Abstract
Because parasite virulence factors target host immune responses, identification and functional characterization of these factors can provide insight into poorly understood host immune mechanisms. The fruit fly Drosophila melanogaster is a model system for understanding humoral innate immunity, but Drosophila cellular innate immune responses remain incompletely characterized. Fruit flies are regularly infected by parasitoid wasps in nature and, following infection, flies mount a cellular immune response culminating in the cellular encapsulation of the wasp egg. The mechanistic basis of this response is largely unknown, but wasps use a mixture of virulence proteins derived from the venom gland to suppress cellular encapsulation. To gain insight into the mechanisms underlying wasp virulence and fly cellular immunity, we used a joint transcriptomic/proteomic approach to identify venom genes from Ganaspis sp.1 (G1), a previously uncharacterized Drosophila parasitoid species, and found that G1 venom contains a highly abundant sarco/endoplasmic reticulum calcium ATPase (SERCA) pump. Accordingly, we found that fly immune cells termed plasmatocytes normally undergo a cytoplasmic calcium burst following infection, and that this calcium burst is required for activation of the cellular immune response. We further found that the plasmatocyte calcium burst is suppressed by G1 venom in a SERCA-dependent manner, leading to the failure of plasmatocytes to become activated and migrate toward G1 eggs. Finally, by genetically manipulating plasmatocyte calcium levels, we were able to alter fly immune success against G1 and other parasitoid species. Our characterization of parasitoid wasp venom proteins led us to identify plasmatocyte cytoplasmic calcium bursts as an important aspect of fly cellular immunity.
The outcome of a parasitic infection is largely determined by the interaction between parasite virulence factors and host immune defenses (1). Therefore, identification of virulence factors can provide insight into host immune mechanisms and, in the case of medically relevant parasites, suggest potential treatments. The fruit fly Drosophila melanogaster is used as a model of humoral innate immune responses in both mammals and insect vectors of human disease (2, 3). Studies of humoral immunity in D. melanogaster have largely focused on the roles of the closely related Toll and Immune deficiency (Imd) signaling pathways in antimicrobial immunity (2, 4–6). Both Toll and Imd signaling culminate in the activation of NF-κB homologs that are required for the induction of humoral immunity (7, 8). Subsequently, a role in mammalian innate immunity has been found for the family of homologous Toll-like receptors (9, 10), and their downstream mechanisms are strikingly conserved (11–13). The study of antimicrobial immunity in flies has therefore allowed for a detailed understanding of these important mammalian immune pathways.
Although these fly responses to microbial pathogens are well characterized, the response to macroparasites is only partially understood. Among the most common macroparasites of Drosophila are parasitoid wasps, which can infect up to 80% of flies in natural populations (14). Larval parasitoids attack fly larvae, simultaneously injecting both an egg and a complex mixture of venom proteins directly into the larval hemocoel. In response to wasp infection, flies mount a cellular immune response against wasp eggs termed melanotic encapsulation (15). Following recognition of the wasp egg, circulating plasmatocytes are activated via an unknown molecular mechanism. Activated plasmatocytes migrate toward and bind to the wasp egg, leading to the formation of a continuous plasmatocyte layer (16, 17). Recognition of the wasp egg also induces the production of specialized immune cells termed lamellocytes that are present at high numbers in the hemolymph within 24 h following infection. Lamellocytes bind to the primary plasmatocyte layer surrounding the wasp egg, forming a dissociation-resistant outer layer (17, 18). The wasp egg is then melanized inside the hemocyte capsule, leading to the death of the developing wasp. This encapsulation response is conserved across arthropods (19). The molecular mechanisms underlying encapsulation are largely unknown, but recent work has revealed a high degree of genetic conservation between fly and mammalian hematopoeisis and other aspects of cellular immunity (20–22).
Parasitoid wasps use virulence factors in their venom to short circuit the fly encapsulation response to protect their developing offspring (17, 23, 24). In nature, Drosophila are targeted by parasitoids from at least four Hymenopteran families with numerous virulence strategies (14, 25, 26), suggesting that wasp venom proteins have evolved to target specific aspects of the encapsulation response. For instance, whereas venom from the parasitoid Leptopilina heterotoma causes lamellocyte cell death (23), the venom of its sister species, Leptopilina victoriae, specifically inhibits protein N-glycosylation of lamellocyte surface proteins (17). The study of naturally coevolving pathogens has allowed for a better understanding of Drosophila humoral immunity (27), and we hypothesize that the examination of coevolved virulence strategies from multiple parasitoid species will similarly provide insight into the mechanisms of fly cellular immunity. Here, we identify the virulence strategy of a previously uncharacterized Figitid larval parasitoid of Drosophila, Ganaspis sp.1 (G1), as a window into Drosophila cellular immunity.
Results
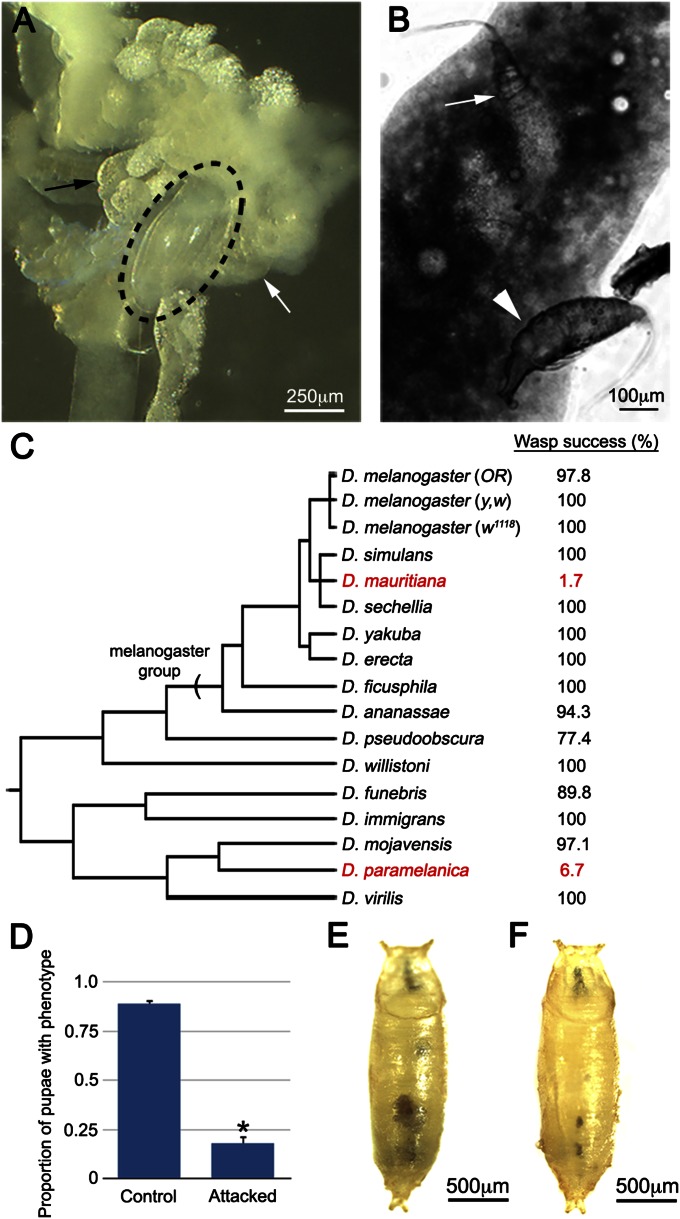
In laboratory trials, we found that G1 readily attacks D. melanogaster larvae (Table S1), laying eggs that attach to internal fly tissues within 12 h post attack (PA) (Fig. 1A). G1 can efficiently escape encapsulation in D. melanogaster (Table S1), and following host pupation, the wasp eggs hatch into larvae that begin to consume host tissues (Fig. 1B), resulting in successful parasitization, with adult wasps emerging from nearly 100% of attacked D. melanogaster hosts (Fig. 1C). Drosophila parasitoid wasp species have widely differing host ranges (26), varying from specialists that target a narrow range of phylogenetically related species to generalists that can successfully infect a wide range of Drosophila species. To determine the host range of G1, we repeated our attack trials on a diverse subset of Drosophila species. We found that G1 is a generalist, successfully parasitizing 13 of the 15 species tested (Fig. 1C), including 7 of 8 species from the melanogaster group and representative species of an additional six species groups. The melanica group member Drosophila paramelanica is G1 resistant and mounts an encapsulation-independent anti-wasp immune response (28), suggesting that G1 uses an antiencapsulation virulence strategy.
Fig. 1.
(A) G1 egg (circled) attached to fly gut (white arrow) and fat body (black arrow) 24 h PA. (B) Hatched G1 larvae dissected from fly pupae, feeding on host fat body (white arrow) or hemolymph (white arrowhead). (C) Wasp eclosion success (by percent) across the genus Drosophila. G1-resistant species shown in red. Branch lengths are approximated. (D) Penetrance of tu(1)Sz phenotype in control and attacked larvae, *P < 0.01 relative to control, error bars indicate SE, n = 6 independent replicates for each treatment. Control (E) and attacked tu(1)Sz pupae (F). Scale bars as indicated.
Parasitoid wasps may be characterized as immune evasive, in which case the egg is not detected by the immune response, or immune suppressive, in which case the immune response is inhibited. To differentiate between these strategies, we tested the ability of G1 to suppress the self-encapsulation phenotype of tu(1)Sz mutant larvae. tu(1)Sz mutants encapsulate their own posterior fat body tissue in a manner analogous to wasp egg encapsulation (29). We predict that an immune evasive wasp would have no effect on the progression of self-encapsulation in tu(1)Sz mutants, whereas an immune suppressive wasp would be expected to inhibit the self-encapsulation phenotype. We found that the tu(1)Sz phenotype is significantly rescued by G1 attack (Fig. 1 D–F), demonstrating that G1 venom has immune suppressive properties.
Because immune suppression has been linked to the destruction of hemocytes, and in particular lamellocytes (25, 26), we assayed the total hemocyte count (THC) and the number of lamellocytes in G1 attacked larvae at 48 h PA. There is a significant increase in both THC (Fig. S1A) and lamellocyte number (Fig. S1B) in G1 attacked larvae compared with unattacked controls, and there was no observable hemocyte death in G1-attacked larvae at 24, 48, or 72 h PA. Furthermore, G1-attacked larvae show increased THC and lamellocyte numbers relative to larvae attacked by the avirulent wasp Leptopilina clavipes (which is encapsulated by D. melanogaster), demonstrating that G1-attacked larvae have a sufficient number of hemocytes for encapsulation of the wasp egg (Fig. S1). These data suggest that G1 venom suppresses the fly immune response by disabling, rather than destroying, host hemocytes.
To understand how G1 disables fly hemocytes, we used our recently described sequencing/bioinformatic approach to identify G1 venom genes (30). We first performed RNA-Seq on mRNAs isolated from dissected female wasp abdomens, and the sequence data were assembled into 234,516 transcripts (Table S2). The transcriptome was filtered by RSEM to remove low quality and low abundance sequences (30), resulting in a final assembly of 27,354 transcripts. We then used mass spectrometry to identify the proteins in purified venom, and the resulting 2,891 peptides were mapped against the transcriptome sequences. The peptides mapped to the predicted ORFs of 166 different transcripts (Datasets S1 and S2) for an average of 17.4 peptides per ORF, with an average protein coverage of 23.5% (Table S3). These venom genes accounted for just 0.61% of the expression-filtered assembly (Table S2), meaning that the identified venoms represent a specific subset of abdomen transcripts. We also found that the venom proteins are more likely to contain secretion signals than nonvenoms (28% of venom genes vs. 6% of nonvenoms; P < 0.01), consistent with the hypothesis that many venom genes encode small, classically secreted proteins (Table S4). Similar to Leptopilina boulardi and Leptopilina heterotoma venoms, Gene Ontology (GO) term enrichment suggests that G1 venom may regulate host physiology (via carbohydrate and nucleotide metabolism and antioxidant activity; Table S4) (30), but provides few clues to potential virulence mechanisms.
To further confirm the specificity of our approach, we performed suppression subtractive hybridization (SSH) (31) on cDNA samples made from G1 venom glands and carcasses (whole wasps with venom glands removed) to select for the most abundant transcripts that are specific to, or overrepresented in, the venom gland sample. Of the 71 SSH clones sequenced, 56 aligned to G1 transcripts and 14 of these clones matched our identified venom genes (Table 1), showing broad overlap across venom identification methods. Furthermore, we found that the most highly represented venom transcript by SSH was also one of the most abundant venom proteins identified by mass spectrometry (Table S5) and showed strong homology to the sarco/endoplasmic reticulum calcium ATPase (SERCA). SERCA plays a conserved role in calcium homeostasis, inhibiting intracellular calcium levels by pumping calcium ions from the cytoplasm into SR/ER stores and is therefore a negative regulator of calcium-mediated signaling pathways. Based on this interesting putative function, and abundance of SERCA in both the proteomic and SSH sequencing projects, we decided to focus on characterizing its role in G1 venom.
Table 1.
Results of SSH sequencing
| Transcript ID | Annotation | No. of SSH hits | Venom peptide hits |
| comp1045_seq1 | SERCA | 5 | 79 |
| comp845_seq1 | Troponin C | 2 | 7 |
| comp755_seq1 | Arginine kinase | 1 | 129 |
| comp181_seq1 | — | 1 | 69 |
| comp844_seq5 | Neprilysin-2 | 1 | 11 |
| comp630_seq1 | Myosin LC alkali | 1 | 11 |
| comp171_seq2 | Cysteine protease | 1 | 6 |
| comp317_seq1 | — | 1 | 5 |
| comp636_seq2 | Erythrose-4-P dehydrogenase | 1 | 4 |
| comp1_seq1 | 28S rRNA | 30 | — |
| comp9199_seq1 | — | 4 | — |
| comp0_seq1 | 18S rRNA | 2 | — |
| comp696_seq1 | Torso | 2 | — |
| comp3562_seq1 | Titan | 1 | — |
| comp25221_seq1 | — | 1 | — |
| comp111_seq1 | — | 1 | — |
| comp1032_seq1 | CG31997 | 1 | — |
Identified transcripts are listed according to number of SSH clones, annotation, and peptide hits from venom proteomics.
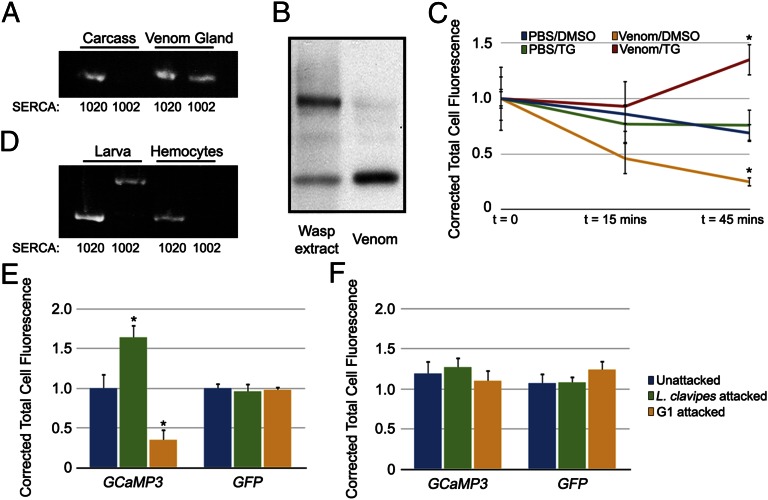
Mammalian genomes have multiple SERCA genes, each encoding at least two protein isoforms. These isoforms differ only in the extreme C terminus with the longer isoform having an additional transmembrane domain and a higher affinity for calcium ions (32). These protein isoforms are conserved in insects; both the D. melanogaster homolog Ca-P60A and the G1 homolog identified in our transcriptome data (comp1045) encode two protein isoforms (referred to as SERCA1020 and SERCA1002 according to size). Interestingly, only SERCA1002 was specifically identified by both venom mass spectrometry and SSH sequencing (Fig. S2), despite the presence of both transcripts in wasp abdomens. To test the idea that SERCA1002 represents a venom-specific isoform, we performed transcript-specific PCR on cDNA samples from venom glands and carcasses. We found SERCA1020 transcript in both carcass and venom gland samples, whereas SERCA1002 was specifically found in venom glands (Fig. 2A). The identification of a single SERCA isoform by venom mass spectrometry despite the coexpression of both isoforms in the venom gland is consistent with SERCA1002 being a venom-specific isoform and supports the specificity of our results. To further confirm that SERCA is found in G1 venom, Western blots were performed on purified venom and total protein extract with anti-SERCA antiserum and we observed distinct bands in the two samples: smaller bands of ∼150 kDa and a larger band of ∼200 kDa that is excluded from the purified venom sample (Fig. 2B). These findings demonstrate that the SERCA found in our sequencing projects represents a true venom protein rather than a contaminant released from unintentionally lysed cells during venom purification.
Fig. 2.
(A) Isoform-specific PCR of wasp carcass and venom gland cDNAs. (B) Western blot of wasp protein extracts and purified venom with anti-SERCA. (C) Corrected total cell fluorescence plotted over time during ex vivo incubation of GCaMP3-expressing hemocytes with the indicated treatment. *P < 0.05 compared with PBS/DMSO control, error bars indicate SE, n = 3 independent replicates per treatment. (D) Isoform-specific PCR on cDNAs from D. melanogaster third instar larvae and hemocytes. (E and F) Corrected total cell fluorescence of GCaMP3 and GFP expressing hemocytes, 6 h PA (E) and 24 h PA (F) with the indicated wasps. *P < 0.01 compared with respective unattacked controls, error bars indicate SE, n = 3 independent replicates for each time point per treatment.
To test whether venom SERCA is active, we designed an ex vivo assay to measure the ability of G1 venom to regulate intracellular calcium levels in fly cells. Purified G1 venom (or PBS control) was pretreated with either thapsigargin (TG), an irreversible and specific inhibitor of SERCA (33), or vehicle control (DMSO) and then dialyzed to remove excess TG. These samples were then incubated with plasmatocytes expressing a genetically encoded fluorescent calcium sensor (GCaMP3; ref. 34) bled from third instar larvae. The intensity of GCaMP3 fluorescence is proportional to intracellular calcium levels (34), and we assayed the ability of each sample to alter calcium levels by measuring GCaMP3 fluorescence during the incubation period. We found that incubation with the PBS samples had no effect on GCaMP3 fluorescence (Fig. 2C, blue and green lines). However, incubation with G1 venom/DMSO resulted in a significant decrease in GCaMP3 fluorescence (Fig. 2C, yellow line), showing that G1 venom is able to manipulate host intracellular calcium levels. This effect of G1 venom was completely blocked by pretreatment with the SERCA inhibitor TG (Fig. 2C, red line), confirming that venom SERCA actively removes calcium ions from the plasmatocyte cytoplasm. The ability of venom SERCA to profoundly affect host plasmatocyte calcium levels suggests that it is functionally distinct from the endogenous fly plasmatocyte SERCA. Isoform-specific PCR shows that in contrast to the SERCA1002 isoform found in G1 venom, fly hemocytes specifically express the SERCA1020 isoform (Fig. 2D). Data from mammalian systems show that the two isoforms have different affinities for calcium and are subject to different modes of regulation, resulting in a higher maximal pumping rate for the shorter SERCA isoform (32, 35). If this difference is conserved in insect SERCA isoforms, it could account for the effect of G1 venom SERCA1002 in fly hemocytes. These findings demonstrate that G1 venom antagonizes host hemocyte calcium levels in a SERCA-dependent manner.
The presence of active SERCA in G1 venom suggests that regulation of intracellular calcium concentration might be important for wasp virulence and, in turn, host resistance. To assay intracellular calcium levels in vivo, we expressed GCaMP3 in larval immune tissues with the Cg-GAL4 driver, which expresses in the plasmatocytes and fat body (36). We attacked these larvae with the avirulent wasp L. clavipes to assay calcium levels in a successful immune response and with G1 to assay the effect of G1 venom on host calcium levels in vivo. We simultaneously attacked larvae expressing calcium-independent GFP under the control of the Cg-GAL4 driver to control for differences in GAL4 expression following wasp attack. We found that plasmatocytes from L. clavipes-attacked larvae showed a significant increase in GCaMP3 fluorescence at 6 h PA, but no change in GFP fluorescence at this time (Fig. 2E), indicating a specific elevation of intracellular calcium levels following wasp attack. Following L. clavipes attack, we did not detect a change in GCaMP3 fluorescence either in plasmatocytes at 24 h PA (Fig. 2F) or in the fat body at either time point, demonstrating that the calcium burst is specific to plasmatocytes and is part of an immediate response to wasp attack. Conversely, plasmatocytes from G1-attacked larvae showed a significant decrease in GCaMP3 fluorescence at 6 h PA, whereas GFP levels remained constant (Fig. 2E). There was no alteration in GCaMP3 fluorescence in plasmatocytes at 24 h following G1 attack (Fig. 2F), or in the fat body at either time point. We also expressed GCaMP3 with the pan-hemocyte driver He-GAL4 (37) and did not detect GCaMP3 fluorescence in lamellocytes following attack by either wasp, confirming that the calcium burst is specific to plasmatocytes. These data show that the calcium regulatory activity of G1 venom demonstrated in our ex vivo assay is also functional in vivo following wasp attack and that G1 venom specifically targets the wasp-induced calcium burst in fly plasmatocytes that occurs immediately following attack.
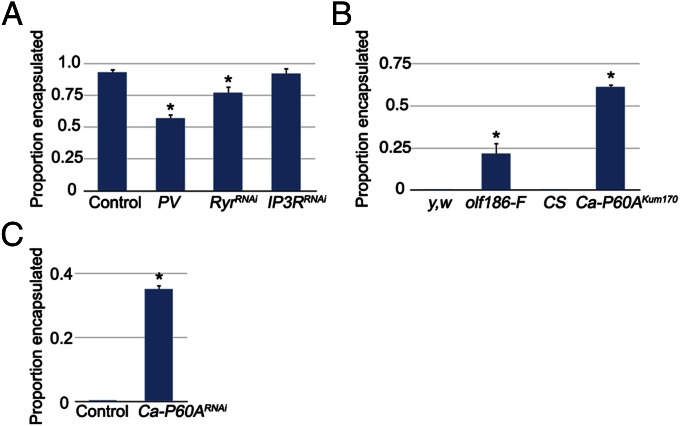
The calcium burst seen in fly plasmatocytes following attack with the avirulent wasp L. clavipes suggests that calcium signaling may be required to activate hemocytes for encapsulation. A similar calcium burst is seen in both fly and mammalian immune cells in response to diverse pathogen stimuli (38, 39). To test for a role of the calcium burst in wasp egg encapsulation, we used He-GAL4 to express parvalbumin (PV), a vertebrate-specific calcium binding protein that negatively regulates calcium levels in D. melanogaster cells (40). At 72 h PA, there was no evidence of capsule formation in a large proportion of fly larvae attacked by the normally avirulent wasp L. clavipes (Fig. 3A), suggesting that He-GAL4–driven PV expression prevented capsule initiation by fly plasmatocytes. Furthermore, increased intracellular calcium is typically mediated by the release of calcium from ER stores by either the IP3 receptor (IP3R) or Ryanodine receptor (Ryr), the major ER calcium release channels in eukaryotic cells (41). Both of these calcium channels have homologs in D. melanogaster [encoded by the Inositol 1,4,5,-tris-phosphate receptor (Itp-r83A) and Ryanodine receptor 44F (Rya-r44F) genes, respectively] (41, 42), and we found that hemocyte-specific knockdown of Rya-r44F, but not Itp-r83A, also resulted in a significant decrease in the proportion of L. clavipes eggs encapsulated by larval hemocytes (Fig. 3A). Ryr is also required for phagocytosis by fly hemocytes (38) and is important for the calcium burst in human B and T cells (39). These data show that the hemocyte calcium burst is important for hemocyte activation during the D. melanogaster encapsulation response and is conserved between mammalian and insect immune responses.
Fig. 3.
(A) Proportion of L. clavipes eggs encapsulated in the indicated genotypes. *P < 0.01 compared with control, error bars indicate SE in all graphs, within each graph, n = 3 independent replicates per genotype. (B) Proportion of G1 eggs encapsulated in the indicated genotypes. *P < 0.01 compared with genetic background controls. (C) Proportion of G1 eggs encapsulated in the indicated genotypes. *P < 0.01 compared with control.
If the ability of G1 venom to antagonize the hemocyte calcium burst is important for G1 virulence, ectopically raising hemocyte calcium levels should allow fly larval hemocytes to encapsulate G1 eggs. To test this hypothesis, we used G1 wasps to attack larvae from two mutant lines with a demonstrated elevation of intracellular calcium levels (olf186-FEY01467, the D. melanogaster homolog of the Orai calcium release-activated calcium channel, and Ca-P60AKum170) (43, 44). We found that hemocytes from both olf186-FEY01467 and Ca-P60AKum170 mutant larvae were able to encapsulate G1 eggs at a significantly higher rate than their genetic background controls [yellow,white (y,w) and Canton S (CS), respectively] (Fig. 3B and Table 2). Hemocyte-specific knockdown of Ca-P60A also conferred larvae with increased encapsulation ability against G1 (Fig. 3C), confirming the cell specificity of the calcium signaling phenotype. These results suggest that elevated calcium levels either block G1 virulence specifically or make fly larvae generally more wasp resistant. To distinguish between these possibilities, we attacked these same flies with the melanogaster subgroup specialist L. boulardi, a wasp whose venom does not contain homologs of any known calcium regulators (30). We found that L. boulardi eggs were not encapsulated by any of these genotypes regardless of calcium level (Table 2), showing that increased hemocyte intracellular calcium specifically affects G1 virulence.
Table 2.
Encapsulation rates of G1 and L. boulardi eggs in the indicated genotypes
| Genotype | Encapsulation rate of G1 eggs, % | n | Encapsulation rate of L. boulardi eggs, % | n |
| y,w | 0 | 82 | 0 | 73 |
| olf186-FEY01467 | 21.7 | 92 | 0 | 69 |
| CS | 0 | 90 | 0 | 67 |
| Ca-P60AKum170 | 61.3 | 80 | 0 | 76 |
Discussion
The identification of wasp venom proteins led us to uncover a unique and important aspect of the Drosophila innate cellular immune response against macroparasites. Fly plasmatocytes undergo a cytoplasmic calcium burst within 6 h of parasitoid wasp infection, which is required for activation of the antiwasp immune response. A similar calcium burst is observed in plasmatocytes before phagocytosis of bacteria or apoptotic cells (38). When the calcium burst is genetically blocked by knockout or knockdown of Rya-r44F, plasmatocytes fail to initiate capsule formation in response to wasp infection (Fig. 3) and are unable to phagocytize invading bacteria (38), demonstrating that calcium signaling is a conserved mechanism among fly immune responses to various pathogens.
Calcium signaling also plays an important role in mammalian immunity. Increased cytoplasmic levels of calcium are observed in lymphocytes following activation of antigen receptors (39), and this increase activates the calcium-dependent phosphatase calcineurin, which is required for the activation of mammalian immune responses via regulation of the nuclear factor of activated T-cells (NFAT) family of transcriptional activators (45). Interestingly, calcineurin is also required in fly hemocytes for the activation of Imd pathway signaling in response to infection with Gram-negative bacteria (46), suggesting that the activation of diverse immune responses likely has a shared genetic basis. G1 wasps use venom SERCA to target this highly conserved plasmatocyte calcium burst and prevent the initiation of the encapsulation response. This finding demonstrates the advantage of studying naturally coevolving host-pathogen/parasite interactions to gain insight into conserved immune mechanisms.
Identifying parasitic wasp venom proteins can enhance our understanding of the delivery and function of immunomodulatory proteins in general. Wasp venom genes were proposed to encode small, classically secreted proteins (47), and this idea appears to be somewhat true of G1 venom proteins; 28% contained a predicted secretion signal, and these proteins had an average size of 44 kDa. However, venom SERCA is a large (110-kDa) transmembrane protein with no classical secretion signal sequence, and this finding was not unusual across G1 and other Hymenopteran venoms; G1 venom proteins range in size from 9 kDa to 379 kDa, and bioinformatic analyses using transmembrane domain prediction software (48) reveals that 7% (12/166) of G1 venom proteins and 20% (176/864) of the previously identified Hymenopteran venoms in GenBank (30) contain predicted transmembrane domains. How a parasite might secrete and deliver transmembrane proteins into hosts is unknown. The venoms of wasps closely related to G1 have been shown to contain “virus-like particles,” thought to act as venom delivery vehicles, that enter host hemocytes to mediate wasp virulence (49), and we hypothesize that G1 SERCA may use a similar mechanism. Understanding the packaging of SERCA in G1 venom, and its delivery to host hemocytes, represents an interesting subject for future study.
Materials and Methods
Insect Strains.
The following D. melanogaster strains were used in this study: the mutant alleles tu(1)Sz1 (29), Ca-P60AKum170 (43), and olf186-FEY01467 (44); the transgenic constructs Cg-GAL4 (36), He-GAL4 (37), UAS-GCaMP3 (34), UAS-GFP{AH2}, UAS-PV (40), Ca-P60ATRiP.JF01948, Itp-r83ATRiP.JF01957, and Rya-r44FTRiP.JF03381; and the control lines y,w, Canton S (CS), Oregon R (OR), and w1118. For the host range experiment, the fly species used were as follows: Drosophila simulans, Drosophila mauritiana, Drosophila sechellia, Drosophila yakuba, Drosophila erecta, Drosophila ficusphila, Drosophila ananassae, Drosophila pseudoobscura, Drosophila willistoni, Drosophila funebris, Drosophila immigrans, Drosophila mojavensis, Drosophila paramelanica, and Drosophila virilis.
This study also used the following wasps: G1, L. clavipes (strain LcNet), and L. boulardi (strain Lb17). The G1 wasps used in this study were caught in Homestead, FL, in 2008. G1 COI and ITS2 sequences have been deposited in GenBank (accession nos. JQ808430 and JQ808406, respectively). LcNet was provided by J. van Alphen (University of Amsterdam, Amsterdam) and Lb17 has been described (26). Laboratory cultures of G1 and Lb17 are maintained on D. melanogaster and LcNet is maintained on D. virilis.
Wasp Attack.
Wasp attacks were performed as described (17). Briefly, three female wasps were allowed to attack 40 second instar fly larvae in 35-mm Petri dishes filled with Drosophila medium for a 72-h period at 25 °C. To assay attack and encapsulation rates, larvae were dissected and scored for the presence of an encapsulated wasp egg or live wasp larva. To assay eclosion rates, 30 fly larvae were recovered from each plate and allowed to eclose at 25 °C. The total number of flies and wasps that eclosed were determined 15 d and 30 d after infection, respectively, times by which all viable flies and wasps should have emerged. All experiments were performed in triplicate.
tu(1)Sz Phenotype Suppression.
The temperature-sensitive tu(1)Sz1 self-encapsulation mutant was used to assay wasp virulence strategy (29). The flies were raised at 28 °C and to assay the ability of wasp venom to suppress the phenotype, 40 second instar larvae were attacked by three female wasps for 3 h at 28 °C. The attacked larvae were raised for a further 96 h at 28 °C. Attacked pupae and age-matched controls were then scored for the tu(1)Sz phenotype and dissected to ensure attack status; unattacked pupae were discarded from analysis.
Imaging.
Images were acquired by using a Leica stereo-dissecting scope with a Moticam MIP 2.0 and Multi-Focus Pro software. Figures were compiled by using Adobe Photoshop.
Hemocyte Counts.
Hemocyte counts were performed in triplicate according to ref. 17. After a 72-h wasp attack period, five larvae from each replicate were washed in Drosophila Ringer’s solution and bled into PBS containing 0.01% phenylthiourea to prevent melanization. Hemocytes were applied to a disposable hemocytometer (Incyto C-Chip DHC-N01) and allowed to adhere for 30 min. Hemocytes of each replicate were counted from 16 0.25 × 0.25 × 0.1 mm squares, and the counts were normalized to a per larva value.
Wasp Transcriptomes.
RNA was extracted from ∼200 female wasp abdomens by using the standard TRIzol (Invitrogen) protocol. Poly(A)RNAs were purified by using the Dynabeads mRNA Direct kit (Invitrogen) according to manufacturer specifications. Double-stranded cDNAs were synthesized by using the SuperScript II ds cDNA Synthesis kit (Invitrogen). The cDNAs were then sequenced by using an Illumina HiSeq 2000. Transcripts were de novo assembled by using Trinity (version r2011-10-29) (50) and filtered by RSEM (51) using a cutoff of one transcript per million. See ref. 30 for more detailed protocols. Tools for secretion signal analysis and GO term annotation and enrichment are described in ref. 30.
Mass Spectrometry.
Venom proteins were purified from venom glands dissected into PBS supplemented with 0.5 mM EDTA and Complete Protease Inhibitor Mixture (Roche). Venom glands were homogenized under nonlysing conditions, and gland cells were pelleted by centrifugation. Venom proteins were run on SDS/PAGE, trypsinized, and subjected to Nano LC-MS(MS)2. The identified peptides were mapped back to the transcriptome data by using SEQUEST software. See ref. 30 for more detailed protocols.
SERCA Isoform-Specific PCRs.
To assay expression of SERCA isoform transcripts, total RNA was made from wasp venom glands or wasp carcasses (wasps with venom gland removed), and whole third instar larvae or dissected larval hemocytes, by standard TRIzol preparations as described above. RNA was reverse transcribed to cDNA by using the QuantiTect reverse transcriptase kit (Qiagen). Isoform specific primers were used to amplify SERCA from each cDNA sample (primer sequences available upon request).
SSH.
SSH was performed essentially as described (31) by using venom gland cDNAs as the tester library and wasp carcass cDNAs as the driver library and with the following alterations: libraries were hybridized at a 30:1 (driver:tester) ratio and then used undiluted for two rounds of PCR amplification. PCR products were cloned were the Strataclone PCR cloning kit (Stratagene) and sequenced.
Western Blotting.
G1 venom was purified as described above. Protein extracts from purified venom and wasp carcasses were run on 9% (vol/vol) polyacrylamide SDS/PAGE gels and transferred to PVDF membranes. Blots were probed with the 809-27 SERCA antibody at a concentration of 1:100,000 (52).
GCaMP3 Calcium Assays.
Ex vivo assay.
We used thapsigargin, a plant-derived sesquiterpene lactone that acts as a specific and irreversible inhibitor of SERCA (33). G1 venom was purified as described above and incubated with 1 μM thapsigargin in DMSO or DMSO control at room temperature for 15 min. Samples were then twice dialyzed against PBS at room temperature (15 min, 1 h), added to dissected GCaMP3-expressing third instar larval hemocytes, and incubated at room temperature. During the incubation period, cells were imaged by using an Olympus BX51 microscope with a FITC filter and Olympus DP2-BSW software. Corrected total cell fluorescence (CTCF) was calculated as described (53).
In vivo assay.
Larvae expressing GCaMP3 or GFP were attacked by wasps as described and dissected at 6 or 24 h PA. Cells were imaged, and CTCFs were calculated as for the ex vivo assay.
Statistics.
All analyses were performed in R version 2.15.0. Fisher’s exact test was used to compare tu(1)Sz phenotype penetrance between G1 attacked and control samples and to compare the results of Signal P analysis on G1 venom and nonvenom genes. Total hemocyte counts and lamellocyte numbers were compared by ANOVA, and Tukey’s HSD test was used for pairwise comparisons. Differences in in vivo GCaMP3 and GFP fluorescence levels were compared by two-way ANOVA at 6 h and 24 h PA to test effects for of fly genotype and wasp attack. Tukey’s HSD test was used for pairwise comparisons within each time point. To test the effect of venom and PBS on GCaMP3 fluorescence throughout the incubation period in the ex vivo assay we used repeated measures ANOVA (from the car R library) and pairwise t tests to compare fluorescence at each time point. Finally, generalized linear models with binomial errors and logit link functions were used to examine differences in encapsulation rate between genotypes. Bonferroni correction was used for multiple comparisons.
Data Access.
RNA-seq is available at the DNA Data Bank of Japan Sequence Read Archive under accession number SRP020527; single-end data under SRA accession number SRX259389 and paired-end data are under SRX259413. All G1 venom transcript and venom sequences are attached as Fasta formatted files.
Supplementary Material
Acknowledgments
We thank Jesper Moller, Loren Looger, Subhabrata Sanyal, the Bloomington Drosophila Stock Center, and the Drosophila Species Stock Center at the University of California San Diego, for stocks and reagents, the Emory Georgia Research Alliance Genome Center, and Erin Keebaugh for technical assistance. This work was supported by the National Institutes of Health Grants R01 AI081879 (to T.A.S.) and R21 HG005133 (to J.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The G1 Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank (accession no. GAIW00000000). The version described in this paper is the first version, GAIW01000000. G1 COI and ITS2 sequences have been deposited in GenBank (accession nos. JQ808430 and JQ808406, respectively).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222351110/-/DCSupplemental.
References
- 1.Kraaijeveld AR, Van Alphen JJ, Godfray HC. The coevolution of host resistance and parasitoid virulence. Parasitology. 1998;116(Suppl):S29–S45. doi: 10.1017/s0031182000084924. [DOI] [PubMed] [Google Scholar]
- 2.Brennan CA, Anderson KV. Drosophila: The genetics of innate immune recognition and response. Annu Rev Immunol. 2004;22:457–483. doi: 10.1146/annurev.immunol.22.012703.104626. [DOI] [PubMed] [Google Scholar]
- 3.Brandt SM, Jaramillo-Gutierrez G, Kumar S, Barillas-Mury C, Schneider DS. Use of a Drosophila model to identify genes regulating Plasmodium growth in the mosquito. Genetics. 2008;180(3):1671–1678. doi: 10.1534/genetics.108.089748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: Differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci USA. 1997;94(26):14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemaitre B, et al. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc Natl Acad Sci USA. 1995;92(21):9465–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86(6):973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 7.Ip YT, et al. Dif, a dorsal-related gene that mediates an immune response in Drosophila. Cell. 1993;75(4):753–763. doi: 10.1016/0092-8674(93)90495-c. [DOI] [PubMed] [Google Scholar]
- 8.Wu LP, Anderson KV. Regulated nuclear import of Rel proteins in the Drosophila immune response. Nature. 1998;392(6671):93–97. doi: 10.1038/32195. [DOI] [PubMed] [Google Scholar]
- 9.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388(6640):394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 10.Rock FL, Hardiman G, Timans JC, Kastelein RA, Bazan JF. A family of human receptors structurally related to Drosophila Toll. Proc Natl Acad Sci USA. 1998;95(2):588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kopp EB, Medzhitov R. The Toll-receptor family and control of innate immunity. Curr Opin Immunol. 1999;11(1):13–18. doi: 10.1016/s0952-7915(99)80003-x. [DOI] [PubMed] [Google Scholar]
- 12.Medzhitov R, Janeway C., Jr Innate immunity. N Engl J Med. 2000;343(5):338–344. doi: 10.1056/NEJM200008033430506. [DOI] [PubMed] [Google Scholar]
- 13.Hayden MS, West AP, Ghosh S. NF-kappaB and the immune response. Oncogene. 2006;25(51):6758–6780. doi: 10.1038/sj.onc.1209943. [DOI] [PubMed] [Google Scholar]
- 14.Carton Y, Bouletreau M, van Alphen JJM, van Lenteren JC. 1986. The Drosophila parasitic wasps. The Genetics and Biology of Drosophila, eds Ashburner M, Carson L, Thompson JN (Academic Press, London), pp 347–394. [Google Scholar]
- 15.Carton Y, Nappi AJ. Drosophila cellular immunity against parasitoids. Parasitol Today. 1997;13(6):218–227. doi: 10.1016/s0169-4758(97)01058-2. [DOI] [PubMed] [Google Scholar]
- 16.Williams MJ, Ando I, Hultmark D. Drosophila melanogaster Rac2 is necessary for a proper cellular immune response. Genes Cells. 2005;10(8):813–823. doi: 10.1111/j.1365-2443.2005.00883.x. [DOI] [PubMed] [Google Scholar]
- 17.Mortimer NT, Kacsoh BZ, Keebaugh ES, Schlenke TA. Mgat1-dependent N-glycosylation of membrane components primes Drosophila melanogaster blood cells for the cellular encapsulation response. PLoS Pathog. 2012;8(7):e1002819. doi: 10.1371/journal.ppat.1002819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russo J, Dupas S, Frey F, Carton Y, Brehelin M. Insect immunity: Early events in the encapsulation process of parasitoid (Leptopilina boulardi) eggs in resistant and susceptible strains of Drosophila. Parasitology. 1996;112(Pt 1):135–142. doi: 10.1017/s0031182000065173. [DOI] [PubMed] [Google Scholar]
- 19.Salt GW. 1970. The cellular defense reactions of insects (Cambridge Univ Press, Cambridge, UK) [Google Scholar]
- 20.Williams MJ. Drosophila hemopoiesis and cellular immunity. J Immunol. 2007;178(8):4711–4716. doi: 10.4049/jimmunol.178.8.4711. [DOI] [PubMed] [Google Scholar]
- 21.Howell L, et al. A directed miniscreen for genes involved in the Drosophila anti-parasitoid immune response. Immunogenetics. 2012;64(2):155–161. doi: 10.1007/s00251-011-0571-3. [DOI] [PubMed] [Google Scholar]
- 22.Evans CJ, Hartenstein V, Banerjee U. Thicker than blood: Conserved mechanisms in Drosophila and vertebrate hematopoiesis. Dev Cell. 2003;5(5):673–690. doi: 10.1016/s1534-5807(03)00335-6. [DOI] [PubMed] [Google Scholar]
- 23.Rizki RM, Rizki TM. Selective destruction of a host blood cell type by a parasitoid wasp. Proc Natl Acad Sci USA. 1984;81(19):6154–6158. doi: 10.1073/pnas.81.19.6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Labrosse C, Carton Y, Dubuffet A, Drezen JM, Poirie M. Active suppression of D. melanogaster immune response by long gland products of the parasitic wasp Leptopilina boulardi. J Insect Physiol. 2003;49(5):513–522. doi: 10.1016/s0022-1910(03)00054-4. [DOI] [PubMed] [Google Scholar]
- 25.Rizki TM, Rizki RM, Carton Y. Leptopilina heterotoma and L. boulardi: Strategies to avoid cellular defense responses of Drosophila melanogaster. Exp Parasitol. 1990;70(4):466–475. doi: 10.1016/0014-4894(90)90131-u. [DOI] [PubMed] [Google Scholar]
- 26.Schlenke TA, Morales J, Govind S, Clark AG. Contrasting infection strategies in generalist and specialist wasp parasitoids of Drosophila melanogaster. PLoS Pathog. 2007;3(10):1486–1501. doi: 10.1371/journal.ppat.0030158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basset A, et al. The phytopathogenic bacteria Erwinia carotovora infects Drosophila and activates an immune response. Proc Natl Acad Sci USA. 2000;97(7):3376–3381. doi: 10.1073/pnas.070357597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carton Y, Frey F, Nappi AJ. Parasite-induced changes in nitric oxide levels in Drosophila paramelanica. J Parasitol. 2009;95(5):1134–1141. doi: 10.1645/GE-2091.1. [DOI] [PubMed] [Google Scholar]
- 29.Rizki TM, Rizki RM. Developmental analysis of a temperature-sensitive melanotic tumor mutant in Drosophila melanogaster. Wilhelm Roux's Archives. 1980;189:197–206. doi: 10.1007/BF00868678. [DOI] [PubMed] [Google Scholar]
- 30.Goecks J, et al. Integrative approach reveals composition of endoparasitoid wasp venoms. PLoS ONE. 2013 doi: 10.1371/journal.pone.0064125. 10.1371/journal.pone.0064125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rebrikov DV. 2003. Identification of differential genes by suppression subtractive hybridization. PCR Primer, eds Dieffenbach CW, Dveksler GS (Cold Spring Harbor Lab Press, Plainview, NY), 2nd Ed, pp 297–325. [Google Scholar]
- 32.Vangheluwe P, Raeymaekers L, Dode L, Wuytack F. Modulating sarco(endo)plasmic reticulum Ca2+ ATPase 2 (SERCA2) activity: Cell biological implications. Cell Calcium. 2005;38(3-4):291–302. doi: 10.1016/j.ceca.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 33.Lytton J, Westlin M, Hanley MR. Thapsigargin inhibits the sarcoplasmic or endoplasmic reticulum Ca-ATPase family of calcium pumps. J Biol Chem. 1991;266(26):17067–17071. [PubMed] [Google Scholar]
- 34.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6(12):875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dode L, et al. Dissection of the functional differences between sarco(endo)plasmic reticulum Ca2+-ATPase (SERCA) 1 and 2 isoforms and characterization of Darier disease (SERCA2) mutants by steady-state and transient kinetic analyses. J Biol Chem. 2003;278(48):47877–47889. doi: 10.1074/jbc.M306784200. [DOI] [PubMed] [Google Scholar]
- 36.Asha H, et al. Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics. 2003;163(1):203–215. doi: 10.1093/genetics/163.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zettervall CJ, et al. A directed screen for genes involved in Drosophila blood cell activation. Proc Natl Acad Sci USA. 2004;101(39):14192–14197. doi: 10.1073/pnas.0403789101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cuttell L, et al. Undertaker, a Drosophila Junctophilin, links Draper-mediated phagocytosis and calcium homeostasis. Cell. 2008;135(3):524–534. doi: 10.1016/j.cell.2008.08.033. [DOI] [PubMed] [Google Scholar]
- 39.Vig M, Kinet JP. Calcium signaling in immune cells. Nat Immunol. 2009;10(1):21–27. doi: 10.1038/ni.f.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harrisingh MC, Wu Y, Lnenicka GA, Nitabach MN. Intracellular Ca2+ regulates free-running circadian clock oscillation in vivo. J Neurosci. 2007;27(46):12489–12499. doi: 10.1523/JNEUROSCI.3680-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clapham DE. Calcium signaling. Cell. 2007;131(6):1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 42.Vázquez-Martínez O, Cañedo-Merino R, Díaz-Muñoz M, Riesgo-Escovar JR. Biochemical characterization, distribution and phylogenetic analysis of Drosophila melanogaster ryanodine and IP3 receptors, and thapsigargin-sensitive Ca2+ ATPase. J Cell Sci. 2003;116(Pt 12):2483–2494. doi: 10.1242/jcs.00455. [DOI] [PubMed] [Google Scholar]
- 43.Sanyal S, et al. Analysis of conditional paralytic mutants in Drosophila sarco-endoplasmic reticulum calcium ATPase reveals novel mechanisms for regulating membrane excitability. Genetics. 2005;169(2):737–750. doi: 10.1534/genetics.104.031930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Venkiteswaran G, Hasan G. Intracellular Ca2+ signaling and store-operated Ca2+ entry are required in Drosophila neurons for flight. Proc Natl Acad Sci USA. 2009;106(25):10326–10331. doi: 10.1073/pnas.0902982106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17(18):2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 46.Dijkers PF, O’Farrell PH. Drosophila calcineurin promotes induction of innate immune responses. Curr Biol. 2007;17(23):2087–2093. doi: 10.1016/j.cub.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Graaf DC, et al. Insights into the venom composition of the ectoparasitoid wasp Nasonia vitripennis from bioinformatic and proteomic studies. Insect Mol Biol. 2010;19(Suppl 1):11–26. doi: 10.1111/j.1365-2583.2009.00914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305(3):567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 49.Rizki RM, Rizki TM. Parasitoid virus-like particles destroy Drosophila cellular immunity. Proc Natl Acad Sci USA. 1990;87(21):8388–8392. doi: 10.1073/pnas.87.21.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li B, Dewey CN. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moller JV, et al. Probing of the membrane topology of sarcoplasmic reticulum Ca2+-ATPase with sequence-specific antibodies. Evidence for plasticity of the c-terminal domain. J Biol Chem. 1997;272(46):29015–29032. doi: 10.1074/jbc.272.46.29015. [DOI] [PubMed] [Google Scholar]
- 53.Burgess A, et al. Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc Natl Acad Sci USA. 2010;107(28):12564–12569. doi: 10.1073/pnas.0914191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.