Abstract
The study of reaction–diffusion processes is much more complicated on general curved surfaces than on standard Cartesian coordinate spaces. Here we show how to formulate and solve systems of reaction–diffusion equations on surfaces in an extremely simple way, using only the standard Cartesian form of differential operators, and a discrete unorganized point set to represent the surface. Our method decouples surface geometry from the underlying differential operators. As a consequence, it becomes possible to formulate and solve rather general reaction–diffusion equations on general surfaces without having to consider the complexities of differential geometry or sophisticated numerical analysis. To illustrate the generality of the method, computations for surface diffusion, pattern formation, excitable media, and bulk-surface coupling are provided for a variety of complex point cloud surfaces.
Keywords: closest point method, embedding method, Laplace–Beltrami
Partial differential equations (PDEs) are widely used to describe continuum processes such as diffusion, chemical reactions, fluid flow, or electrodynamics. In standard 3D settings, these take a familiar PDE form, such as a reaction–diffusion equation:
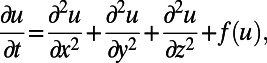 |
and the ways to numerically solve such equations are well-developed. The basic approach can be quite simple, such as laying down a uniform Cartesian grid of points, 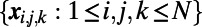 , and using simple, familiar approximations of the differential terms on this grid, such as:
, and using simple, familiar approximations of the differential terms on this grid, such as:
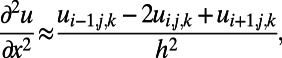 |
where 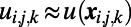 and h is the uniform grid spacing. As a result, it is very easy to implement methods for the numerical solution of such equations to study the phenomena of interest. To achieve efficiency for large-scale computations, more advanced methods are required, such as implicit discretization and solvers for systems of equations. These solvers, while internally complex, have been implemented in standard, well-validated numerical routines and are accessible through numerical subroutine libraries.
and h is the uniform grid spacing. As a result, it is very easy to implement methods for the numerical solution of such equations to study the phenomena of interest. To achieve efficiency for large-scale computations, more advanced methods are required, such as implicit discretization and solvers for systems of equations. These solvers, while internally complex, have been implemented in standard, well-validated numerical routines and are accessible through numerical subroutine libraries.
Important physical processes also arise in complex geometrical settings, such as on complicated surfaces in three dimensions. The abstract form of differential operators on surfaces remains the same as in 3D, however, when explicitly expressed in coordinates, the formulas for the operators and the corresponding discretized expressions are relatively complicated and have received much less attention. Moreover, in practical settings, surfaces are often defined simply as a set of points—a point cloud—sampled from the underlying surface. Because the connectivity of the points is not provided, this adds further complexity to methods that need to reconstruct the geometric properties of the surface, such as the metric distance.
Here we present a method for solving reaction–diffusion equations on a point cloud that represents the underlying surface—or any other geometric object—in a way that reduces the problem to working with entirely standard classical 3D discretizations and solver libraries. Our approach is fundamentally different from other recent methods for computing point clouds (e.g., refs. 1, 2), in that it does not make assumptions on the codimension of the set upon which we compute, nor does it ever construct the local connectivity between points (cf. ref. 2). A more detailed comparison with other methods is provided in the Discussion section.
The article unfolds as follows: We begin by introducing the problem and reviewing some recent results that are relevant for our method. An algorithm for solving diffusion on point cloud surfaces is developed by formulating the surface PDE problem in the embedding space and by giving a convolutional discretization of the corresponding equations. Numerical experiments are provided to explore the accuracy and stability of the method. An extension to reaction–diffusion equations is developed, and some experiments on systems are provided. The article concludes by contrasting the method to some of the others available, and outlining generalizations and directions for future work.
Diffusion Intrinsic to a Surface
We develop the method first for the fundamental case of pure diffusion.
Consider evolving a scalar diffusion process on a smooth surface 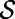 . The corresponding PDE is, formally:
. The corresponding PDE is, formally:
where the right-hand side involves the Laplacian intrinsic to the surface, also known as the Laplace–Beltrami operator. This generalizes the standard Laplacian, an operator with a well-known Cartesian coordinate formula, to a surface. We assume that in practice we do not know 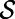 ; instead, we are given a discrete, unconnected set of points
; instead, we are given a discrete, unconnected set of points  which sample
which sample 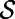 (perhaps even including noise; i.e., error in point positions relative to the true geometry). We seek a method that solves Eq. 1 without explicitly constructing an approximation to
(perhaps even including noise; i.e., error in point positions relative to the true geometry). We seek a method that solves Eq. 1 without explicitly constructing an approximation to 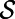 from the point cloud
from the point cloud  , and which uses only knowledge of how to express and solve the standard 3D diffusion equation in standard 3D Cartesian space.
, and which uses only knowledge of how to express and solve the standard 3D diffusion equation in standard 3D Cartesian space.
Continuous Formulation in the Embedding Space.
Our first step to formulating a method is to replace the surface PDE by a related equation posed on the surrounding 3D space that can be solved using standard Cartesian grid methods in 3D. When restricted to the surface, this 3D embedding equation will give the solution to the original surface problem. To obtain an intuition for the design of this equation, consider a function u defined on a smooth surface. The function u may be extended to a 3D neighborhood of the surface by extending its value constant along the normal directions to the surface. The 3D gradients of this extended function agree with the surface gradients of the surface function at the surface, since the only 3D variation is along the surface. Expressing this intuition mathematically leads to the following fundamental principle (3):
1. Equivalence of gradients.
Suppose u is any function defined on 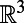 that is constant along directions normal to the surface. Then, at the surface:
that is constant along directions normal to the surface. Then, at the surface:
where 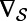 denotes the intrinsic surface gradient [specifically,
denotes the intrinsic surface gradient [specifically, 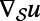 is the tangent vector to the surface defined by
is the tangent vector to the surface defined by 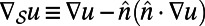 , where
, where 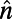 is a unit normal to the surface].
is a unit normal to the surface].
Similarly, a flux that is everywhere directed along the surface can only spread out within the surface directions. This gives us our second fundamental principle (3):
2. Equivalence of divergence.
Suppose v is any vector field on 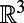 that is tangent at
that is tangent at 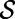 , and also tangent at all surfaces displaced by a fixed distance from
, and also tangent at all surfaces displaced by a fixed distance from 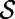 (i.e., all surfaces defined as level sets of the distance function to
(i.e., all surfaces defined as level sets of the distance function to 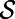 ). Then, at the surface:
). Then, at the surface:
In fact, a version of this principle is true even if the vector field v is not tangent to 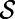 (4).
(4).
In both instances, we observe that derivatives intrinsic to the surface may be computed in 3D by extending quantities off the surface along the normal directions emanating from the surface. Our approach for numerically approximating PDEs on surfaces, the closest point method (3), evaluates derivatives using these fundamental principles and a “closest point function” representation of the surface 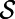 : for a point
: for a point 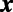 in space, let
in space, let 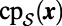 be the point belonging to
be the point belonging to 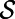 that is closest to
that is closest to 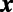 . Notice that the constant normal extension may be efficiently evaluated by interpolation at the closest point on the surface, and that the closest point representation makes sense for any point set
. Notice that the constant normal extension may be efficiently evaluated by interpolation at the closest point on the surface, and that the closest point representation makes sense for any point set 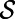 , not just a continuous surface. The latter property is what will give our approach its geometric generality to point clouds.
, not just a continuous surface. The latter property is what will give our approach its geometric generality to point clouds.
Combining these results gives the following theorem for evaluating the Laplace–Beltrami operator using only the classical 3D Cartesian Laplacian and the closest point function (3):
Theorem 1.
Let
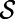 be a smooth surface in
be a smooth surface in
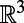 and
and
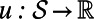 be a smooth function. Let
be a smooth function. Let
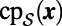 be the closest point function for
be the closest point function for
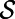 . Then:
. Then:
In addition, this remains true in the general case where 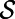 is a smooth object of dimensions
is a smooth object of dimensions 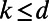 in
in 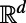 , such as for a filament (
, such as for a filament (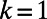 ) in
) in 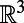 (3, 4).
(3, 4).
The right-hand side of Eq. 2 is well-defined for a surface 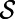 because
because 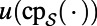 is a function on
is a function on 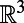 over which the standard Laplacian is defined. A consequence of this is that Eq. 2 will provide a generalization of the Laplace operator to point clouds, as well as a practical means of computing the corresponding diffusion flows.
over which the standard Laplacian is defined. A consequence of this is that Eq. 2 will provide a generalization of the Laplace operator to point clouds, as well as a practical means of computing the corresponding diffusion flows.
Applying Eq. 2 to the surface PDE (Eq. 1) yields the explicit closest point method for solving the surface PDE in the embedding space. In its simplest form, the method alternates two steps. First, quantities on the surface are extended into the embedding space using the closest point operator:
Next, a short-time evolution of the corresponding PDE in 3D is carried out:
starting from the extended variable, v. For a small time, the solution of this embedding equation gives the desired approximation of the underlying surface flow (Eq. 1) on the surface 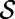 .
.
To illustrate the intuition behind the method, consider a surface process that is driven by diffusion. Suppose the evolving function is artificially extended to all of 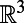 by making it constant along the normal directions off the surface. We then apply standard 3D diffusion to the extended function for a short time
by making it constant along the normal directions off the surface. We then apply standard 3D diffusion to the extended function for a short time 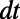 . Intuitively, the fully 3D diffusion is driven by in-surface gradients since the extended function does not vary normal to the surface. This implies that the method will carry out the desired in-surface flow, at least for the short time scale of interest. Note that the leading order term in the error is
. Intuitively, the fully 3D diffusion is driven by in-surface gradients since the extended function does not vary normal to the surface. This implies that the method will carry out the desired in-surface flow, at least for the short time scale of interest. Note that the leading order term in the error is 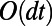 since the second derivative of u normal to the surface is only guaranteed to be zero initially; at later times, it will vary linearly with
since the second derivative of u normal to the surface is only guaranteed to be zero initially; at later times, it will vary linearly with 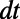 .
.
As noted, the closest point method is valid for smooth surfaces of any codimension embedded in 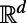 . In particular, in
. In particular, in 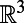 , it applies to 2D surface diffusion as well as 1D diffusion along a filament. It is even valid for solid regions since Eqs. 3 and 4 reduce to the standard heat equation in
, it applies to 2D surface diffusion as well as 1D diffusion along a filament. It is even valid for solid regions since Eqs. 3 and 4 reduce to the standard heat equation in 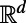 when applied to a d-dimensional subset of
when applied to a d-dimensional subset of 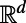 . Because the extension procedure is well-defined and easily performed even if the surface is simply represented as a point cloud, and because the evolution takes place in standard 3D, the method retains the same simplicity no matter how complex the surface or point set on which the problem is posed. The discretization of the method that we develop preserves this geometric flexibility, and ultimately yields methods that are independent of the dimensionality of the underlying point cloud set.
. Because the extension procedure is well-defined and easily performed even if the surface is simply represented as a point cloud, and because the evolution takes place in standard 3D, the method retains the same simplicity no matter how complex the surface or point set on which the problem is posed. The discretization of the method that we develop preserves this geometric flexibility, and ultimately yields methods that are independent of the dimensionality of the underlying point cloud set.
Discretization of the Embedding Equation.
The closest point method must be discretized in time and space. To construct the discretization, it is crucial to note that we do not have 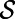 ; instead, we have a point cloud approximation
; instead, we have a point cloud approximation  of the surface. Consequently, we may assume that the easily computed
of the surface. Consequently, we may assume that the easily computed 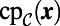 is available, but not
is available, but not 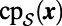 . Specifically, let
. Specifically, let 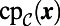 be the point belonging to the set
be the point belonging to the set  that is closest to
that is closest to 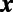 .
.
To carry out the first step in the closest point method, we make a direct replacement of 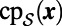 by
by 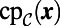 in Eq. 3. That is, the constant normal extension is carried out by replacing u in the embedding space by the value at the closest point in the point cloud,
in Eq. 3. That is, the constant normal extension is carried out by replacing u in the embedding space by the value at the closest point in the point cloud, 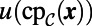 . Similar to refs. 3, 5, we carry out this step on a uniform grid with spacing h in a dimension-by-dimension fashion using standard polynomial interpolation of degree
. Similar to refs. 3, 5, we carry out this step on a uniform grid with spacing h in a dimension-by-dimension fashion using standard polynomial interpolation of degree 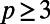 .
.
In refs. 3, 5 standard finite differences are used in the second step of the closest point method to discretize the 3D diffusion flow (Eq. 4). Unfortunately, this gives poor accuracy and is not recommended except for very dense point clouds. To see this, observe that 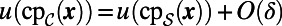 on the surface for a smooth
on the surface for a smooth 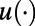 where δ is the maximum distance between an arbitrary point on the surface and its closest point in the point cloud. Forming the discrete Laplacian amplifies the error by a factor of
where δ is the maximum distance between an arbitrary point on the surface and its closest point in the point cloud. Forming the discrete Laplacian amplifies the error by a factor of 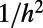 , where h is the mesh spacing, potentially leading to an
, where h is the mesh spacing, potentially leading to an 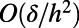 error at each time step.
error at each time step.
To avoid the amplification of errors arising in a standard finite difference approach, we discretize the 3D diffusion flow (Eq. 4) convolutionally; that is, we convolve the extended function with a kernel corresponding to diffusion for a short time 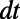 . The convolution is evaluated in Fourier space via the fast Fourier transform (FFT). We remark that to apply our discretization the problem must have a convolutional form and the corresponding convolution kernel should be accurately approximated using the discrete Fourier basis. This article focuses on flows involving diffusion and we note that there will be situations where our discretizations do not apply (e.g., pure convection).
. The convolution is evaluated in Fourier space via the fast Fourier transform (FFT). We remark that to apply our discretization the problem must have a convolutional form and the corresponding convolution kernel should be accurately approximated using the discrete Fourier basis. This article focuses on flows involving diffusion and we note that there will be situations where our discretizations do not apply (e.g., pure convection).
In a typical point cloud calculation, we are given a cloud of points and choose a suitable time step-size 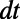 . Normally, the time step-size should be chosen so that the effective width of the diffusion kernel (namely
. Normally, the time step-size should be chosen so that the effective width of the diffusion kernel (namely 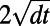 ) exceeds the distance between points in the cloud. If this condition is violated, diffusion is insufficiently evolved to cause the desired coupling between neighboring points and the method assumes that there is a hole in the surface. We may quantify this condition by assigning a measure μ to the point cloud spacing:
) exceeds the distance between points in the cloud. If this condition is violated, diffusion is insufficiently evolved to cause the desired coupling between neighboring points and the method assumes that there is a hole in the surface. We may quantify this condition by assigning a measure μ to the point cloud spacing:
where the points 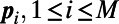 define a cloud of M points. With this measure of spacing, we see that we want to choose
define a cloud of M points. With this measure of spacing, we see that we want to choose 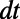 such that
such that 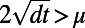 . The grid spacing h is determined from
. The grid spacing h is determined from 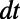 . Specifically, we select a mesh that is sufficiently refined to ensure that the error arising from truncating the corresponding Fourier series is exponentially small. This leads us to the requirement
. Specifically, we select a mesh that is sufficiently refined to ensure that the error arising from truncating the corresponding Fourier series is exponentially small. This leads us to the requirement 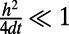 , which is a standard choice in spectral convolution (6).
, which is a standard choice in spectral convolution (6).
Estimates of the errors arising from one step of the method may now be formally computed. We have already seen that alternating diffusion for a time 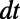 with constant normal extension leads to an
with constant normal extension leads to an 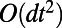 error after one time step of size
error after one time step of size 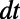 . The replacement of
. The replacement of 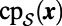 by
by 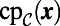 in Eq. 3 introduces an
in Eq. 3 introduces an 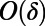 error into the approximation of v. Interpolation with degree-p polynomials introduces an additional
error into the approximation of v. Interpolation with degree-p polynomials introduces an additional 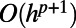 error into the approximation of v. Using
error into the approximation of v. Using 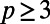 and
and 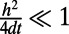 , we find that this interpolation error is
, we find that this interpolation error is 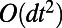 . Finally, errors arise from truncating the Fourier series. These are exponentially small relative to the other errors induced by the method provided
. Finally, errors arise from truncating the Fourier series. These are exponentially small relative to the other errors induced by the method provided 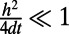 .
.
Methods.
The closest point function is constructed using the k-nearest neighbors algorithm. An implementation of this step in the high-level language Matlab is given by:
 |
where PT_CLOUD is an 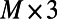 matrix defining a cloud of M points in 3D; X, Y, Z are matrices specifying the
matrix defining a cloud of M points in 3D; X, Y, Z are matrices specifying the 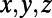 coordinates of the grid nodes; and CPx, CPy, CPz are the corresponding coordinates of the closest points in the point cloud.
coordinates of the grid nodes; and CPx, CPy, CPz are the corresponding coordinates of the closest points in the point cloud.
To evolve diffusion on point cloud surfaces, we alternate two steps. The first step extends values off the surface into a 3D region surrounding the surface using tricubic interpolation. This step can be implemented in Matlab using:
The second step simply evolves the corresponding 3D PDE in a neighborhood of the surface. In Matlab, this can be accomplished on a uniform lattice using:
where convolution is carried out using K_hat, the discrete Fourier transform of the diffusion kernel. Note that fftn and ifftn are the standard 3D Fourier transform and inverse Fourier transform in Matlab. Thus, the only difference between this code and a standard one for solving diffusion equations in 3D is the constant normal extension of the quantity U preceding each time step. In our code we compute the K_hat matrix analytically; for example, on a cube of side length L with a grid of 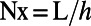 points in each direction, diffusion coefficient D, and step size dt, we have:
points in each direction, diffusion coefficient D, and step size dt, we have:
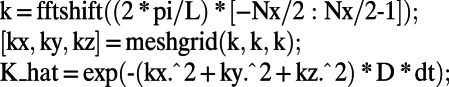 |
In this work, we use a uniform grid of the embedding space 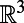 . This approach is simple and makes direct use of the built-in Matlab functions for cubic interpolation and FFTs. For improved efficiency, interpolation can be carried out in a band around the surface since the diffusion kernel becomes exponentially small outside of its effective width (see refs. 3, 5 for localization techniques for the finite difference case). Furthermore, the convolution step may be accelerated using the fft() and ifft() functions on a graphics processing unit. Even with these enhancements, some computations will remain computationally expensive. For example, it is expensive to apply this discretization to a PDE on a surface that is embedded in four or more dimensions. It may also be inefficient to compute on a uniform discretization of space if the point cloud sampling of the underlying surface is highly nonuniform. For such problems, alternative methods for evaluating the convolutions are needed. A particularly promising approach is to evaluate convolutions by a suitable spatial quadrature formula, and compute the subsequent discrete sums by the fast Gauss transform (7, 8). This fast convolution technique requires only
. This approach is simple and makes direct use of the built-in Matlab functions for cubic interpolation and FFTs. For improved efficiency, interpolation can be carried out in a band around the surface since the diffusion kernel becomes exponentially small outside of its effective width (see refs. 3, 5 for localization techniques for the finite difference case). Furthermore, the convolution step may be accelerated using the fft() and ifft() functions on a graphics processing unit. Even with these enhancements, some computations will remain computationally expensive. For example, it is expensive to apply this discretization to a PDE on a surface that is embedded in four or more dimensions. It may also be inefficient to compute on a uniform discretization of space if the point cloud sampling of the underlying surface is highly nonuniform. For such problems, alternative methods for evaluating the convolutions are needed. A particularly promising approach is to evaluate convolutions by a suitable spatial quadrature formula, and compute the subsequent discrete sums by the fast Gauss transform (7, 8). This fast convolution technique requires only 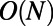 operations per time step, where N is the number of grid nodes forming the discretization.
operations per time step, where N is the number of grid nodes forming the discretization.
Numerical Experiment.
We consider an initial value problem for diffusion on the unit sphere with exact solution 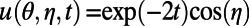 in spherical coordinates
in spherical coordinates 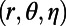 .
.
The evolution is carried out on a point cloud defined by the method of Saff and Kuijlaars (9). This method places nodes along a spiral in such a way that the distance between nodes along the spiral is approximately equal to the distance between the coils of the spiral. Using cubic interpolation, the max norm relative errors at 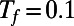 are computed for a variety of point clouds. This yields the results reported in Table 1. Our experiments measure the point cloud spacing using the value μ given by Eq. 5. This convergence test indicates a first order error in μ, and shows that the regularity of the error improves as the discretization parameters
are computed for a variety of point clouds. This yields the results reported in Table 1. Our experiments measure the point cloud spacing using the value μ given by Eq. 5. This convergence test indicates a first order error in μ, and shows that the regularity of the error improves as the discretization parameters 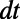 and h are refined.
and h are refined.
Table 1.
Max norm relative errors for the heat equation on a sphere.
| μ | 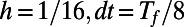 |
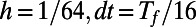 |
| 0.11025 | 0.0622 | 0.0728 |
| 0.05585 | 0.0346 | 0.0326 |
| 0.02792 | 0.0154 | 0.0152 |
| 0.01401 | 0.0087 | 0.0075 |
| — | — | — |
| 0 | 0.0021 | 0.0010 |
Reaction–Diffusion Intrinsic to a Surface
Many surface processes are modeled by systems of reaction–diffusion equations. For illustrative purposes, this article considers systems of two equations:
Applying a forward Euler discretization for the reaction terms and a backward Euler discretization for the diffusion terms yields:
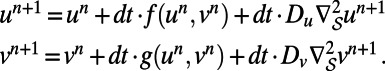 |
This decouples the equations for 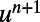 and
and 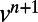 . Similar to the scalar diffusion case, we alternate between a convolutional discretization and a reextension using tricubic interpolation. In Matlab, the system can be evolved to time
. Similar to the scalar diffusion case, we alternate between a convolutional discretization and a reextension using tricubic interpolation. In Matlab, the system can be evolved to time 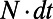 using:
using:
 |
where Ku_hat and Kv_hat are the discrete Fourier transforms of the diffusion kernels for 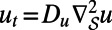 and
and 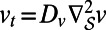 , respectively. As in the scalar diffusion case, X, Y, Z are matrices specifying the
, respectively. As in the scalar diffusion case, X, Y, Z are matrices specifying the 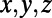 coordinates of the grid nodes, and CPx, CPy, CPz are the corresponding coordinates of the closest points in the point cloud.
coordinates of the grid nodes, and CPx, CPy, CPz are the corresponding coordinates of the closest points in the point cloud.
Patterns on Surfaces.
Fig. 1 displays the result of a computation for the Gray–Scott model (10) for pattern formation (11). This model has reaction terms: 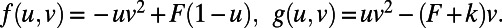 The parameters are chosen to be
The parameters are chosen to be 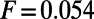 ,
, 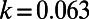 ,
, 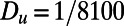 , and
, and 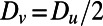 . The computation starts with a perturbation of the steady-state values of u and v localized near the right of the Drosophila embryo, and continues until
. The computation starts with a perturbation of the steady-state values of u and v localized near the right of the Drosophila embryo, and continues until 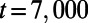 . By choosing this as our final time, we stop the evolution before the entire surface is covered in stripes. The Drosophila embryo surface (12) consists of 5,709 cells, which form a point cloud about four units long. We discretize using a
. By choosing this as our final time, we stop the evolution before the entire surface is covered in stripes. The Drosophila embryo surface (12) consists of 5,709 cells, which form a point cloud about four units long. We discretize using a 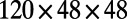 lattice on a
lattice on a 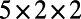 box and select a time step-size of
box and select a time step-size of 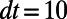 . In our visualization, each point in the point cloud is represented by a small sphere colored according to the value of the solution u.
. In our visualization, each point in the point cloud is represented by a small sphere colored according to the value of the solution u.
Fig. 1.
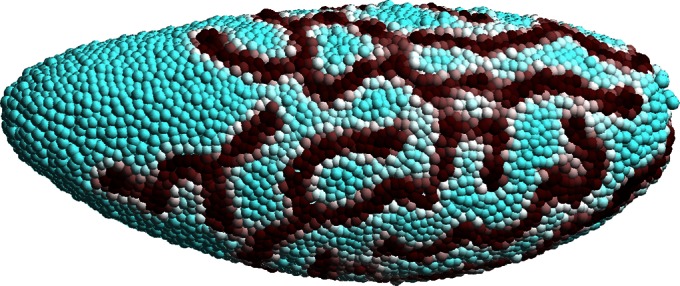
A Turing pattern on a point cloud Drosophila embryo surface (5,709 points).
Fig. 2 also shows pattern formation, in this case for the Brusselator (13), a model that has reaction terms: 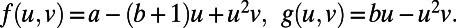 To give interesting steady patterns, the parameters are chosen to be
To give interesting steady patterns, the parameters are chosen to be 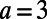 ,
, 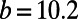 ,
, 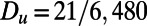 , and
, and 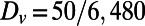 . The initial conditions are perturbations around the steady state. The sea shell is a surface represented by a cloud of 39,733 points (14), and is about 2.2 units wide. A lattice of
. The initial conditions are perturbations around the steady state. The sea shell is a surface represented by a cloud of 39,733 points (14), and is about 2.2 units wide. A lattice of 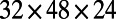 points and a time step-size of 0.1 is used. In this example and the excitable media example below, visualizations are obtained using Matlab’s trisurf() command and a known triangulation. We emphasize, however, that a triangulation is not used in the PDE solver itself. Note that effective visualization methods are available when surface triangulations are unavailable (see ref. 15 for some examples). It is also possible to perform real-time ray-casting on the closest point representation of the surface
points and a time step-size of 0.1 is used. In this example and the excitable media example below, visualizations are obtained using Matlab’s trisurf() command and a known triangulation. We emphasize, however, that a triangulation is not used in the PDE solver itself. Note that effective visualization methods are available when surface triangulations are unavailable (see ref. 15 for some examples). It is also possible to perform real-time ray-casting on the closest point representation of the surface  (16), although we have not explored this option for point clouds using
(16), although we have not explored this option for point clouds using  .
.
Fig. 2.
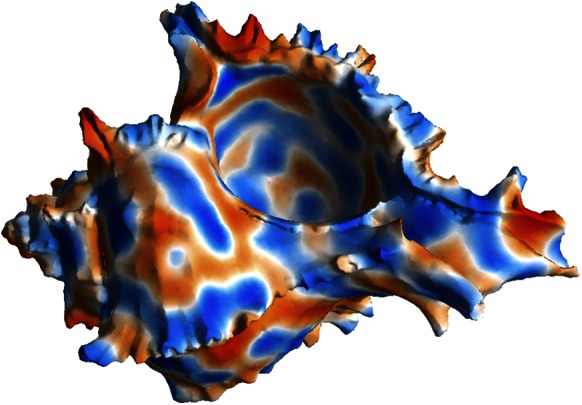
A Turing pattern on a point cloud sea shell (39,733 points).
Patterns on Solid Objects.
Fig. 3 displays another result for the Brusselator model with these same choices of parameters and initial conditions. However, the point cloud in this example consists of points selected randomly between spheres of radii 0.5 and 1. The same code is used for this 3D solid region computation as for our 2D surface computations. Zero flux boundary conditions naturally arise, an effect that leads to stripes intersecting the boundaries at about 90°. Dirichlet boundary conditions may alternatively be imposed; for such problems, the known boundary values are propagated out as part of the extension step. The analytical form of the shape is not used to compute the solution; it is only used as part of the visualization. A 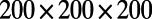 lattice is used with a time step-size of 0.1.
lattice is used with a time step-size of 0.1.
Fig. 3.

A Turing pattern on a point cloud representing the 3D region between spheres of radii 0.5 and 1 (∼800,000 randomly placed points form the point cloud). To visualize the result, we have cut away one-fourth of the region.
This example considers a reaction–diffusion equation with homogeneous Neumann boundary conditions on a densely sampled 3D spherical shell. Suppose we consider the corresponding family of problems on solid spherical shells that vary by thickness. In the limit as the thickness tends to zero, we obtain a reaction–diffusion equation intrinsic to a sphere. It is interesting to note that our approach can compute the solution to this entire family of problems, including the limiting case on a sphere, with the same code and without imposing any assumptions on the dimensionality of the object upon which we compute.
Excitable Media.
Fig. 4 is the result of a computation for the Fitzhugh–Nagumo model (17, 18), which is a model for excitable media (19). This model has reaction terms: 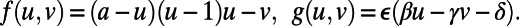 To obtain traveling waves, appropriate parameters and initial conditions must be used. We take
To obtain traveling waves, appropriate parameters and initial conditions must be used. We take 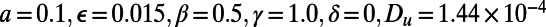 and
and 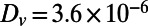 . For initial conditions, the variable u is set equal to 1 for
. For initial conditions, the variable u is set equal to 1 for 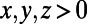 and 0 elsewhere, while v is set equal to 1 for
and 0 elsewhere, while v is set equal to 1 for 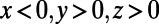 and 0 elsewhere. The surface, the iconic heart (20), is represented by a point cloud of 57,346 points. It is centered at the origin and is about 2.15 units in height. A lattice of
and 0 elsewhere. The surface, the iconic heart (20), is represented by a point cloud of 57,346 points. It is centered at the origin and is about 2.15 units in height. A lattice of 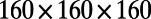 points with a time step-size of 0.875 is used.
points with a time step-size of 0.875 is used.
Fig. 4.

Fitzhugh–Nagumo waves of excitation (dark green) computed on the surface of the iconic heart (defined by 57,346 points).
Bulk Coupling.
Our method also solves surface reaction–diffusion equations that are coupled to processes occurring in the bulk surrounding space. Typically, the surface is coupled to the bulk through the boundary conditions of the bulk equation.
Suppose we have two species, each of which is present both in the bulk and on the surface. Further, suppose that each species can transfer between the surface and bulk via adsorption and desorption. Such situations arise, for example, when studying surfactants (21) or cell-level processes (22). We denote the concentrations of the species on the surface by u and v and the corresponding concentrations in the bulk by U and V. A possible model describing this type of behavior is:
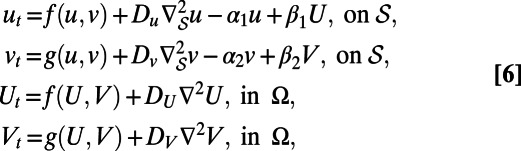 |
with coupling boundary conditions:
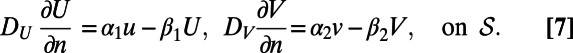 |
Here 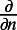 represents the normal derivative outward from the bulk at the surface. The adsorption and desorption between the surface and bulk is controlled by the α and β parameters.
represents the normal derivative outward from the bulk at the surface. The adsorption and desorption between the surface and bulk is controlled by the α and β parameters.
Fig. 5 shows an example computation where the surface is the Drosophila embryo from before but now coupled to an inner bulk process. Portions of the surface have been cut away in the visualization to show the bulk solution in the interior. Here the dynamics are the Schnakenberg process (23) for spot formation: 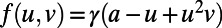 and
and 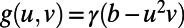 . For parameter values, we use
. For parameter values, we use 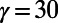 ,
, 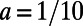 ,
, 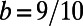 ,
, 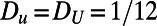 ,
, 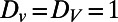 ,
, 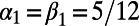 , and
, and 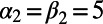 . The coupled Drosophila system is embedded inside a
. The coupled Drosophila system is embedded inside a 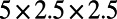 box that we discretize using a
box that we discretize using a 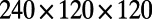 grid. Note that because the surface process is implemented on a regular Cartesian grid in 3D, we reuse this grid for the bulk process.
grid. Note that because the surface process is implemented on a regular Cartesian grid in 3D, we reuse this grid for the bulk process.
Fig. 5.
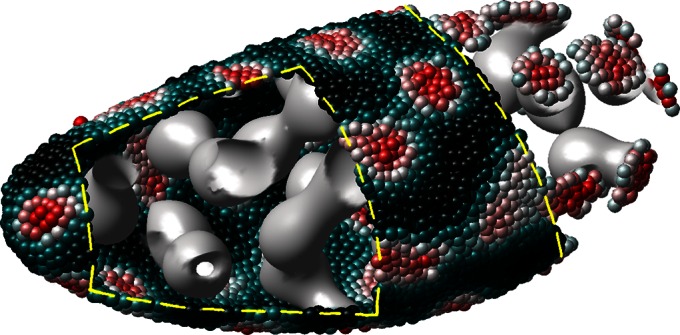
Coupling a diffusion equation in the bulk to a Turing surface pattern formation problem. Some of the point cloud  has been hidden to see the interior bulk.
has been hidden to see the interior bulk.
As in all of our examples, the numerical computation consists of alternating between two steps. The second step simply evolves 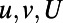 and V for one time-step using the same convolution-based technique as our previous examples. The first step extends the various quantities throughout the computational domain. For the quantities u and v, this extension is simply
and V for one time-step using the same convolution-based technique as our previous examples. The first step extends the various quantities throughout the computational domain. For the quantities u and v, this extension is simply 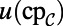 and
and 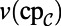 as before. For the first two equations in Eq. 6, we also need the value of the bulk quantities evaluated on the surface, but extended to the computational domain; these are
as before. For the first two equations in Eq. 6, we also need the value of the bulk quantities evaluated on the surface, but extended to the computational domain; these are 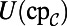 and
and 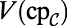 . For the latter two equations in Eq. 6, no extension is required for grid points in the interior of the Drosophila embryo. For grid points outside the Drosophila embryo, the values must be extended in such a way as to satisfy the coupling boundary conditions (Eq. 7). Using the extensions already computed above, we approximate the surface values and the normal derivative as follows:
. For the latter two equations in Eq. 6, no extension is required for grid points in the interior of the Drosophila embryo. For grid points outside the Drosophila embryo, the values must be extended in such a way as to satisfy the coupling boundary conditions (Eq. 7). Using the extensions already computed above, we approximate the surface values and the normal derivative as follows:
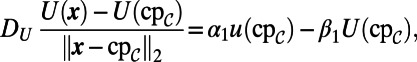 |
where we have approximated 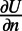 using a finite difference in the normal direction. We then rearrange this formula for
using a finite difference in the normal direction. We then rearrange this formula for 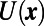 to define our extension for U when
to define our extension for U when  is an exterior point (U is left unchanged for interior points). This process is essentially an extrapolation based on the value of the normal derivative given on the surface to points off the surface. Unlike in the previous examples, this model requires knowledge of which grid points are inside of the surface and which are outside. For a simple surface such as the Drosophila embryo, we determine the interior by comparing the grid point
is an exterior point (U is left unchanged for interior points). This process is essentially an extrapolation based on the value of the normal derivative given on the surface to points off the surface. Unlike in the previous examples, this model requires knowledge of which grid points are inside of the surface and which are outside. For a simple surface such as the Drosophila embryo, we determine the interior by comparing the grid point  , its closest point
, its closest point 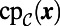 , and the centroid of the point cloud. More sophisticated procedures to determine orientation may be necessary for complex geometry.
, and the centroid of the point cloud. More sophisticated procedures to determine orientation may be necessary for complex geometry.
More generally, there could be bulk concentrations both inside and outside the surface. In such models, knowledge of the orientation of the surface is crucial to correctly approximate the coupling boundary conditions. Our approach to such problems assigns an orientation by splitting a bulk concentration U into two (global) quantities: 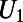 (the inner concentration) and
(the inner concentration) and 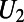 (the outer concentration).
(the outer concentration).
Discussion
We now contrast the present approach with the other methods that have been developed for solving PDEs on surfaces. While most of these other methods do not immediately extend to point clouds, they could generally be used if the point cloud were first reconstructed into a suitable surface representation, a process that can be highly complex.
The first, and potentially most efficient, broad class of numerical methods for solving PDEs on surfaces are parameterization methods. These methods explicitly parameterize the surface by a smooth function of two Cartesian coordinates, α and β. Writing out the PDEs in terms of α- and β-derivatives and metric coefficients results in complicated formulas and, typically, singularities that require special care (24). To handle complex geometries and avoid such singularities, local patch approximations, such as splines, must be constructed that cover the entire surface as a collection of local smooth coordinate systems. This results in substantial organizational complexity. Note, however, there are recent methods that work on point clouds by performing the parameterization locally for each point in the cloud, using the nearest neighbors and a moving least squares problem (1). The second broad class of methods for solving PDEs on complex surfaces is to approximate the surface by a polyhedral triangulation, and use a finite element or finite volume type of method to discretize the equations (25, 26). In the case where only a point cloud is provided, these methods require an intermediate step of constructing a numerically well-behaved triangulation of the point cloud, which can be a considerable challenge for complex surfaces. Furthermore, discretizing and solving PDEs on such polyhedral grids can be deceptively difficult to implement, as described in ref. 26. The third and final broad class of methods are the embedding methods, which solve the surface PDEs in some 3D region encompassing the surface, using standard Cartesian-grid methods (3, 27–32). These methods do not use the metric tensor on the surface, thus eliminating one of the major technical complexities of working with PDEs on surfaces. However, embedding methods typically alter the surface PDEs and introduce artificial boundary conditions in sophisticated ways to extend the problem to a 3D neighborhood of the surface. Traditional embedding methods also require some continuous surface representation, typically the zero-level set of a smooth function defined on all of space (as in the level set method), rather than a point cloud. The accurate and efficient construction of such representations of the surface is a challenge that remains a subject of current research interest, especially for open surfaces, or surfaces of codimension-two or higher (33, 34).
The method presented here, the closest point method, belongs to the general class of embedding methods, but distinguishes itself in several major respects. In particular, the extended PDE for the closest point method is just the classical 3D analog of the surface PDE, a fact that enables the use of standard 3D discretizations. Also, the representation of the surface (i.e., the closest point function) is simple and robust, and it immediately extends beyond surfaces to arbitrary point clouds, and thus directly to the input data format. In the special case of pure diffusion, the present method bears some similarity to the embedding method introduced by Schwartz et al. (27). In particular, both methods rely on the fact that under suitable conditions the solution to a standard 3D diffusion equation provides a close approximation to the solution of the surface diffusion equation. However, ref. 27 formulates the problem using “cut cell” discretizations, the introduction of Neumann boundary conditions, and sophisticated level-set PDE methods for reconstructing the surface of interest as a level set function. In contrast, the closest point method uses only a standard Cartesian grid discretization, no boundary condition discretizations, and is based on the easily computed closest point function. The last of these properties ensures that no surface reconstruction is needed, and allows the method to be immediately generalized from surfaces to point clouds, and other more general geometric objects (e.g., objects of mixed dimensions).
As we have seen, our method solves systems of reaction–diffusion equations on surfaces. The surface representation can be given as an unorganized point set, a property that makes the method easy to implement for complex surfaces such as those acquired from laser range scanners. Note, however, that the fundamental principles underlying the closest point method apply to much more general PDEs. See, for example, refs. 3, 5, 35, where the closest point method is used to solve an interesting variety of PDEs on complex continuous surfaces. Our ongoing work for the closest point method investigates solving general PDEs on general point cloud surfaces using only classical 3D numerical methods and solution routines. We anticipate that the development of methods of this type should encourage powerful models of continuum processes on surfaces to be applied more widely, across diverse sciences and in new research areas.
Acknowledgments
C.B.M. thanks Dr. Chandrasekhar Venkataraman (University of Sussex) for useful discussions on bulk-coupled reaction–diffusion models. The work of C.B.M. was supported by Award KUK-C1-013-04 from King Abdullah University of Science and Technology (KAUST). The work of S.J.R. was partially supported by a Natural Sciences and Engineering Research Council of Canada Discovery Grant and by Award KUK-C1-013-04 from KAUST.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
References
- 1.Liang J, Zhao H-K. Solving partial differential equations on point clouds. UCLA CAM Reports. 2012:12–25. [Google Scholar]
- 2.Lai R-J, Liang J, Zhao H-K. A local mesh method for solving PDEs on point clouds. UCLA CAM Report. 2012:12–60. [Google Scholar]
- 3.Ruuth SJ, Merriman B. A simple embedding method for solving partial differential equations on surfaces. J Comput Phys. 2008;227(3):1943–1961. [Google Scholar]
- 4.März T, Macdonald CB. Calculus on surfaces with general closest point functions. SIAM J Numer Anal. 2012;50(6):3303–3328. [Google Scholar]
- 5.Macdonald CB, Ruuth SJ. The implicit closest point method for the numerical solution of partial differential equations on surfaces. SIAM J Sci Comput. 2009;31(6):4330–4350. [Google Scholar]
- 6.Boyd JP. Chebyshev and Fourier Spectral Methods 2nd Rev Ed. New York: Courier Dover Publications; 2001. [Google Scholar]
- 7.Spivak M, Veerapaneni SK, Greengard L. The fast generalized Gauss transform. SIAM J Sci Comput. 2010;32(5):3092–3107. [Google Scholar]
- 8.Greengard L, Strain J. The fast Gauss transform. SIAM J Sci Statist Comput. 1991;12(1):79–94. [Google Scholar]
- 9.Saff EB, Kuijlaars ABJ. Distributing many points on a sphere. The Mathematical Intelligencer. 1997;19(1):5–11. [Google Scholar]
- 10.Pearson JE. Complex patterns in a simple system. Science. 1993;261(5118):189–192. doi: 10.1126/science.261.5118.189. [DOI] [PubMed] [Google Scholar]
- 11.Turing AM. The chemical basis of morphogenesis. Philos Trans R Soc Lond. 1952;B237(641):37–72. [Google Scholar]
- 12.Berkeley Drosophila Transcription Network Project 2010. http://bdtnp.lbl.gov. Accessed January 1, 2010.
- 13.Prigogine I, Lefever R. Symmetry breaking instabilities in dissipative systems. II. J Chem Phys. 1968;48(4):1695–1700. [Google Scholar]
- 14. NextEngine (2009) NextEngine “shell”. www.nextengine.com/gallery/shell. Accessed May 2, 2009.
- 15.Kobbelt L, Botsch M. A survey of point-based techniques in computer graphics. Comput Graph. 2004;28(6):801–814. [Google Scholar]
- 16.Auer A, Macdonald CB, Treib M, Schneider J, Westermann R. Real-time fluid effects on surfaces using the Closest Point Method. Comput Graph Forum. 2012;31(6):1909–1923. [Google Scholar]
- 17.Fitzhugh R. Impulses and physiological states in theoretical models of nerve membrane. Biophys J. 1961;1(6):445–466. doi: 10.1016/s0006-3495(61)86902-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagumo J, Arimoto S, Yoshizawa S. Active pulse transmission line simulating nerve axon. Proc IRE. 1962;50(10):2061–2070. [Google Scholar]
- 19.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117(4):500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cbspicer (2008) “Iconic Heart” from the TurboSquid 3D stock catalog. www.turbosquid.com/FullPreview/Index.cfm/ID/389728. Accessed January 20, 2011.
- 21.Teigen KE, Li X, Lowengrub J, Wang F, Voigt A. A diffuse-interface approach for modeling transport, diffusion and adsorption/desorption of material quantities on a deformable interface. Commun Math Sci. 2009;4(7):1009–1037. doi: 10.4310/cms.2009.v7.n4.a10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novak IL, et al. Diffusion on a curved surface coupled to diffusion in the volume: Application to cell biology. J Comput Phys. 2007;226(2):1271–1290. doi: 10.1016/j.jcp.2007.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schnakenberg J. Simple chemical reaction systems with limit cycle behaviour. J Theor Biol. 1979;81(3):389–400. doi: 10.1016/0022-5193(79)90042-0. [DOI] [PubMed] [Google Scholar]
- 24.Floater MS, Hormann K. Surface parametrization: a tutorial and survey. In: Dodgson N, Floater M, Sabin M, editors. In Advances in Multiresolution for Geometric Modelling. Heidelberg, Germany: Springer; 2005. pp. 157–186. [Google Scholar]
- 25.Dziuk G, Elliott CM. Eulerian finite element method for parabolic PDEs on implicit surfaces. Interf Free Bound. 2008;10:119–138. [Google Scholar]
- 26.Reuter M, Wolter FE, Peinecke N. Laplace-Beltrami spectra as ‘Shape-DNA’ of surfaces and solids. Comput Aided Des. 2006;38(4):342–366. [Google Scholar]
- 27.Schwartz P, Adalsteinsson D, Colella P, Arkin AP, Onsum M. Numerical computation of diffusion on a surface. Proc Natl Acad Sci USA. 2005;102(32):11151–11156. doi: 10.1073/pnas.0504953102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bertalmio M, Cheng L-T, Osher S, Sapiro G. Variational problems and partial differential equations on implicit surfaces. J Comput Phys. 2001;174(2):759–780. [Google Scholar]
- 29.Xu J-J, Zhao H-K. An Eulerian formulation for solving partial differential equations along a moving interface. J Sci Comput. 2003;19(1-3):573–594. [Google Scholar]
- 30.Leung S-Y, Zhao H-K. A grid based particle method for moving interface problems. J Comput Phys. 2009;228(8):2993–3024. [Google Scholar]
- 31.Leung S-Y, Lowengrub J, Zhao H-K. A grid based particle method for solving partial differential equations on evolving surfaces and modeling high order geometrical motion. J Comput Phys. 2011;230(7):2540–2561. [Google Scholar]
- 32.Greer JB. An improvement of a recent Eulerian method for solving PDEs on general geometries. J Sci Comput. 2006;29(3):321–352. [Google Scholar]
- 33.Zhao H-K, Osher S, Fedkiw R. In Proceedings of the IEEE Workshop on Variational and Level Set Methods. Vancouver, Canada: IEEE Computer Society; 2001. Fast surface reconstruction using the level set method; pp. 194–201. [Google Scholar]
- 34.Goldstein T, Bresson X, Osher S. Geometric applications of the split Bregman method: segmentation and surface reconstruction. J Sci Comput. 2010;45(1-3):272–293. [Google Scholar]
- 35.Macdonald CB, Ruuth SJ. Level set equations on surfaces via the Closest Point Method. J Sci Comput. 2008;35(2-3):219–240. [Google Scholar]


