Despite their different lifestyles, animals and plants share the dependence on small molecules, hormones, for systemic regulation of development and other cellular processes. The hormones act through specific receptors, either through triggering secondary signaling cascades or through direct effect of the nuclear receptor complexes on gene transcription (1). Plant hormones, however, use different mechanisms of action: their nuclear receptors are not transcription factors but act through protein–protein interactions, resulting usually in degradation of their interacting partners (2). In PNAS, Park et al. (3) describe a very different hormone receptor with an alternative localization and a mode of action. Not only is the receptor, cyclophilin 20-3 (CYP20-3), found in the chloroplast, but its hormone complex binds a metabolic enzyme and results in increased production of cysteine. The newly synthesized cysteine alters the redox state of the cell, resulting in activation of TGA transcription factors (3). The action of this receptor, thus, connects retrograde signaling, sulfur metabolism, and redox regulation.
In plants, jasmonic acid (JA) is the best known member of the oxylipin phytohormone family, with its main function in regulation of defense (4). JA is conjugated to isoleucine by JASMONATE RESISTANT1 (5), and the conjugate is perceived by the receptor CORONATINE INSENSITIVE1 (COI1) (6). The F-Box COI1 is part of ubiquitin E3 ligase complex, which, upon binding of JA-Ile, targets a number of JASMONATE-ZIM DOMAIN repressor proteins for degradation (7–9). This relieves inhibition of a specific set of transcription factors and results in rapid activation of a large number of genes (10). The mechanism of JA signaling, thus, seems well understood, except for a small problem: not all JA-regulated genes are COI1-dependent (11). In their search for alternative JA receptors, Park et al. (3) discovered another JA-binding protein, CYP20-3. Instead of being a straightforward alternative to COI1, however, this receptor presents a few surprises and provides interesting links from oxylipin signaling to other areas of plant metabolism.
Why is the CYP20-3 so interesting? Firstly, although found in a screen for proteins interacting with JA, its physiological ligand is actually the intermediate in JA synthesis, 12-oxo-phytodieonic acid (OPDA) (3). Indeed, a set of genes was shown previously to be specifically regulated by OPDA and not JA (12), and these genes were also independent from COI1. The results of Park et al. (3), thus, provide the mechanistic explanation for this observation and place OPDA onto the growing list of phytohormones for which the receptor has been identified (2).
Importantly, CYP20-3 is a hormone receptor localized not at the plasma membrane or nucleus but in the plastids. Although this seems to make sense for a receptor of a hormone synthesized in the chloroplast (13), it involves the inconvenience of transmitting the signal across the plastid envelope to the nucleus, the retrograde signaling (14, 15). Retrograde signaling is essential to enable the cell to react to signals perceived in the plastids, such as high light, drought, or reactive oxygen species and readjust homeostasis (15). A number of such signals acting through diverse pathways have been proposed, such as Mg protoporphyrin IX, haem, singlet oxygen, 3′-phosphoadenosine 5′-phosphate (PAP), methylerythritol cyclodiphosphate, or plant homeodomain-type transcription factors with transmembrane domain, which are normally present in plastid envelopes and upon stress migrate to the nucleus (reviewed in ref. 15). So, why is addition of OPDA on this list remarkable? The answer is in the detailed dissection of the mechanism of action of CYP20-3 revealed by Park et al. (3). The authors were able to find the interaction partner of the OPDA receptor, to measure the redox changes triggered by the signal, to identify the transcription factors controlled by these redox changes, and to record the resulting changes in gene expression (Fig. 1). In contrast, for the other proposed signals, the molecular mechanisms of their action and the interacting proteins and/or molecules are largely unknown. The detailed dissection of OPDA signaling by Park et al. (3), thus, forms a benchmark for characterizations of other retrograde-signaling pathways.
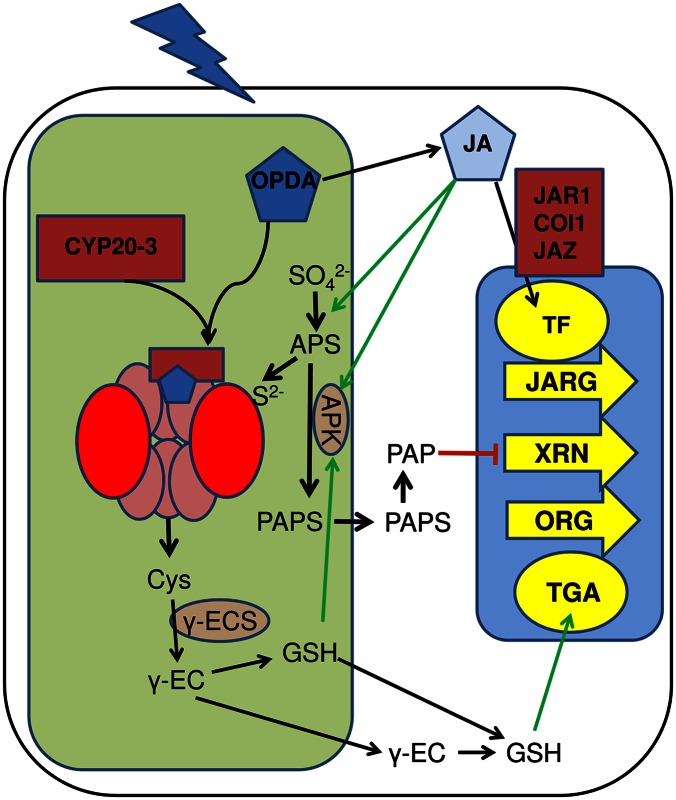
Fig. 1.
Scheme of the interaction between OPDA signaling and sulfur metabolism. Park et al. (3) show that the OPDA synthesized in the chloroplast in reaction to stress binds to cyclophilin CYP20-3, and the hormone–receptor complex interacts with SAT to stabilize formation of cysteine synthase and increase cysteine synthesis. Cysteine is metabolized to glutathione (GSH), resulting in changes of redox homeostasis in plastids, as well as in the cytosol. The cytosolic GSH migrates to the nucleus to activate TGA transcription factors and induce transcription of OPDA-responsive genes (ORG). In parallel, OPDA is metabolized to JA, which up-regulates set of genes responsive to JA (JARG). Among JARGs are genes for components of sulfate assimilation, providing the reduced sulfur needed for increased cysteine production. The redox signaling may be coupled with PAP retrograde signaling: reduced GSH in plastids activates adenosine 5′-phosphosulfate (APS) kinase, synthesizing PAPS, which is converted in the cytosol to PAP. Increased PAP blocks XRN ribonucleases and triggers changes in transcript levels of another subset of genes.
The OPDA receptor, CYP20-3, has previously been shown to bind serine acetyltransferase (SAT), an enzyme essential for sulfate assimilation and a component of cysteine synthase complex (16, 17). The binding increased SAT activity, leading to higher production of cysteine, because SAT is limiting for its synthesis (18). The interaction of CYP20-3 with SAT seemed to be important for abiotic stress signaling [e.g., the cyp20-3 mutants were sensitive to salt stress (16)]. The results of Park et al. (3) add another layer to these observations, showing that the interaction of CYP20-3 and SAT is facilitated by OPDA and is a part of the OPDA-signaling mechanism. The OPDA–CYP20-3 complex promotes the formation of cysteine synthase complex, which is necessary for SAT activity. However, for the resulting cysteine to work as a signal or second messenger for regulation of gene expression, it has to move to the nucleus. The mediator of such OPDA retrograde signaling, however, seems to be not the cysteine per se but a change in redox potential caused by increased concentration of thiol groups. Increased cysteine synthesis leads to synthesis of glutathione that can convey the redox signal to the TGA transcription factors, which have been shown previously to be redox-sensitive (19). Thus, sulfur metabolites are necessary for the OPDA-triggered retrograde signaling.
The involvement of sulfur merits emphasis because this is not the first case in the retrograde-signaling field. A typical example of such signaling is the induction of APX2 gene for ascorbate peroxidase by high light. This process obviously requires the transmission of a signal generated in chloroplast to the nucleus. Interestingly, in a genetic screen for mutants impaired in this signaling, only two mutants with the same lesion in γ-glutamylcysteine synthetase (γECS), the first enzyme in glutathione synthesis from cysteine, were identified (20). It is tempting to speculate that γECS is involved in the transmission of the redox signal generated by ODPA as well, but to prove it will require some additional work. A different link between retrograde signaling and sulfur metabolism isone of the newest signals, PAP. PAP is a byproduct of biological sulfations, which uses an
The identification of mechanisms of OPDA signaling can form a starting point for the dissection of the interactions between the multiple retrograde-signaling pathways.
activated sulfate in the form of 3′-phosphoadenosine 5′-phosphosulfate (PAPS) (17). PAP is dephosphorylated in the plastids, because its accumulation inhibits the sulfation reactions and ribonucleases and triggers large changes in gene transcription (21, 22). The role of PAP as a retrograde signal was inferred from the observation that PAP accumulates in both plastids and the nucleus of plants lacking the enzyme-degrading PAP and also in plants subjected to drought stress resulting in similar expression alterations (21). Interestingly, both branches of sulfate assimilation, the reduction of sulfate to sulfide for cysteine synthesis, as well as formation of PAPS, are under control of JA and are also redox-regulated on a posttranslational level (17, 23, 24), indicating a crosstalk of the signals. The identification of mechanisms of OPDA signaling (3) can form a starting point for the dissection of the interactions between the multiple retrograde-signaling pathways. This will help to unravel the complex way in which plant cells react to signals localized in organelles and understand the fundamental basis of control of plant homeostasis.
Footnotes
The author declares no conflict of interest.
See companion article on page 9559.
References
- 1.Evans RM. The nuclear receptor superfamily: A rosetta stone for physiology. Mol Endocrinol. 2005;19(6):1429–1438. doi: 10.1210/me.2005-0046. [DOI] [PubMed] [Google Scholar]
- 2.Lumba S, Cutler S, McCourt P. Plant nuclear hormone receptors: A role for small molecules in protein-protein interactions. Annu Rev Cell Dev Biol. 2010;26:445–469. doi: 10.1146/annurev-cellbio-100109-103956. [DOI] [PubMed] [Google Scholar]
- 3.Park S-W, et al. Cyclophilin 20-3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proc Natl Acad Sci USA. 2013;110:9559–9564. doi: 10.1073/pnas.1218872110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballaré CL. Jasmonate-induced defenses: A tale of intelligence, collaborators and rascals. Trends Plant Sci. 2011;16(5):249–257. doi: 10.1016/j.tplants.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 5.Staswick PE, Tiryaki I. The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell. 2004;16(8):2117–2127. doi: 10.1105/tpc.104.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan J, et al. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell. 2009;21(8):2220–2236. doi: 10.1105/tpc.109.065730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chini A, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448(7154):666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 8.Thines B, et al. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature. 2007;448(7154):661–665. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 9.Yan Y, et al. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell. 2007;19(8):2470–2483. doi: 10.1105/tpc.107.050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pauwels L, Goossens A. The JAZ proteins: A crucial interface in the jasmonate signaling cascade. Plant Cell. 2011;23(9):3089–3100. doi: 10.1105/tpc.111.089300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devoto A, et al. Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol Biol. 2005;58(4):497–513. doi: 10.1007/s11103-005-7306-5. [DOI] [PubMed] [Google Scholar]
- 12.Taki N, et al. 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol. 2005;139(3):1268–1283. doi: 10.1104/pp.105.067058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell E, Creelman RA, Mullet JE. A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc Natl Acad Sci USA. 1995;92(19):8675–8679. doi: 10.1073/pnas.92.19.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan KX, Crisp PA, Estavillo GM, Pogson BJ. Chloroplast-to-nucleus communication: Current knowledge, experimental strategies and relationship to drought stress signaling. Plant Signal Behav. 2010;5(12):1575–1582. doi: 10.4161/psb.5.12.13758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estavillo GM, Chan KX, Phua SY, Pogson BJ. Reconsidering the nature and mode of action of metabolite retrograde signals from the chloroplast. Front Plant Sci. 2012;3:300. doi: 10.3389/fpls.2012.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominguez-Solis JR, et al. A cyclophilin links redox and light signals to cysteine biosynthesis and stress responses in chloroplasts. Proc Natl Acad Sci USA. 2008;105(42):16386–16391. doi: 10.1073/pnas.0808204105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi H, Kopriva S, Giordano M, Saito K, Hell R. Sulfur assimilation in photosynthetic organisms: Molecular functions and regulations of transporters and assimilatory enzymes. Annu Rev Plant Biol. 2011;62:157–184. doi: 10.1146/annurev-arplant-042110-103921. [DOI] [PubMed] [Google Scholar]
- 18.Blaszczyk A, Brodzik R, Sirko A. Increased resistance to oxidative stress in transgenic tobacco plants overexpressing bacterial serine acetyltransferase. Plant J. 1999;20(2):237–243. doi: 10.1046/j.1365-313x.1999.00596.x. [DOI] [PubMed] [Google Scholar]
- 19.Böttcher C, Pollmann S. Plant oxylipins: Plant responses to 12-oxo-phytodienoic acid are governed by its specific structural and functional properties. FEBS J. 2009;276(17):4693–4704. doi: 10.1111/j.1742-4658.2009.07195.x. [DOI] [PubMed] [Google Scholar]
- 20.Ball L, et al. Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell. 2004;16(9):2448–2462. doi: 10.1105/tpc.104.022608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Estavillo GM, et al. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell. 2011;23(11):3992–4012. doi: 10.1105/tpc.111.091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee BR, et al. Effects of fou8/fry1 mutation on sulfur metabolism: Is decreased internal sulfate the trigger of sulfate starvation response? PLoS ONE. 2012;7(6):e39425. doi: 10.1371/journal.pone.0039425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ravilious GE, Nguyen A, Francois JA, Jez JM. Structural basis and evolution of redox regulation in plant adenosine-5′-phosphosulfate kinase. Proc Natl Acad Sci USA. 2012;109(1):309–314. doi: 10.1073/pnas.1115772108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jost R, et al. Expression profiling of metabolic genes in response to methyl jasmonate reveals regulation of genes of primary and secondary sulfur-related pathways in Arabidopsis thaliana. Photosynth Res. 2005;86(3):491–508. doi: 10.1007/s11120-005-7386-8. [DOI] [PubMed] [Google Scholar]