, which is the endogenous receptor for HDLs in the liver. Interestingly, SR-BI is also an HCV receptor, and HDLs were shown to facilitate HCV entry via SR-BI and to attenuate virus neutralization, suggesting that HCV has developed an advantageous strategy to hijack the physiological interaction of HDL with SR-BI (6). Our finding that apoA-I is exposed on the surface of virions may provide a link to explaining the complex interplay between HCV, HDL, and SR-BI. ApoB-100 is the longer isoform of the apoB protein, specifically produced in the liver, required for VLDL biogenesis and found on VLDL, intermediate density lipoprotein (IDL), and LDL. Its role in the assembly and infectivity of HCV produced in cell culture systems is still being debated (4). Nevertheless, HCV RNA–containing particles purified from low-density fractions (i.e., VLDLs, LDLs) of chronically infected patients were found to be positive for apoB, supporting the incorporation of apoB in LVPs (4). Huh-7.5 cells were thought to be impaired in the formation of apoB-containing VLDLs and therefore unable to produce apoB-associated HCV virions (4). Notably, here we show that extracellular virions purified from Huh-7.5 cultures expose apoB on their envelope. ApoB is considered a nonexchangeable apolipoprotein; therefore, we can infer that viral particles acquire it in the liver cell and not from serum lipoproteins. In line with this hypothesis, HCV produced in primary human hepatocytes, unlike HCVcc, are secreted in serum-free media and yet contains apoB. Interestingly, most HCVcc- and HFLC-derived virions reacted with only one apoB immunogold particle, which might indicate a similar stoichiometry to VLDL, which assembles around a single molecule of apoB.
In comparing the ultrastructure of HCV from cell culture media with that of purified samples with narrower density and higher specific infectivity, two differences were noticed. First, exosome-like structures were observed in HCV-containing supernatant but not in preparations enriched in infectious particles. These vesicles were captured via tag-specific methods, indicating that they exposed at least the E2 envelope glycoprotein on their surface. They were characterized by a relatively smooth surface compared with the particles found in highly infectious samples that were heavily covered by electron-dense globular material, suggesting a different composition and likely separate biogenesis. Second, when we looked at highly infectious, purified HCV, we observed an enrichment in particles with a diameter of 81–85 nm displaying internal structures (Fig. 6). However, it was not possible to determine whether these particles accounted for the increased specific infectivity.
Interestingly, as one would expect for an enveloped virus, the cryo-ET images did not show evidence of a continuous bilayer. This could be due to the low resolution of the images or may indicate that the particle is embedded in a nonunique amount of lipoprotein, therefore delimited it by a lipid monolayer. In this context, the HCV envelope glycoproteins may position their transmembrane domains parallel to the lipid monolayer, as suggested by Bartenschlager et al. (4), unless the capsid is in close proximity of the lipoprotein-delimiting monolayer. In that case, a partial bilayer might form and E1/E2 would be able to acquire a transmembrane topology, thus anchoring the lipoprotein to the capsid. This hypothesis would be consistent with some of our images (Figs. 6C and 7).
In summary, our results reveal the hybrid nature of HCV, assembled as a mixed particle with a thick shell of host-derived apolipoproteins coating the viral envelope that presumably aid both release and entry of the virus as well as escape from circulating neutralizing antibodies, ultimately allowing this veiled pathogen to fly under the radar.
Materials and Methods
Virus Purification.
Virus-containing media was harvested every 4 h for 4 d after switching electroporated cells to low-serum media [1.5% (vol/vol) FBS]. High-titer HCVcc stocks were obtained by concentration of the infectious supernatant in a stirred ultrafiltration cell (Model 8400 with 100-kDa MWCO membranes; Millipore). Concentrated samples were purified over heparin column (GE Hitrap Heparin) according to the manufacturer’s instructions. Heparin-eluted virus was fractionated over a 10–40% (wt/vol) iodixanol buoyant density gradient (Optiprep; Sigma) to isolate fractions with the highest infectivity (range 1.12–1.16 g/mL).
Cryo-EM and Cryo-ET: Sample Preparation and Data Collection.
Holey carbon grids (400 mesh, Ted Pella) were coated with 20% (vol/vol) Ni-NTA lipid mesh to generate affinity grids suitable for cryo-EM. Ten-nanometer gold particles (Aurion Gold Sol, EMS) were added to the virus suspension to serve as fiduciary markers for tomography. Grids were floated carbon side down on a 50-μL drop of virus solution containing 20 mM imidazole for 30 min, blotted for 2.0 s in a Gatan Cryoplunge Cp3 with 70–80% chamber humidity, and plunged into liquid ethane. Cryo-EM images were collected using a Titan Krios electron microscope (FEI) at 300 keV under low-dose conditions, using an Ultrascan 950 4k CCD (Gatan). For cryo-ET, imaging was done on a JEOL 3200FSC electron microscope (JEOL USA) operating at 300 KeV under control of SerialEM software using low-dose conditions. Images were collected on a Gatan Ultrascan 4k camera at 50,000× nominal magnification and 2× binning, with a final pixel size of 4.40A and dose per frame of 2.4e−/A2. An energy filter was inserted for all recorded images with slit width set to 20 eV. Tilt series were collected in 2° increments at the maximum range allowed by the grid: from −62° to +30° in the best case and −64° to +20° in the worst. The tilt range was limited by the mesh size of the grids. Tilt series were aligned and reconstructed using Protomo software. Back-projected reconstructions were viewed using Imod.
Detailed methods and the associated references can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
The authors thank Dr. Thomas Walz and Daniel Zachs (Harvard Medical School), and Dr. Zheng Liu and Guimei Yu (Purdue University) for assistance with the preparation of affinity grids, Dr. Mansun Law (The Scripps Research Institute) for providing the AR4A and B6 antibodies, and Dr. Cynthia de la Fuente for editing the manuscript. This work was supported by National Institutes of Health (NIH) Grants AI072613 and AI075099 (to C.M.R.), the Greenberg Medical Research Institute, The Starr Foundation, and The Rockefeller University Women and Science Fellowship (to M.T.C.). The cryo-ET facility at New York Structural Biology Center is supported by Research Facilities Improvement Program Grant C06 RR017528-01-CEM from the National Center for Research Resources and NIH and the JEOL 3200FSC electron microscope was purchased with funds from NIH Grant S10 RR17291.
Footnotes
Conflict of interest statement: This paper discusses hepatitis C virus research and development tools that were developed in academia and licensed to Apath, LLC, a company in which C.M.R. has equity interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307527110/-/DCSupplemental.
References
- 1.Lavanchy D. Evolving epidemiology of hepatitis C virus. Clin Microbiol Infect. 2011;17(2):107–115. doi: 10.1111/j.1469-0691.2010.03432.x. [DOI] [PubMed] [Google Scholar]
- 2.Delang L, et al. Hepatitis C virus-specific directly acting antiviral drugs. Curr Top Microbiol Immunol. 2013;369:289–320. doi: 10.1007/978-3-642-27340-7_12. [DOI] [PubMed] [Google Scholar]
- 3.Moradpour D, Penin F. Hepatitis C virus proteins: From structure to function. Curr Top Microbiol Immunol. 2013;369:113–142. doi: 10.1007/978-3-642-27340-7_5. [DOI] [PubMed] [Google Scholar]
- 4.Bartenschlager R, Penin F, Lohmann V, André P. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 2011;19(2):95–103. doi: 10.1016/j.tim.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Lindenbach BD. Virion assembly and release. Curr Top Microbiol Immunol. 2013;369:199–218. doi: 10.1007/978-3-642-27340-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeisel MB, Felmlee DJ, Baumert TF. Hepatitis C virus entry. Curr Top Microbiol Immunol. 2013;369:87–112. doi: 10.1007/978-3-642-27340-7_4. [DOI] [PubMed] [Google Scholar]
- 7.Kuhn RJ, et al. Structure of dengue virus: Implications for flavivirus organization, maturation, and fusion. Cell. 2002;108(5):717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukhopadhyay S, Kim BS, Chipman PR, Rossmann MG, Kuhn RJ. Structure of West Nile virus. Science. 2003;302(5643):248. doi: 10.1126/science.1089316. [DOI] [PubMed] [Google Scholar]
- 9.Gastaminza P, et al. Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture. J Virol. 2010;84(21):10999–11009. doi: 10.1128/JVI.00526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merz A, et al. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J Biol Chem. 2011;286(4):3018–3032. doi: 10.1074/jbc.M110.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andrus L, et al. Expression of paramyxovirus V proteins promotes replication and spread of hepatitis C virus in cultures of primary human fetal liver cells. Hepatology. 2011;54(6):1901–1912. doi: 10.1002/hep.24557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones CT, et al. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat Biotechnol. 2010;28(2):167–171. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roingeard P. Viral detection by electron microscopy: Past, present and future. Biol Cell. 2008;100(8):491–501. doi: 10.1042/BC20070173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choo QL, et al. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244(4902):359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 15.Steinmann E, Pietschmann T. Cell culture systems for hepatitis C virus. Curr Top Microbiol Immunol. 2013;369:17–48. doi: 10.1007/978-3-642-27340-7_2. [DOI] [PubMed] [Google Scholar]
- 16.Giang E, et al. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc Natl Acad Sci USA. 2012;109(16):6205–6210. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Catanese MT, et al. Different requirements for SR-BI in hepatitis C virus cell-free versus cell-to-cell transmission. J Virol. 2013 doi: 10.1128/JVI.01102-13. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly DF, Abeyrathne PD, Dukovski D, Walz T. The Affinity Grid: A pre-fabricated EM grid for monolayer purification. J Mol Biol. 2008;382(2):423–433. doi: 10.1016/j.jmb.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomssen R, et al. Association of hepatitis C virus in human sera with beta-lipoprotein. Med Microbiol Immunol (Berl) 1992;181(5):293–300. doi: 10.1007/BF00198849. [DOI] [PubMed] [Google Scholar]
- 20.Prince AM, Huima-Byron T, Parker TS, Levine DM. Visualization of hepatitis C virions and putative defective interfering particles isolated from low-density lipoproteins. J Viral Hepat. 1996;3(1):11–17. doi: 10.1111/j.1365-2893.1996.tb00075.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.