Abstract
Recognizing and quantifying specific biomolecules in aqueous samples are constantly needed in research and diagnostic laboratories. As the typical detection procedures are rather lengthy and involve the use of labeled secondary antibodies or other agents to provide a signal, efforts have been made over the last 10 y to develop alternative label-free methods that enable direct detection. We propose and demonstrate an extremely simple, low-cost, label-free biodetector based on measuring the intensity of light reflected by the interface between a fluid sample and an amorphous fluoropolymer substrate having a refractive index very close to that of water and hosting various antibodies immobilized in spots. Under these index-matching conditions, the amount of light reflected by the interface allows straightforward quantification of the amount of antigen binding to each spot. Using antibodies targeting heterologous immunoglobulins and antigens commonly used as markers for diagnoses of hepatitis B and HIV, we demonstrate the limit of detection of a few picograms per square millimeter of surface-bound molecules. We also show that direct and real-time access to the amount of binding molecules allows the precise extrapolation of adhesion rates, from which the concentrations of antigens in solution can be estimated down to fractions of nanograms per milliliter.
Keywords: immunoassay, optical biosensor, protein microarray, biomolecular detection, reflective phantom interface
The increasing importance of biomarker detection in medical diagnostics (1) and the expected demand for distributed diagnostic devices in healthcare systems (2) have stimulated the search for novel technical solutions to rapidly detect specific proteins or nucleic acid fragments in biological fluids (3, 4), and, over the last decade, a number of new methods of detecting and characterizing biomolecular interactions have been proposed (5, 6).
The well-established and extensively used methods, such as ELISA (7, 8), which is used in research and diagnostic laboratories, and the so-called “lateral flow tests”, which are more suitable for point-of-care diagnostics (9–11), are based on indirect detection by means of “labeled” secondary probes that also bind to the target and provide a measurable optical, electrical, or radioactive signal. When the molecular interaction and the instrumental detection of the labeling moieties are optimized, these approaches can be very sensitive and specific. Arguably the most powerful ELISA format is the sandwich assay, in which the target molecule is recognized by a couple of antibodies: One is immobilized on a surface and provides the capture function; and the other is added to the solution and enables the detection, optionally through the use of a third secondary antibody carrying the labeling moiety. Although the recognition by multiple antibodies at the same time can provide an effective increase of specificity, it requires additional steps that further complicate and slow down the procedure and effectively limit the capability of parallel detection of multiple targets in the same sample, because of the increased chances of cross-reactivity between different targets and probes (12).
These limitations can in principle be overcome by “label-free” methods, which are based on the direct detection of target molecules by means of their intrinsic effect on some of the physical properties of the sensing surface as they interact with immobilized probes (13–16). The most widely used of these methods are those based on surface plasmon resonance (SPR), in which the optical reflectance of a surface coated with a thin layer of gold is modified by the presence of biomolecules (17–19). Other optical approaches with potentially similar characteristics are those based on measuring the spectral changes in the light reflected by a suitably structured surface (14, 20, 21). However, despite their ability to provide multiplex, real-time detection, the diagnostic use of label-free methods is still limited by their relatively lower sensitivity and specificity and the fact that they typically require more sophisticated measuring equipment than that used in the case of indirect detection (22).
We describe an extremely simple and potentially low-cost, multiplex, label-free detection method based on measuring the weak intensity of the light reflected by the functionalized surface of a plastic material whose refractive index is very close to that of water. The target molecules interacting with the probes immobilized on the sensing surface are directly detected and quantified on the basis of the local increase in optical reflectivity. We name this detection method reflective phantom interface (RPI). Its effectiveness is here demonstrated in the context of protein microarrays. Remarkably, the analysis of something as simple as the reflection images of the functionalized surface can detect picomolar concentrations of immunoglobulins and hepatitis B and HIV antigens. Real-time monitoring of the interactions provides intrinsic time-to-result optimization, thus enabling the multiplex detection of nanograms per milliliter of proteins in a few minutes and of even lower concentrations with longer measuring times.
Results
RPI Method.
Perfluorinated polymeric materials can be optimized to ensure high transparency within the visible range and a refractive index close to that of water. In previous studies, we have used these properties to create suspensions of functionalized nanocolloids whose scattering cross-section greatly depended on the amount of carbon-based molecules adhering to their surfaces and can therefore be used to signal nonspecific hydrophobic interactions or specific ligand/receptor bindings (6, 23, 24). The drawbacks of that approach were the tendency of the particles to cluster and the large number of molecules involved in the surface interactions—proportional to the always very large total interface of nanoparticle suspensions.
Based on the same class of materials, the much more sensitive RPI method exploits Fresnel basic laws of optical reflection (25): As the reflectivity of a flat interface separating two transparent materials with refractive indexes n1 and n2 is proportional to (n1 – n2)2, the intensity of reflected light can be made arbitrarily small by matching n1 and n2. In this study, we used Hyflon AD (trademarked product of Solvay Specialty Polymers Italy), a perfluorinated amorphous copolymer of tetrafluoroethylene (Materials and Methods) with a refractive index of n1 = 1.327 for red light. When immersed in water (n2 = 1.333), a polished surface of Hyflon AD has a reflectivity at normal incidence of less than 10−5, basically becoming invisible to the naked eye. Under these conditions, the interface accumulation of even a small quantity of molecules with a significantly different refractive index nL leads to an increase of the intensity of reflected light that is small in absolute terms, but significant compared with the reflectivity of the bare surface. The RPI method uses this increase to measure the amount of target molecules interacting with specific probes at the interface.
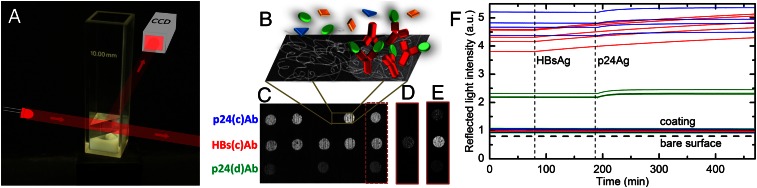
Fig. 1 shows an extremely simple means of using this approach based on widely available low-cost components. The sensing surface is provided by the diagonal face of a right angle prism of Hyflon AD, which is placed into a standard 1-cm cuvette and submerged in an aqueous solution constantly stirred by a magnetic bar (Fig. 1A). The prism is coated with a multifunctional copolymer of dimethylacrylamide (DMA), N-acryloyloxysuccinimide (NAS), and 3-(trimethoxysilyl) propyl methacrylate (MAPS)—copoly(DMA-NAS-MAPS)—that provides reactive groups suitable for immobilizing the probe molecules and simultaneously prevents nonspecific adsorption of the components of biological fluids (26). Different antibodies are covalently immobilized by the copolymer on the diagonal surface of the prism in 200-μm spots (Fig. 1B). The spotted surface of the prism is illuminated by the light of an inexpensive light-emitting diode (LED), and the reflected intensity is imaged by a standard CCD camera. The experiments are performed by acquiring a sequence of images while adding a solution containing the target molecules to the cuvette. Each pixel of the acquired image records the local reflectivity u(t) of the sensing surface and its variations over time t as more molecules adhere to the surface.
Fig. 1.
Optical setup and measured intensity of reflected light. (A) A standard 1-cm cuvette containing a prism of Hyflon AD is shown. The diagonal face of the prism has been functionalized with DMA-NAS-MAPS copolymer and spotted antibodies. The prism is held by a plastic support that also houses a stirring magnetic bar. The light emitted by a LED is reflected by the sensing surface and acquired by a CCD camera. The temperature of the cuvette is maintained at 37 °C ± 0.1 °C. (B) Schematic illustration of the functionalized surface showing the copolymer as gray threads, the immobilized antibodies, and the target molecules, some of them bound to antibodies. (C) Image of the light reflected by the prism surface spotted with antibodies targeting proteins HBsAg and p24Ag acquired before the addition of the antigens in solution. Multiple spots of the same antibody are placed in the same row, as shown. The image of the three spots on the right-hand side of C is subtracted from the image of the same area acquired 110 min after the addition of 50 ng/mL HBsAg in solution and 280 min after the subsequent addition of 50 ng/mL p24Ag. The resulting brightened images are, respectively, shown in D and E. (F) Reflected light intensity of the spots of HBs(c)Ab (red), p24(c)Ab (blue), and p24(d)Ab (green) shown in C, measured before and after the addition of the corresponding target molecules in solution (injection times are indicated by the vertical dashed lines). The colored lines around the unit value of reflected light represent the intensity of the copolymer-coated surface around each spot. The horizontal dashed line at the bottom indicates the intensity of light reflected by the bare surface.
In the experiments described here, antibodies with the function of coating and detection probes in commercially available ELISA sandwich assays, targeting viral proteins that are commonly used as biomarkers for the diagnosis of hepatitis B (hepatitis B surface antigen, HBsAg) and HIV (p24 capsid protein, p24Ag), were immobilized in rows of spots on the same prism surface. Fig. 1C is a reflection image u(0) recorded after spotting one antibody targeting HBsAg [HBs(c)Ab] and two targeting p24Ag [p24(c)Ab and p24(d)Ab], but before adding the antigens. When the proteins targeted by the antibodies are added to the solution, the brightness of the related spots starts to increase. Fig. 1 D and E shows brightened images of the difference u(t) − u(0) relating to the three spots on the right-hand side of Fig. 1C, acquired after the addition of HBsAg and after the subsequent addition of p24Ag, respectively. Only the spot with HBs(c)Ab antibodies brightens after the addition of the corresponding target proteins (Fig. 1D), whereas the other spots and background area remain basically unchanged; in Fig. 1E, both the p24(c)Ab and the p24(d)Ab spots are lit up and the brightness of the HBs(c)Ab spot is also further increased, as expected because the binding of the previously added HBsAg molecules continues over time. Movie S1 shows the sequence of the difference images of the sensing surface acquired during the experiment.
Fig. 1F shows the time dependence of the brightness of each spot shown in Fig. 1C and the background area surrounding the spots containing only the copolymer coating. The HBsAg and p24Ag antigens were added to the solutions at the times corresponding to the vertical dashed lines. The differences in intensity at t = 0 indicate that the immobilization procedure yields different amounts of antibodies in each spot. Growth in u(t) reflects an increase in the molecular mass on the top of the spots, thus revealing antigen binding. The flatness of the background and the spot regions before the addition of antigen and the absence of extra kinks in the curves at the times the antigens were injected demonstrate that the measurements are free of nonspecific or cross-interactions, thus confirming the effectiveness of the coating copolymer.
Quantification of Bound Molecules.
The reflectivity of a thin molecular layer with a refractive index of nL at the interface between two bulk media can be easily evaluated by means of standard optics (25). This enables us to extract the amount of molecular mass at the interface from the local reflectivity in a simple and straightforward manner. Specifically, the absolute mass of material per unit surface σ(t) can be obtained from the pixel values u(t) as
 |
where u0 is the brightness of the bare surface, which can be directly measured by including an uncoated portion of the sensor surface within the field of view. σ0 is a value that depends on the relevant physical parameter of the system. Assuming nL ∼ 1.42, and given the values of n1 and n2, we obtain σ0 ∼ 4.9 ng/mm2 (more details are given in SI Materials and Methods). Eq. 1 indicates that a doubling in reflectivity in comparison with the bare interface corresponds to σ(t) = σ0 of molecular mass per unit surface. As medium-performance imaging detectors discriminate 0.1% variations in light intensity, Eq. 1 shows that the limit of detection (LOD) of the RPI is in the order of a few picograms per square millimeter, typical of the best-performing label-free instruments.
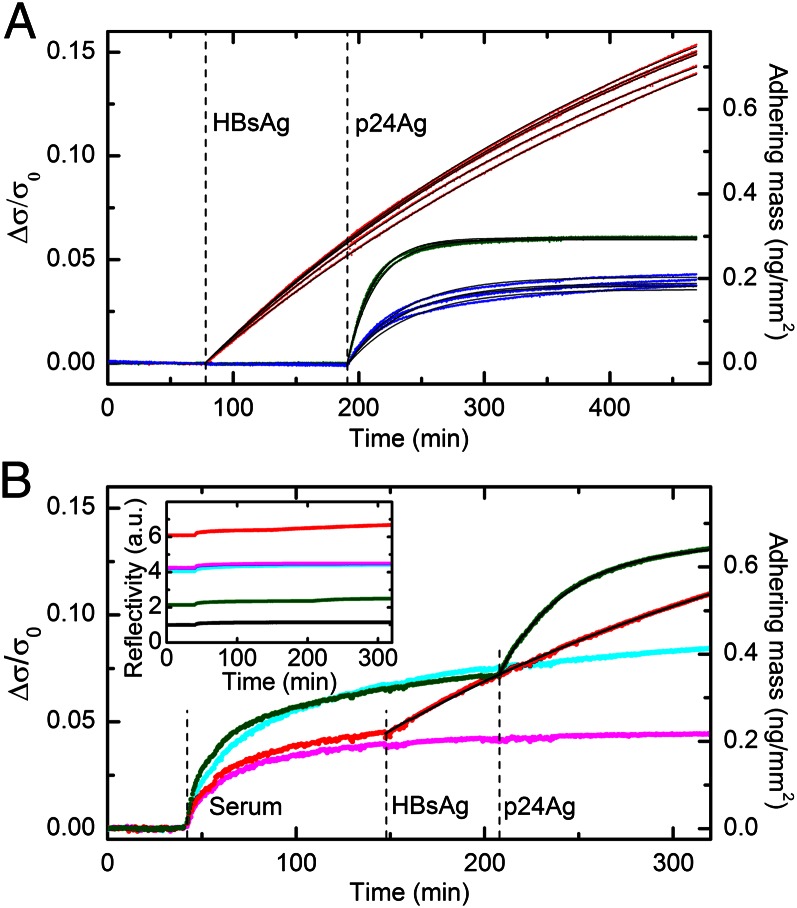
Fig. 2A shows the plot of the data shown in Fig. 1F after their conversion into the corresponding normalized surface density σ/σ0 by means of Eq. 1 and after subtracting the initial value of each curve. The resulting Δσ/σ0 gives the normalized surface density of antigen bound to each spot. Fig. 2A shows two important results: (i) Once converted into Δσ/σ0, the measured reflectivity of each interaction collapses into a narrow bundle of curves; and (ii) the Δσ/σ0 growth curves are generally very well fitted by exponentials (black lines), having similar rates and asymptotic values for each interaction. Multiplying Δσ/σ0 by σ0 provides a direct quantification of the surface mass density of the bound molecules (Fig. 2A, right axis). The plateau Δσ/σ0 values extracted from the exponential fit of the growth curves make it possible to estimate the amount of bound targets at saturation, which in turn enables an estimate of the surface density of binding sites. The asymptotic values of surface mass density of the antigens on the corresponding spots were about 1.2 ng/mm2 for HBs(c)Ab, 0.29 ng/mm2 for p24(c)Ab, and 0.19 ng/mm2 for p24(d)Ab, roughly corresponding to a fractional surface coverage relative to a protein monolayer of about 30% for HBsAg and less than 10% for p24Ag.
Fig. 2.
Quantification of bound target proteins. (A) The data shown in Fig. 1F are converted into the increase of normalized surface density Δσ/σ0 due to the adhesion of target molecules to spots of HBs(c)Ab (red), p24(c)Ab (blue), and p24(d)Ab (green). The right axis shows the corresponding surface density scale of the bound targets, assuming that σ0 = 4.9 ng/mm2 (SI Materials and Methods). The black lines represent single exponential fits yielding rates of 3.8 × 10−5 (±3 × 10−6) s−1 for HBs(c)Ab, 7.8 × 10−4 (±5 × 10−5) s−1 for p24(d)Ab, and 3.6 × 10−4 (±3 × 10−5) s−1 for p24(c)Ab. (B) The amount of adhering mass Δσ/σ0 is shown for two single spots of HBs(c)Ab (red) and p24(d)Ab (green) antibodies and for two control spots, CTR1 (cyan) and CTR2 (magenta). Bovine fetal serum was first added to the incubation buffer (dilution 1:10), and then HBsAg and p24Ag were injected to a final concentration of 52 ng/mL. The times of the additions are indicated by the vertical dashed lines. The right axis shows the corresponding scale of surface density at the top of the antibody spots. The signal contribution due to antigen binding is separated from that of nonspecific adsorption of serum by subtracting the CTR1 and CTR2 signals from the p24(d)Ab and HBs(c)Ab curves, respectively. The resulting curves are then fitted with single exponential functions (Fig. S1). The extracted rates are 4.5 × 10−4 s−1 and 5.8 × 10−5 s−1 for p24Ag and HBsAg, respectively. The black lines represent the exponential fitting curves added to the corresponding smoothened control signals. (Inset) Raw data of reflected light intensity corresponding to the curves shown in B. The black curve represents the signal obtained from the unspotted region.
By means of a similar analysis made by comparing the reflectivity of the spots before the interaction with antigen with that of the copolymer background, it is possible to determine that the amount of immobilized antibodies was 9 (±3) ng/mm2, the variability being due to the differences in functionalization density between the spots. Given the molecular size of IgG antibodies (27), this value is consistent with a tightly packed 2D layer of antibodies on the copolymer coating. Comparing this result with the measured surface densities of target binding sites, antigen:antibody molar ratios of about 1:1 and 1:8 are obtained for HBsAg and p24Ag, respectively. As further discussed below, this difference can be attributed to the reported tendency of HBsAg to self-assemble to virus-like structures (28), thus forming oligomers whose size may depend on the experimental conditions.
Detection in Serum.
To estimate the effect of complex media on the measured interactions and test our technique in a situation in which there are more nonspecific interactions, we repeated the experiment of detecting HBsAg and p24Ag in the presence of a 1:10 dilution of bovine fetal serum. Fig. 2B, Inset shows the measured reflectivity of the spots and of the copolymer coating. When the serum was injected into the cuvette, we observed a very small jump due to the limited refractive index change of the solution, followed by a much slower, nonexponential increase due to the nonspecific adsorption of serum components (SI Materials and Methods and Fig. S1). However, the increases were of limited intensity and, more importantly, control spots of suitable antibodies insensitive to target addition provided a reference signal with kinetics and amplitudes similar to those of the spots of probe antibodies. Therefore, the contribution to the signal due to the subsequent addition of HBsAg and p24Ag can be recovered by subtracting the corresponding control spot curves. Remarkably, the binding of antigen is quantitatively similar to that observed in buffer, despite the very high concentration (>10 mg/mL) and the diversity of some of the biomolecular species present in blood, which, in principle, could lead to significant nonspecific binding (29) and compromise the detection method. Using the same estimate for σ0 as that used before, the amount of nonspecific binding saturated at values of surface densities between 200 and 500 pg/mm2, depending on the spotted antibody (Fig. 2B, right axis). The subsequent addition of target antigens led to a further increase of the signal due to specific binding that was well fitted by exponential functions with saturation levels similar to those obtained in buffer.
Binding Kinetics.
The clean exponential binding shown in Fig. 2 offers a means of directly measuring the concentration of antigens in solution by determining adhesion rates. Provided that the sample solution is efficiently and constantly mixed, when the quantity of target molecules in solution is much larger than the number of available binding sites on the surface, the time evolution of the fraction of surface sites bound to a target molecule φ(t) is described by a simple differential equation,
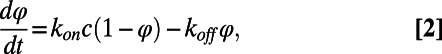 |
where c is the antigen concentration and kon and koff are the association and dissociation rate constants, respectively. The general solution of Eq. 2 is an exponential growth with rate Γ(c) = konc + koff and asymptotic plateau φ∞(c) = (1 + Kd/c)−1, where Kd = koff/kon is the equilibrium dissociation constant of the interaction. Accordingly, the analysis of a suitable set of binding curves obtained from a single spot enables determining the rate constants characterizing the interactions or, alternatively, unknown concentrations of target molecules in solution can be determined from the growth rate of the reflected intensity, when the kinetics parameters for a specific probe–target couple are known.
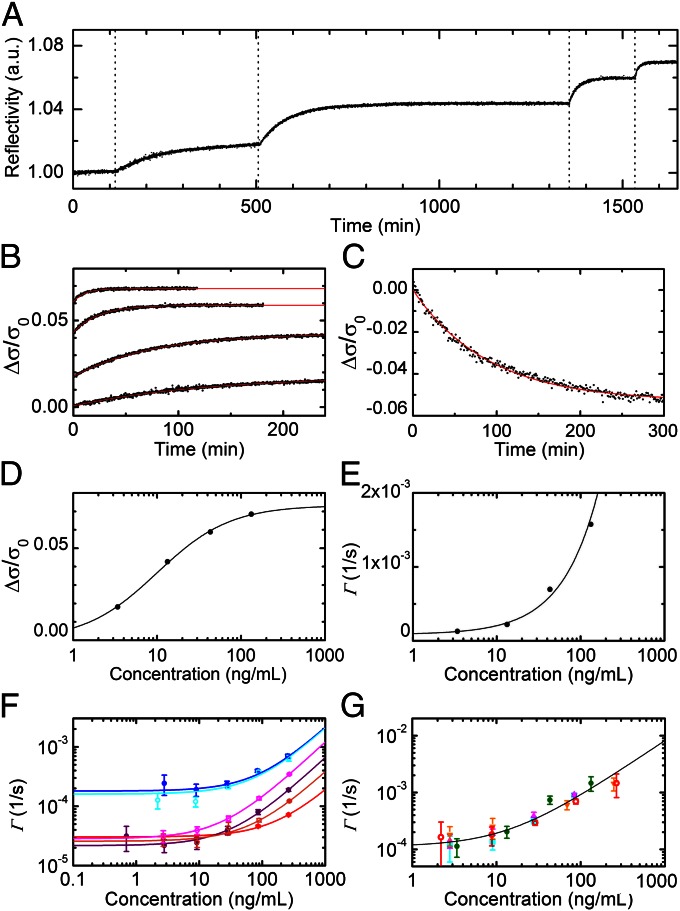
To investigate these possibilities, we first experimentally explored the validity of Eq. 2 on our system by measuring the binding of p24Ag on a single spot of p24(d)Ab as increasing quantities of the antigen were progressively added to the cuvette containing the incubation buffer. The results of this study are presented in Fig. 3 A–E and a similar analysis carried out on p24(c)Ab is reported in Fig. S2. Fig. 3A shows the relative reflectivity of a single spot of p24(d)Ab antibodies measured after the addition of p24Ag in buffer, from 3 to 130 ng/mL. Increasing the concentration of antigen in solution increases the rate of the exponential. The asymptotic value is reached at shorter times with a larger concentration of antigen in solution. Fig. 3B shows the data of Fig. 3A after conversion into bound surface mass by means of Eq. 1. To show the different growth rates more clearly, the data in Fig. 3B are divided into sections corresponding to the different concentrations of antigen in solution, with t = 0 indicating the injection times. The red lines are the exponential fittings to the curves, from which the plateaus φ∞ and the characteristic rates Γ are obtained for each antigen concentration. An independent measure of koff is also obtained, monitoring the reflectivity decrease as a function of time after replacing the target solution with the incubation buffer (Fig. 3C). As shown in Fig. 3 D and E, the observed behavior of both φ∞(c) and Γ(c) is well fitted by the solutions of Eq. 2 (solid lines), whose validity is also demonstrated by the remarkable agreement between the values of Kd independently extracted from the fits of φ∞(c) and Γ(c), being 416 and 301 pM, respectively.
Fig. 3.
Kinetics of spotted antibodies recognition. (A) The time dependence of the reflected intensity of a single spot of p24(d)Ab antibodies was measured while the solution concentration of target protein was brought to 3.4, 13.3, 43.1, and 133 ng/mL at the times indicated by the vertical dotted lines. (B) The reflectivity curves of A are converted into the normalized mass of target molecules adhering to the spot. The curves are shown as a function of the time after each addition (black dots) together with their exponential fits (red lines). (C) A dissociation curve is measured for a p24(d)Ab spot after the experiment reported in A. The normalized mass on the spot decreases after the target solution is replaced with the incubation buffer. The red line represents an exponential fit yielding a koff of 1.8 × 10−4 (±4 × 10−5) s−1. (D) The asymptotic values extracted from the exponential fits reported in B are shown as a function of the target concentration (black circles) and are fitted, according to Eq. 2, by Δσ/σ0 = (Δσmax/σ0)/(1 + Kd/c) (black line), where Δσmax/σ0 = 0.073 (±0.001) and Kd = 416 (±26) pM. (E) The rates of the exponential fits reported in B are shown as a function of the target concentration (black circles) and are fitted by Γ(c) = konc + koff (black line), yielding kon = 2.9 × 105 (±2 × 104) M−1⋅s−1 and koff = 8.6 × 10−5 (±4.8 × 10−5) s−1. (F) Association rates measured for different interactions when increasing concentrations of the corresponding target are progressively added to the solution: p24(c)Ab, HBs(c)Ab, and HBs(d)Ab interactions in incubation buffer are reported as solid blue, red, and purple circles, respectively, whereas the same interactions in diluted bovine serum are represented by open cyan, orange, and magenta circles, respectively. Each value represents the average rate obtained from four to eight spots, and the error bars indicate the corresponding SDs. Each solid line represents a linear fit of the Γ(c) points with the corresponding color. All of the extracted parameters are reported in Table 1. (G) Association rates of p24Ag on spots of p24(d)Ab measured in five experiments similar to those shown in A: The average rate obtained from four to eight spots is reported for four repetitions in incubation buffer (green circles, magenta up triangles, orange down triangles, and cyan squares) and one in diluted bovine serum (open red dots). The error bars indicate the corresponding SDs. The solid line represents a linear fit of all of the reported values of Γ(c), yielding kon = 1.9 × 105 (±2 × 104) M−1⋅s−1 and koff = 1.3 × 10−4 (±2 × 10−5) s−1.
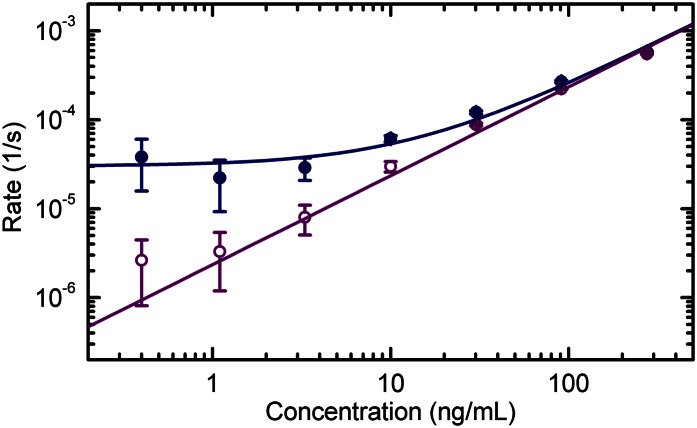
Similar experiments were performed using different antibodies recognizing p24Ag and HBsAg, both in incubation buffer and in diluted bovine serum. An antibody–antibody interaction was also studied in buffer. Remarkably, the validity of Eq. 2 was confirmed for all of the studied interactions over a wide range of concentrations. The measured association curves and the corresponding exponential fits are reported in Figs. S3–S5. For those experiments not sufficiently long to approach equilibrium, we inferred the asymptotic plateaus from the behavior of φ∞(c) observed at the highest concentrations. The determination of such plateau values enabled obtaining the Γ(c) reported in Fig. 3 F and G and Fig. 4 (blue circles and line). The extracted kinetic parameters are reported in Table 1. The values measured for HBsAg and p24Ag in buffer are consistent with those reported for SPR measurements on HBsAg fragment peptides (30) and on different monoclonal antibodies targeting p24Ag (31), respectively. The kinetic constants observed in serum are very close to those measured in buffer, with the partial exception of the kon values of HBsAg, which are found to be slightly higher.
Fig. 4.
Association rate and initial slope of the binding curves. The rates obtained from the exponential fits of the binding curves (solid blue circles) and the slopes of the initial linear growth of φ(t) (open violet circles) are reported for spots of anti-mouse IgG antibodies when increasing concentrations of mouse IgG were progressively added in buffer solution. Each value represents the average rate obtained from six spots, and the error bars indicate the corresponding SDs. The solid lines represent linear fits with Γ(c) = konc + koff (blue) and Γi(c) = konc (violet), being the values of kon obtained from the two analyses substantially identical. The obtained values of kon and koff are reported in Table 1.
Table 1.
Measured kinetic parameters
| Method | Antibody | Solution | kon (104 M−1⋅s−1) | koff (10−5 s−1) |
| RPI | p24(c)Ab | Buffer | 4.7 ± 0.5 | 18 ± 3 |
| Serum | 4.6 ± 0.5 | 16 ± 4 | ||
| p24(d)Ab | Buffer | 19.4 ± 2 | 11 ± 2 | |
| Serum | 14.4 ± 2 | 14 ± 2 | ||
| HBs(c)Ab | Buffer | 0.37 ± 0.2 | 3.0 ± 1 | |
| Serum | 0.82 ± 0.1 | 2.6 ± 1 | ||
| HBs(d)Ab | Buffer | 1.5 ± 0.6 | 2.2 ± 1 | |
| Serum | 2.7 ± 0.1 | 2.8 ± 0.5 | ||
| Anti-IgG | Buffer | 35 ± 8 | 3.0 ± 0.5 | |
| SPR | p24(d)Ab | Buffer | 5.0 ± 0.14 | 17 ± 9.7 |
| HBs(c)Ab | Buffer | 0.5 ± 0.02 | — |
The reproducibility of the results obtained with RPI was tested by performing five detection experiments of p24Ag with different prisms spotted with p24(d)Ab. The rates Γ(c) obtained in such repeated experiments, shown in Fig. 3G, display a limited variability, being the relative SDs of the extracted values of kon and koff less than 20%.
To further validate the results obtained with the proposed RPI method, we performed kinetics experiments on p24(d)Ab and HBs(c)Ab in buffer, using a widely used SPR instrument, as detailed in SI Materials and Methods (Figs. S6 and S7). As reported in Table 1, the SPR measurements yielded a slightly slower kon in the case of p24(d)Ab, whereas we found a remarkable agreement on the koff of the same interaction and on the kon of HBs(c)Ab. Interestingly, with none of the two approaches was it possible to directly observe a dissociation of HBsAg after washing with incubation buffer. This phenomenon can be ascribed to the formation of HBsAg oligomers in solution (28), which can bind to multiple immobilized antibodies, and thus the interaction can be strongly stabilized. This is consistent with the observation of high surface densities of HBsAg saturating both the RPI and SPR functionalized surfaces and with the anomalously low values of the measured association rate constants. In fact, the presence of oligomers reduces the molar concentration of independent antigen particles in solution, thus affecting the analysis based on the total mass concentration of antigen.
Discussion
The simplicity of the method and the use of label-free detection make it possible to control all of the stages in the development and optimization phases of the assay and open up a clear means of optimizing the functionalization protocols and antibody selection. First of all, the RPI method enables one to quantify the deposition of the copolymer. Indeed, the difference between the reflectivity of the bare surface (dashed line in Fig. 1F) and that of the surface after copolymer deposition enabled us to determine the average amount of the coating mass to be about 1 ng/mm2, which is consistent with previous estimates performed on the same copolymer by a different method (32). Additionally, we could quantify the amount of unwanted, nonspecific adsorption when the sensing surface is exposed to a complex medium such as serum. The quantity of serum components adhering to the surface slightly differs in the case of the unspotted copolymer coating and for spots of different antibodies, being the measured surface density for 1:10 dilution of bovine serum never larger than 500 pg/mm2. This value is remarkably low and does not prevent the detection of the target, as shown in Fig. 2B, although with slightly worse performance than in buffer solution. The extraordinary low levels of nonspecific adsorption on the copolymer coating used are also confirmed by experiments carried out by SPR and by dual polarization interferometry (Fig. S8). In particular, the amount of adhering serum components on a SPR chip functionalized with the widely used carboxyl-methyl-dextran coating is found to be much larger than that observed for the copoly(DMA-NAS-MAPS) in the same experimental conditions.
One important finding is that measuring the rate of increase in reflectivity is a means of quantifying unknown target concentrations in solution. This is due to the clean exponential kinetics found to be consistent with Eq. 2 and merits further discussion. The described kinetic behavior is due to some of the characteristics of the experiment: (i) the small number of available surface probes confined in submillimeter spots, (ii) the relatively large sample volume, and (iii) the use of a magnetic stirrer to ensure efficient mixing of the sample (Fig. S9 and SI Materials and Methods). The combination of i and ii guarantees that the target molecules in solution greatly outnumber the binding sites at a wide range of target concentrations, thus preventing depletion effects on c, and the use of the stirrer keeps target concentration uniform even in the proximity of the sensor surface. Together, these three aspects grant first-order kinetics and reaction-limited binding conditions, which in turn ensure the validity of Eq. 2. Under these circumstances, the shortest time required to perform an estimate of an unknown target concentration in solution is limited by the recognition process itself through the kinetic rate constant kon, as can be inferred from Eq. 2, whose solution for t << Γ(c)−1 is a linear growth function with slope konc. This dependence is experimentally confirmed, as shown in Fig. 4 (violet open circles and line), where the initial slope of the binding curves measured from anti-mouse IgG spots is reported as a function of the concentration of mouse IgG progressively injected, from 0.4 to 300 ng/mL. Given the current instrumental performance and the measured values of kon, and assuming targets with a molecular weight of 20–100 kDa, the RPI method can detect target concentrations of 10 ng/mL in a few minutes, concentrations of 1 ng/mL in about 0.5 h, and concentrations of 0.1 ng/mL in about 1–2 h. Further improvements in detection can be made by selecting probes with higher kon values. The kinetic rate constants for antibody/antigen interactions span a wide range of values, typically between 103 and 107 M−1⋅s−1 (31, 33–35). Although the kon values of the examined interactions fall within this range, they do not reach the largest possible values. There is therefore significant potential for improving the time-to-result performance.
Conclusions
The RPI method detects the intensity of the reflected light from the molecular layer at the interface between the perfluorinated substrate and the aqueous solution. Ultimately, the signal is provided by molecular electric polarizability at optical frequencies, which is greater for the detected carbon-based molecules than water or the perfluorinated polymer. This makes the method fully label-free and extendable to the detection of any kind of target molecular species, including nucleic acids, carbohydrates, or hormones. The basic strengths of the RPI method are as follows:
• the straightforward, multiplex determination of the amount of biomolecules adhering to the surface;
• the simplicity of the instrumental setup, which can be constructed using low-cost components;
• the sensing plastic substrate can be easily shaped, thus providing unprecedented flexibility in the design of biosensors for various applications;
• the clean kinetics make it possible to determine unknown target concentrations directly from the increase in the reflectivity of a single spot down to fractions of nanograms per milliliter.
Materials and Methods
Substrate Preparation.
The fluorinated material used in this study (Hyflon AD80; Solvay Solexis) was an optically transparent, amorphous, glassy copolymer of tetrafluoroethylene and 2,2,4-trifluoro-5- trifluoromethoxy-1,3-dioxole containing 80 mol% of the cyclic comonomer. Modulation of the comonomer content makes it possible to tailor the refractive index in a range that is very close to 1.33 (36). The bulk material was machined and mechanically lapped to obtain prisms with optical quality faces. The prisms were cleaned with distilled water and plasma treatment and immersed for 30 min in a water solution of ammonium sulfate at 20% (wt/vol) saturation, containing 1% wt/vol of copoly(DMA-NAS-MAPS) (26, 37, 38). The coated prisms were then rinsed with water and dried under vacuum at 80 °C.
Antibody and Immobilization Strategy.
After coating with the copoly(DMA-NAS-MAPS), the sensing surface of the prism was spotted with the different antibodies, using an automated noncontact dispensing system (sciFLEXARRAYER S5; Scienion AG): anti-rabbit IgG, F(ab′)2 fragments specifically developed in goat (Jackson Immuno Research Laboratories), and monoclonal mouse antibodies recognizing p24Ag and HBsAg (Dia.Pro Diagnostic Bioprobes). Two anti-p24 and anti-HBsAg antibodies were used: the coating [p24(c)Ab, corresponding to the clone 114; and HBs(c)Ab, clone S3] and the corresponding detection antibody without the conjugation [p24(d)Ab, clone 6; and HBs(d)Ab, clone S2] of ELISA kits (product codes IVP24 and HBE.CE).
Negative controls were provided by anti-β-Lactoglobulin developed in rabbit (CTR1; Bethyl Laboratories) and mouse antibodies recognizing Toxoplasma gondii antigens (CTR2; Dia.Pro Diagnostic Bioprobes product code TOXOG.CE). The antibodies were spotted at 1 mg/mL in PBS (100 mM PBS, 300 mM NaCl, pH 7.4; Sigma Aldrich), and then the prisms were incubated overnight in a humid chamber at room temperature. Afterward, the spotted prisms were placed in a solution of 50 mM ethanolamine, 150 mM NaCl, and 10 mM Tris⋅HCl buffer, pH 8 (Sigma Aldrich) to block the unreacted succinimide esters, and then washed with distilled water and dried.
Assay Procedure.
The prisms were inserted into a standard glass cuvette and immersed in an incubation buffer consisting of 0.15 M NaCl, 0.02% Tween 20, 1% BSA, and 0.05 M Tris⋅HCl, pH 7.6 (Sigma Aldrich). A LED (HLMP-ED18-UX000; Avago Technologies) emitting in the spectral band 628–644 nm illuminated the sensing surface, and the reflected light was acquired by a CCD camera (Stingray F-145B/C; Allied Technology). The serum experiments were carried out by adding 10% (vol/vol) of bovine fetal serum (Sigma Aldrich; EU origin). Antigen aliquots of Ig from rabbit serum (Life Line Lab), p24 (in this work called p24Ag) and HBsAg recombinant proteins (Dia.Pro Diagnostic Bioprobes) with a molecular mass of, respectively, 24 kDa and 23 kDa (monomer weight), were added directly to the cuvette.
Supplementary Material
Acknowledgments
We thank Antonella Bugatti for performing SPR analyses and Dia.Pro Diagnostic Bioprobes for providing the antibodies and antigens. This work was supported by the Cariplo Foundation (Grant 2006-9869), Italy.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1214589110/-/DCSupplemental.
References
- 1.Zolg JW, Langen H. How industry is approaching the search for new diagnostic markers and biomarkers. Mol Cell Proteomics. 2004;3(4):345–354. doi: 10.1074/mcp.M400007-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Luppa PB, Müller C, Schlichtiger A, Schlebusch H. Point-of-care testing (POCT): Current techniques and future perspectives. Trends Analyt Chem. 2011;30:887–898. doi: 10.1016/j.trac.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Myers FB, Lee LP. Innovations in optical microfluidic technologies for point-of-care diagnostics. Lab Chip. 2008;8(12):2015–2031. doi: 10.1039/b812343h. [DOI] [PubMed] [Google Scholar]
- 4.Rapp BE, Gruhl FJ, Länge K. Biosensors with label-free detection designed for diagnostic applications. Anal Bioanal Chem. 2010;398(6):2403–2412. doi: 10.1007/s00216-010-3906-2. [DOI] [PubMed] [Google Scholar]
- 5.Walker JM. Biosensors and BioDetection. New York: Humana Press; 2009. [Google Scholar]
- 6.Prosperi D, et al. Phantom nanoparticles as probes of biomolecular interactions. Small. 2006;2(8–9):1060–1067. doi: 10.1002/smll.200600106. [DOI] [PubMed] [Google Scholar]
- 7.Engvall E, Perlmann P. Enzyme-linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry. 1971;8(9):871–874. doi: 10.1016/0019-2791(71)90454-x. [DOI] [PubMed] [Google Scholar]
- 8.Nickerson DA, et al. Automated DNA diagnostics using an ELISA-based oligonucleotide ligation assay. Proc Natl Acad Sci USA. 1990;87(22):8923–8927. doi: 10.1073/pnas.87.22.8923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warsinke A. Point-of-care testing of proteins. Anal Bioanal Chem. 2009;393(5):1393–1405. doi: 10.1007/s00216-008-2572-0. [DOI] [PubMed] [Google Scholar]
- 10.Ngom B, Guo Y, Wang X, Bi D. Development and application of lateral flow test strip technology for detection of infectious agents and chemical contaminants: A review. Anal Bioanal Chem. 2010;397(3):1113–1135. doi: 10.1007/s00216-010-3661-4. [DOI] [PubMed] [Google Scholar]
- 11.Martinez AW, Phillips ST, Whitesides GM. Three-dimensional microfluidic devices fabricated in layered paper and tape. Proc Natl Acad Sci USA. 2008;105(50):19606–19611. doi: 10.1073/pnas.0810903105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michaud GA, et al. Analyzing antibody specificity with whole proteome microarrays. Nat Biotechnol. 2003;21(12):1509–1512. doi: 10.1038/nbt910. [DOI] [PubMed] [Google Scholar]
- 13.Ramachandran N, Larson DN, Stark PRH, Hainsworth E, LaBaer J. Emerging tools for real-time label-free detection of interactions on functional protein microarrays. FEBS J. 2005;272(21):5412–5425. doi: 10.1111/j.1742-4658.2005.04971.x. [DOI] [PubMed] [Google Scholar]
- 14.Qavi AJ, Washburn AL, Byeon JY, Bailey RC. Label-free technologies for quantitative multiparameter biological analysis. Anal Bioanal Chem. 2009;394(1):121–135. doi: 10.1007/s00216-009-2637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rich RL, Myszka DG. Higher-throughput, label-free, real-time molecular interaction analysis. Anal Biochem. 2007;361(1):1–6. doi: 10.1016/j.ab.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 16.Fan X, et al. Sensitive optical biosensors for unlabeled targets: A review. Anal Chim Acta. 2008;620(1–2):8–26. doi: 10.1016/j.aca.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liedberg B, Nylanderand C, Lundstrom I. Surface plasmon resonance for gas detection and biosensing. Sens Actuators. 1983;4:299–304. [Google Scholar]
- 18.Homola J, Yee SS, Gauglitz G. Surface plasmon resonance sensors. Sens Actuators B Chem. 1999;54(1–2):3–15. [Google Scholar]
- 19.Boozer C, Kim G, Cong S, Guan H, Londergan T. Looking towards label-free biomolecular interaction analysis in a high-throughput format: A review of new surface plasmon resonance technologies. Curr Opin Biotechnol. 2006;17(4):400–405. doi: 10.1016/j.copbio.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 20.Ozkumur E, et al. Label-free and dynamic detection of biomolecular interactions for high-throughput microarray applications. Proc Natl Acad Sci USA. 2008;105(23):7988–7992. doi: 10.1073/pnas.0711421105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gauglitz G. Direct optical detection in bioanalysis: An update. Anal Bioanal Chem. 2010;398(6):2363–2372. doi: 10.1007/s00216-010-3904-4. [DOI] [PubMed] [Google Scholar]
- 22.Zhu H, Snyder M. Protein chip technology. Curr Opin Chem Biol. 2003;7(1):55–63. doi: 10.1016/s1367-5931(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 23.Ghetta A, et al. Light scattered by model phantom bacteria reveals molecular interactions at their surface. Proc Natl Acad Sci USA. 2005;102(44):15866–15870. doi: 10.1073/pnas.0505877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morasso C, et al. Towards a universal method for the stable and clean functionalization of inert perfluoropolymer nanoparticles: Exploiting photopolymerizable amphiphilic diacetylenes. Adv Funct Mater. 2010;20(22):3932–3940. [Google Scholar]
- 25.Pedrotti FL, Pedrotti LS. Introduction to Optics. 2nd Ed. Upper Saddle River, NJ: Prentice Hall; 1993. pp. 392–396. [Google Scholar]
- 26.Cretich M, Pirri G, Damin F, Solinas I, Chiari M. A new polymeric coating for protein microarrays. Anal Biochem. 2004;332(1):67–74. doi: 10.1016/j.ab.2004.05.041. [DOI] [PubMed] [Google Scholar]
- 27.Harris LJ, Larson SB, Hasel KW, McPherson A. Refined structure of an intact IgG2a monoclonal antibody. Biochemistry. 1997;36(7):1581–1597. doi: 10.1021/bi962514+. [DOI] [PubMed] [Google Scholar]
- 28.Rushi L, et al. Expression, purification, and characterization of hepatitis B virus surface antigens (HBsAg) in yeast, Pichia pastoris. Appl Biochem Biotechnol. 2009;158:432–444. doi: 10.1007/s12010-009-8527-x. [DOI] [PubMed] [Google Scholar]
- 29.Brash JL. Behavior of proteins at interfaces. Curr Opin Colloid Interface Sci. 1996;1:682–688. [Google Scholar]
- 30.Pizarro JC, Vulliez-le Normand B, Riottot MM, Budkowska A, Bentley GA. Structural and functional characterization of a monoclonal antibody specific for the preS1 region of hepatitis B virus. FEBS Lett. 2001;509(3):463–468. doi: 10.1016/s0014-5793(01)03190-8. [DOI] [PubMed] [Google Scholar]
- 31.Karlsson R, Michaelsson A, Mattsson L. Kinetic analysis of monoclonal antibody-antigen interactions with a new biosensor based analytical system. J Immunol Methods. 1991;145(1–2):229–240. doi: 10.1016/0022-1759(91)90331-9. [DOI] [PubMed] [Google Scholar]
- 32.Yalçin A, et al. Direct observation of conformation of a polymeric coating with implications in microarray applications. Anal Chem. 2009;81(2):625–630. doi: 10.1021/ac801954x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drake AW, Myszka DG, Klakamp SL. Characterizing high-affinity antigen/antibody complexes by kinetic- and equilibrium-based methods. Anal Biochem. 2004;328(1):35–43. doi: 10.1016/j.ab.2003.12.025. [DOI] [PubMed] [Google Scholar]
- 34.Karlsson R, Katsamba PS, Nordin H, Pol E, Myszka DG. Analyzing a kinetic titration series using affinity biosensors. Anal Biochem. 2006;349(1):136–147. doi: 10.1016/j.ab.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 35.Myszka DG. Kinetic analysis of macromolecular interactions using surface plasmon resonance biosensors. Curr Opin Biotechnol. 1997;8(1):50–57. doi: 10.1016/s0958-1669(97)80157-7. [DOI] [PubMed] [Google Scholar]
- 36.Groh W, Zimmermann A. What is the lowest refractive index of an organic polymer? Macromol. 1991;24:6660–6663. [Google Scholar]
- 37.Pirri G, Damin F, Chiari M, Bontempi E, Depero LE. Characterization of a polymeric adsorbed coating for DNA microarray glass slides. Anal Chem. 2004;76(5):1352–1358. doi: 10.1021/ac0352629. [DOI] [PubMed] [Google Scholar]
- 38.Cretich M, et al. Functionalization of poly(dimethylsiloxane) by chemisorption of co-polymers: DNA microarrays for pathogen detection. Sens Actuators B Chem. 2008;132:258–264. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.