Abstract
Malignant melanoma is an aggressive form of skin cancer whose incidence continues to increase worldwide. Increased exposure to sun, ultraviolet radiation and the use of tanning beds can increase the risk of melanoma. Early detection of melanomas is the key to successful treatment mainly through surgical excision of the primary tumor lesion. But in advanced stage melanomas, once the disease has spread beyond the primary site to distant organs, the tumors are difficult to treat and quickly develop resistance to most available forms of therapy. The advent of molecular and cellular techniques has led to a better characterization of tumor cells revealing the presence of heterogeneous melanoma subpopulations. The discovery of gene mutations and alterations of cell-signaling pathways in melanomas has led to the development of new targeted drugs that show dramatic response rates in patients. Single agent therapies generally target one subpopulation of tumor cells while leaving others unharmed. The surviving subpopulations will have the ability to repopulate the original tumors that can continue to progress. Thus, a rational approach to target multiple subpopulations of tumor cells with a combination of drugs instead of single agent therapy will be necessary for long-lasting inhibition of melanoma lesions. In this context, the recent development of immune checkpoint reagents provides an additional armor that can be used in combination with targeted drugs to expand the presence of melanoma reactive T-cells in circulation to prevent tumor recurrence.
Keywords: Melanoma, tumor, heterogeneity, subpopulations, therapy, resistance
I. Introduction
The American Cancer Society (ACS) predicts an increased incidence of all cancers in the United States for the current year (Siegel, Naishadham, & Jemal, 2012). This is also true for malignant melanoma which continues to rise worldwide. According to recent ACS estimates ~ 76,000 new cases of melanomas (~5% of all cancers) will be diagnosed in the United States in 2012 and about 9000 patients will die of metastatic disease (Siegel, et al., 2012). Thus far, the reasons for the higher incidence of melanoma remain unclear but increased exposures to sun or ultraviolet radiation are some of the major risk factors. Family history of melanoma, genetic susceptibility, environmental factors and age related immune-suppressions are also some of the contributing factors that could influence the incidence rates (reviewed in (de Souza, Morais, & Jasiulionis, 2012; Miller & Mihm, 2006)).
In many cases, melanoma begins with the transformation of a benign nevus that develops into a dysplastic lesion before progressing into a radial- and vertical-growth phase (RGP and VGP [primary melanoma]) that can invade into the dermis, regional lymph nodes and from there disseminate to distant organs, leading to metastatic melanoma (reviewed in (Koh, 1991; Miller & Mihm, 2006)). However, not all melanomas arise from nevus and many arise through direct transformation of normal skin cells (de Souza, et al., 2012).
In the last decade, a number of important genetic alterations have been identified during various stages of melanoma progression leading to a better understanding and molecular classification of the disease (reviewed in (Chin, Garraway, & Fisher, 2006; de Souza, et al., 2012; Fecher, Cummings, Keefe, & Alani, 2007; Vidwans, et al., 2011)). These studies have also provided in-depth analysis of cell cycle regulation and alterations in signaling pathways during the progression of the disease. Unlike the older histological classification (Chin, et al., 2006; Koh, 1991; Miller & Mihm, 2006), newer molecular approaches define melanoma as a more heterogeneous and rather complex neoplasm (de Souza, et al., 2012; Koh, 1991; Miller & Mihm, 2006; Vidwans, et al., 2011). Additionally, a better understanding of the aberrant signaling pathways in melanoma has led to the discovery of targeted therapies with drugs such as vemurafenib and a host of others that are either awaiting approval by the US Food and Drug Administration (FDA) or are in various stages of phase I-III clinical trials (Flaherty, Puzanov, et al., 2010; Friedlander & Hodi, 2010; Vidwans, et al., 2011).
Although a large number of primary melanomas can be successfully treated through surgery, therapy of advanced stage metastatic melanoma patients remains challenging (de Souza, et al., 2012; Fecher, et al., 2007; Miller & Mihm, 2006). Melanoma patients undergoing chemotherapy or targeted therapy with small molecule inhibitors aimed at blocking the most frequently mutated oncogene (BRAFV600E) are known to develop drug resistance and experience tumor recurrence (Flaherty, Hodi, & Bastian, 2010; Flaherty, Puzanov, et al., 2010; J. Villanueva, Vultur, & Herlyn, 2011). Several molecular mechanisms underlying acquired drug resistance have been recently described (Johannessen, et al., 2010; Nazarian, et al., 2010; J. Villanueva, et al., 2011; J. Villanueva, et al., 2010); however, tumor recurrence can also be due in part to the presence and potential enrichment of tumor subpopulations that are inherently resistant to therapy (Frank, et al., 2005; Monzani, et al., 2007; Roesch, et al., 2010). Like other malignancies, melanoma is a highly heterogeneous neoplasm, composed of subpopulations of tumor cells with distinct molecular and biological phenotypes (Boiko, et al., 2010; Dick, 2009; Fang, et al., 2005; Monzani, et al., 2007; Roesch, et al., 2010; Schatton, et al., 2008; Zabierowski & Herlyn, 2008). These distinct subpopulations provide the cellular basis for the complex biology of the disease including phenomena such as self-renewal, differentiation, tumor initiation, progression, tumor maintenance and therapy resistance.
Here we discuss the heterogeneous nature of melanoma subpopulations, possible reasons of heterogeneity, its role in therapy resistance and future approaches to targeted therapy.
II. Molecular overview of melanoma
Melanoma arises through the transformation of melanocytes, a melanin producing cell (Koh, 1991; Miller & Mihm, 2006). These cells share a common origin with neural crest cells and during embryonic development migrate towards the skin where they reside in the basal layer of the epidermis (Koh, 1991; Miller & Mihm, 2006). Melanocytes are closely associated with epidermal keratinocytes, dermal fibroblasts, endothelial cells and inflammatory cell types which regulate their functional homeostasis and controlled proliferation; any alteration in the function of these cells due to biological or genetic events can give rise to melanocytic nevi (Satyamoorthy & Herlyn, 2002). Benign nevi (comprised of neval melanocytes) are biologically stable precursor lesions of melanoma (Miller & Mihm, 2006). BRAF is a member of the mitogen-activated protein kinase (MAPK) pathway. It is mutated in about 50% of melanomas, with a glutamic acid for valine substitution at codon 600 (V600E) being the most frequent mutation (Davies, et al., 2002; de Souza, et al., 2012; Fecher, et al., 2007; Vidwans, et al., 2011). Mutant BRAFV600E is also found in ~80% of benign nevi (Davies, et al., 2002; de Souza, et al., 2012; Fecher, et al., 2007; Vidwans, et al., 2011). Cells expressing BRAFV600E usually have increased MAPK activity (Fecher, et al., 2007). The oncogene NRAS, mutated in ~20% of melanomas, can also cause hyper-activation of the MAPK pathway (Fecher, et al., 2007; Vidwans, et al., 2011). BRAF or NRAS mutations are more commonly present in non-chronic sun exposed lesions and less common in chronic sun exposed lesions or lesions of mucosal or acral or familial melanomas (de Souza, et al., 2012; Friedlander & Hodi, 2010). Melanomas that do not express mutant BRAFV600E or mutant NRAS can have alterations in cell cycle regulatory genes or proteins including Cyclin D1 [CCND1] (de Souza, et al., 2012; Fecher, et al., 2007), Cyclin-dependent kinases (CDK1, CDK2, CDK4, and CDK5) (Abdullah, Wang, & Becker, 2011) or mutations in the proto-oncogene C-KIT (Fecher, et al., 2007; Flaherty, Hodi, et al., 2010; Vidwans, et al., 2011). However, a single oncogene cannot transform human melanocytes and additional genetic events are needed for malignant transformation (Bloethner, et al., 2007; de Souza, et al., 2012; Miller & Mihm, 2006). During the course of development and progression into melanoma, melanocytes tend to acquire additional genetic alterations (See Figure 1). These alterations include loss or mutation of certain tumor suppressor genes such as phosphatase and tensin homolog (PTEN), p16INK4A (also known as cyclin-dependent kinase inhibitor [CDKN2a]), and inositol polyphosphate 4-phosphatase type II (INPP4b). Alterations in these genes are associated with activation of the phosphoinositide (PI)-3 kinase (PI3K) pathway, increased proliferation, disease progression, and resistance to therapy (de Souza, et al., 2012; Fecher, et al., 2007; Gewinner, et al., 2009; Miller & Mihm, 2006; Vidwans, et al., 2011; Yuan & Cantley, 2008). Mutations in the p53 tumor suppressor gene, up regulation of the anti-apoptotic factors BCL-2 or MCL-1 or amplification of microphthalmia associated transcription factor (MITF) are frequently observed in metastatic melanoma and have also been associated with chemoresistance (de Souza, et al., 2012; Fecher, et al., 2007; Vidwans, et al., 2011).
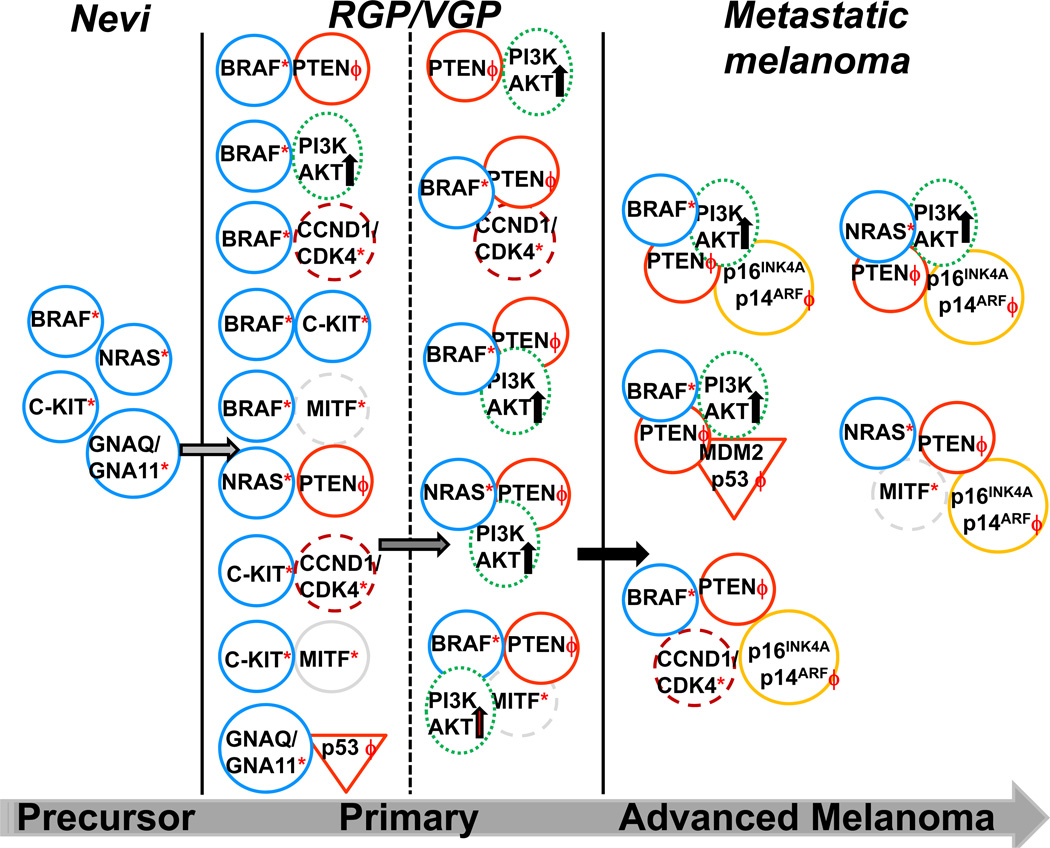
Figure 1. Molecular heterogeneity of melanomas.
Precursor melanocytic lesions frequently harbor single gene mutations (*) such as BRAF, NRAS, C-KIT or GNAQ/GNA11 with a potential for neoplastic transformation. Additional oncogenic events (ϕ) such as deletions, mutations or loss of tumor suppressor genes (PTEN, p16INK4A/p14ARF, p53), alterations in genes associated with cell-cycle regulation (CCND1/CDK4, MITF [dashed circle]) or activation (black arrow) of signaling pathways (PI3K/AKT [dotted oval]; sometimes PI3K/AKT mutations can also be found in low frequency) are needed for malignant transformation of benign nevi to primary tumor and then to progressive metastatic melanoma. The most frequent genetic alterations are depicted for simplicity. Mutations of tumor suppressor genes (p16 INK4A, p14 ARF and p53) may happen very early in the process of malignant transformation but there is no concrete evidence of their exact occurrence. Genomic instability further contributes to genetic heterogeneity.
III. Therapeutic overview
For many decades metastatic melanoma was treated as a single disease entity; dacarbazine (DTIC), an alkylating agent was the standard of care with temporary objective response rates below 15% (Koh, 1991; Miller & Mihm, 2006). Treatment of melanoma patients with temozolomide, a second-generation alkylating agent, also resulted in low response rates of about 10–12% (Fecher, et al., 2007; Miller & Mihm, 2006; Vidwans, et al., 2011). The use of adjuvant therapies such as interferon (IFN)-α or interleukin (IL)-2 has provided a modest improvement in patient survival (de Souza, et al., 2012; Miller & Mihm, 2006). Additionally, these therapeutic modalities were associated with lingering toxicities, frequently leading to discontinuation of treatment. Many other forms of biological and immunological therapies have failed to go beyond the experimental stage. The recent FDA approval of anti-CTLA4 (also known as Ipilimumab or Yervoy), an immune checkpoint agent, has shown some improvement in survival of melanoma patients and has created renewed interest in immunological therapies (Hodi, et al., 2010). Another immune modulating agent, anti-program cell death (PD)-1, has provided favorable response rates in clinical trials (Brahmer, et al., 2010; Kline & Gajewski, 2010). Additionally, recent advances developing engineered T cells designed to express chimeric-antigen receptor (CAR) with specificity against melanoma tumor cells has shown some promising response rates in a clinical trial involving adoptive T-cell therapies (Schmidt, et al., 2009). The discovery of mutations such as BRAFV600E or NRAS and defects in cell cycle regulatory genes or proteins has led to a more personalized targeted therapy approach for the treatment of melanoma. In this context, vemurafenib, a BRAF-selective kinase inhibitor recently approved by the FDA, has shown dramatic regression of metastatic melanoma lesions. Over 50% of BRAF-mutant melanoma patients respond to vemurafenib with a median progression-free survival of about 7 months (Chapman, et al., 2011; Flaherty, Puzanov, et al., 2010; Sosman, et al., 2012). Unfortunately, responses are transient and most patients develop resistance to treatment in the long run.
IV. Therapy resistance
Multiple mechanisms can mediate therapy resistance and the readers are referred to reviews that provide an excellent overview on drug resistance pathways (Dean, Fojo, & Bates, 2005; Tredan, Galmarini, Patel, & Tannock, 2007). Drug resistance in tumor cells could be due to one or more distinct mechanisms, including some briefly described below:
A. Increased drug efflux activity
Multidrug resistance in cancer is frequently linked to over expression of the ABC (ATP-binding cassette) transporters, P-glycoprotein (ABCB1), multidrug resistance-associated proteins (MRP11/ABCC1 and MRP2/ABCC2), and breast cancer resistance protein (ABCG2/BCRP). Enhanced expression of MDR or ABC transporter proteins on the membrane of tumor cells can result in increased drug efflux activity resulting in lower than required intracellular concentration of drugs than is needed for inhibition of tumor cell growth. Several tumor cell types including leukemias, melanomas and carcinoma cells obtained from brain, breast, colon, lungs, ovaries, pancreas, prostate and renal express high levels of ABC transporter proteins (Dean, et al., 2005; Szakacs, et al., 2004), which can collectively pump a multitude of chemical compounds and which lead to chemoresistance. For example, tumor initiating cells or subpopulations in melanoma that express ABCB5 or ABCG2 proteins are highly resistant to chemotherapeutic agents and immune mediated lysis (Schatton, et al., 2008; Taghizadeh, et al., 2011). These subpopulations are described in greater detail in section V.
B. Increased DNA repair activity
In vitro studies have shown that a subset of melanoma cell lines resistant to chemotherapeutic agents have increased or altered DNA repair mechanisms (Bradbury & Middleton, 2004; Kauffmann, et al., 2008; Sarasin & Kauffmann, 2008). There are multiple pathways of DNA repair mechanisms, including direct repair, mismatch repair (MMR), base excision repair (BER), nucleotide excision repair (NER) and double-strand break recombination repair, which include both non-homologous end joining (NHEJ) and homologous recombination repair (HHR) (Bradbury & Middleton, 2004; Sarasin & Kauffmann, 2008). Polyadenosine diphosphate-ribose polymerase (PARP), a BER DNA repair enzyme, is frequently up regulated in melanoma cells (Bradbury & Middleton, 2004; Kauffmann, et al., 2008). Several reports have shown that melanoma cells resistant to temozolomide or DTIC have elevated levels of O6-methylguanine-DNA methyltransferase (MGMT), a protein that removes drug-induced alkylguanine adducts from DNA (Augustine, et al., 2009; Bradbury & Middleton, 2004; Kauffmann, et al., 2008; Rastetter, et al., 2007). Similar to MGMT, BER plays an important role in repairing the cytotoxic methyl DNA adducts created by temozolomide, and consequently, high BER activity can confer tumor resistance to temozolomide (Augustine, et al., 2009; Bradbury & Middleton, 2004; Kauffmann, et al., 2008; Runger, et al., 2000). Some clinical studies indicate that better response rates can be achieved in melanoma patients treated with a combination of PARP inhibitors and DTIC (Jones & Plummer, 2008; Plummer, et al., 2008), further suggesting that DNA repair mechanisms are associated with chemoresistance.
C. Increased existence of slow cycling cells or tumor side population
The presence within a tumor of non-proliferating cells or cells that proliferate very slowly (slow cycling cells) or a population of cells that excludes the DNA-binding dye Hoechst 33342, called ‘side population’, has also been linked to therapy resistance (Addla, Brown, Hart, Ramani, & Clarke, 2008; Dembinski & Krauss, 2009; Hadnagy, Gaboury, Beaulieu, & Balicki, 2006; Ho, Ng, Lam, & Hung, 2007; Nishimura, et al., 2002; Roesch, et al., 2010; Scharenberg, Harkey, & Torok-Storb, 2002). This is likely due to the fact that chemotherapeutic agents are effective on fast dividing cells as they generally cause DNA alkylation or adduct formation and therefore are less effective on slow cycling or non-proliferating cells.
D. Tumor microenvironment (TME) induced drug resistance
It is well established that therapy can induce changes in the TME; certain chemotherapeutic agents such as paclitaxel or carboplatin cause preferential accumulation of macrophages or other leukocytes in the tumor stroma, which can influence disease outcome (Zitvogel, Apetoh, Ghiringhelli, & Kroemer, 2008; Zitvogel, Kepp, & Kroemer, 2011). Tumor stromal-derived fibroblasts as well as tumor associated macrophages (TAMs) can play a role in resistance to treatment by modulating the tumor phenotype (Brennen, Isaacs, & Denmeade, 2012; Denardo, et al., 2011; van Kempen, Ruiter, van Muijen, & Coussens, 2003). Inflammatory cytokines produced by the infiltrating cells can induce tumor phenotypic changes; they can induce changes in the surface expression of human leukocyte antigen class I or class II molecules and costimulatory molecules that are necessary for interactions with immune cells (Zitvogel, et al., 2011). In addition, infiltrating inflammatory cells are a source of reactive oxygen species (ROS) and reactive nitrogen intermediates (RNI) that can cause epigenetic changes, DNA strand breaks, point mutations, and aberrant DNA cross-linking leading to genomic instability (Grivennikov, Greten, & Karin, 2010; Schetter, Heegaard, & Harris, 2009). Furthermore, chronic inflammatory conditions promote tumor initiation and increase tumor survival by activating anti-apoptotic pathways and inducing the expression of anti-apoptotic factors such as BCL-2, MCL-1, and survivin that are frequently associated with therapy resistant cells (Grivennikov, et al., 2010; Schetter, et al., 2009). These findings have spurred new therapeutic combinatorial approaches targeting both tumor and stroma-derived macrophages or fibroblasts to curtail the negative influence of inflammatory cells on neoplastic growth. Recent pilot trials aimed at targeting both tumor cells and the infiltrating macrophages or fibroblasts have shown improved therapy responses, indicating the beneficial effects of this new treatment strategy (Brennen, et al., 2012; Denardo, et al., 2011; Korkaya, Liu, & Wicha, 2011a, 2011b). Additional clinical trials will be needed to confirm these findings.
E. Epigenetic changes after therapy
Patients with small cell lung carcinoma show transient resistance to certain targeted drugs such as tyrosine kinase inhibitors (TKIs) (Sharma, et al., 2010). Patients who acquire resistance to TKIs respond to re-treatment after a ‘drug-holiday’, indicating the transient nature of drug resistance (Sharma, et al., 2010). This phenomenon is known as adaptive resistance due to drug-induced stress. Certain tumor subpopulations undergo epigenetic changes and acquire transient resistance to escape the effect of drugs. Upon drug withdrawal, the residual subpopulations can revert and become drug sensitive again. Settleman’s group has shown that the histone demethylase JARID1A is responsible for transient drug resistance. In melanoma, in vitro studies have shown that tumor cells can undergo epigenetic changes leading to increased resistance to chemotherapeutic agents (Sharma, et al., 2010). Likewise, methylation of certain DNA regions can alter signaling pathways, activating survival mechanisms in the tumor cells. For example, increased expression of BCL-2/MCL-1, activation of β-catenin/MITF and silencing of tumor suppressor genes such as p53 or the invasive suppressor CD82 are some of the mechanisms that are known to occur following DNA methylation (Chung, 2011; Dean, et al., 2005; Halaban, et al., 2009; Howell, et al., 2009; Taylor, Hickman, & Dive, 2000). Studies using tumor specimens obtained before and after therapy have confirmed the above in vitro results. Both chemotherapeutic agents and targeted drugs can indirectly recruit inflammatory cells that can cause epigenetic changes via cytokine mediators, which also stimulate increased expression or activation of anti-apoptotic proteins and alterations in cell signaling mechanisms promoting tumor cell survival.
F. Activation of alternative signaling mechanisms after therapy
Melanoma patients treated with newly discovered targeted drugs frequently develop resistance to therapy (Vidwans, et al., 2011). Several studies have shown that tumor cells chronically treated with targeted drugs, such as BRAF-selective inhibitors, can activate alternate signaling pathways to promote proliferation and survival, and thus develop therapy resistance (Fecher, et al., 2007; Vidwans, et al., 2011; J. Villanueva, et al., 2011; J. Villanueva, et al., 2010). Multiple studies suggest that reactivation of the MAPK pathway in a BRAF-V600E-independent manner is commonly associated with resistance to BRAF-selective inhibitors. In addition to others, we have demonstrated that BRAF-V600E-mutant melanoma cells express somewhat increased levels of CRAF or ARAF after prolonged exposure to BRAF inhibitors (Montagut, et al., 2008; J. Villanueva, et al., 2010). Furthermore, BRAF-V600E melanoma cells which acquire resistance to BRAF inhibitors no longer rely on BRAF for MAPK activation but rather use one of the other two RAF isoforms to sustain the MAPK signaling pathway (J. Villanueva, et al., 2010). Some melanoma cells resistant to BRAF inhibitors also displayed increased NRAS activity or mutations in NRAS (Poulikakos, et al., 2011), which can promote signaling via the MAPK and PI3K pathways (Atefi, et al., 2011; Vidwans, et al., 2011). Reactivation of the MAPK pathway can also be mediated by over expression or amplification of the serine threonine kinase COT/MAPK8 (Johannessen, et al., 2010). More recently, Poulikakos et al discovered that resistance to BRAF inhibitors and reactivation of the MAPK pathway can be mediated through the expression of a truncated form of BRAF, which lacks the RAS activation domain (Poulikakos, et al., 2011). In addition, resistance to BRAF inhibitors has also been linked to enhanced expression of receptor tyrosine kinases (RTK), including insulin-dependent growth factor (IGF)-1 or platelet derived growth factor (PDGF) receptors, leading to altered receptor activity and signaling via the PI3k/AKT pathway (Nazarian, et al., 2010; Vidwans, et al., 2011; J. Villanueva, et al., 2011; J. Villanueva, et al., 2010). Activation of the MAPK and PI3K/AKT pathways results in increased expression of anti-apoptotic proteins such as MCL-1 that increases survival of tumor cells (Vidwans, et al., 2011). Interestingly, although about 10% of colon carcinoma patients express BRAFV600E only 5% of this patient cohort responds to vemurafenib (M. T. Villanueva, 2012). Resistant tumors from these patients exhibit upregulation of the epidermal growth factor (EGF)-receptor pathway following inhibition of the MAPK pathway after treatment with BRAF inhibitors. In these patients a combination strategy using vemurafenib and the EGFR inhibitor Erlotinib or the monoclonal EGFR antibody Cetuximab increased tumor response (Prahallad, et al., 2012). The reported resistant mechanisms have been validated in tumor samples obtained from patients after tumor recurrence.
V. Tumor heterogeneity and melanoma subpopulations - their role in therapy resistance
Some patients with metastatic melanoma treated with chemo-, targeted- or immunological therapies show mixed responses to treatment. While some lesions undergo dramatic responses to therapy, even complete regression, other lesions in the same patient continue to progress or in some cases, new lesions develop, indicating the emergence of drug resistant clones (See Figure 2). Genotypic and phenotypic analyses of melanoma cells have revealed that the tumors are more heterogeneous than the original lesions. As melanoma progresses from primary to metastatic disease, the tumor acquires additional genetic and biologic properties that support tumor growth, invasion, and metastasis (See Figure 1). It is known that some of these acquired properties are profoundly influenced by the TME. Using laser micro-dissection, Yancovitz et al described both intra and inter-tumor variability in BRAFV600E expression in tumor cells isolated from different regions of the primary lesions (Yancovitz, et al., 2012). In that study, the primary melanoma lesion likely harbored mutation positive BRAFV600E cells as well as mutation negative or wild-type (WT) BRAF; tumor cells with either genotype have equal ability to develop metastasis (Yancovitz, et al., 2012). This finding is further confirmed by the work of Sensi and colleagues on the heterogeneous genotypic expression of BRAFV600E/WT-NRAS and WT-BRAF/NRASQ61R in individual tumor cells (isolated after single cell cloning) from the same lesion was shown (Sensi, et al., 2006). Furthermore, Yancovitz et al reported the presence of NRAS and BRAF mutations in different cells within the same primary lesion (Yancovitz, et al., 2012). BRAFV600E and NRAS mutations have long been considered as mutually exclusive (Fecher, et al., 2007). However, recently Nazarian et al have shown the presence of two different NRAS (Q61K and Q61R) mutations co-existent with BRAF-V600E in a nodal metastasis of a melanoma patient after therapy (Nazarian, et al., 2010). In each study, the TME niche appears to play a critical role mediating the emergence of selective melanoma subpopulations with distinct genetic mutations. Similar observations were made in patients with advanced metastatic melanoma treated with immunological therapies. In patients who experienced mixed responses to therapy, their tumors had inter-and intra-lesional heterogeneity in the expression of melanoma associated antigens (MAA), resulting in poor ability of T cells to bind and lyse the cancer cells (Campoli, Ferris, Ferrone, & Wang, 2009). Furthermore, many metastatic melanoma cells with low MITF expression have similar down modulation of MAA (Dissanayake, et al., 2008). Given the implications of tumor heterogeneity in melanoma therapy, a better understanding of tumor subpopulations and their role in therapy resistance is required. In the following section, we will describe the most common melanoma subpopulations described thus far by us or others, which can mediate chemo, targeted- or immune-therapy resistance.
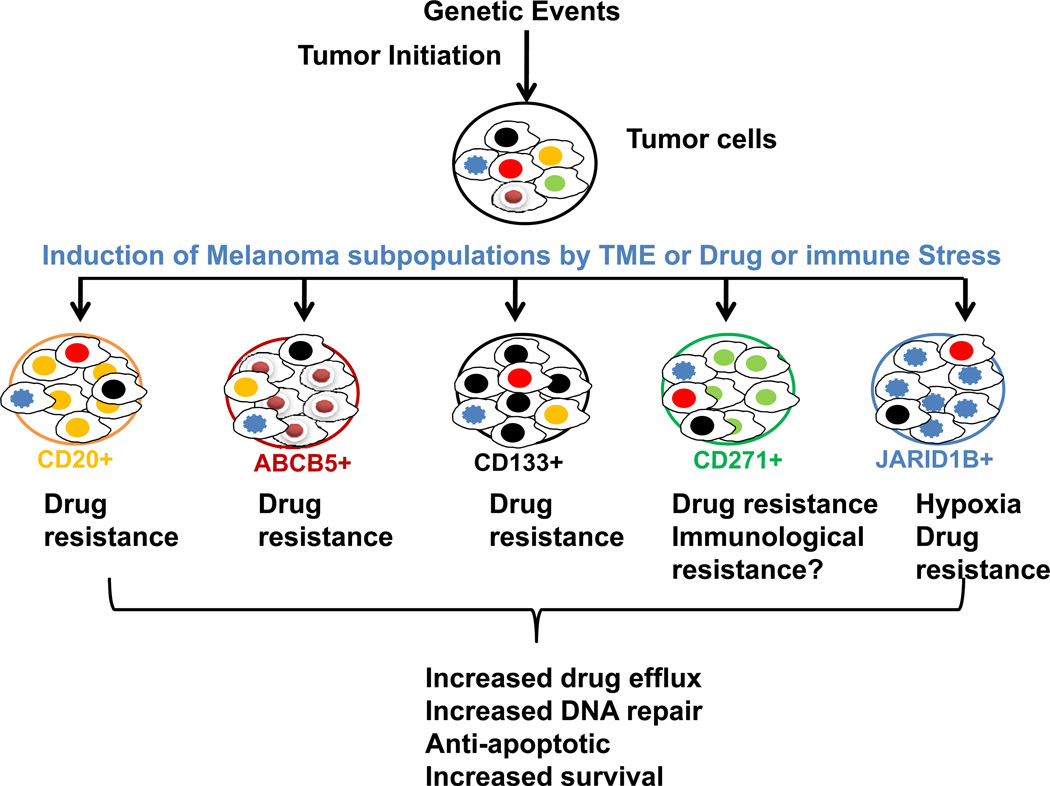
Figure 2. Induction of melanoma subpopulations: the role of TME, chemo- or targeted-therapy and immune related stress.
TME niche and therapy-induced infiltration of leukocytes supports and promotes the induction of tumor subpopulations which express increased levels of drug efflux proteins, DNA repair enzymes and anti-apoptotic proteins resulting in activation of pro-tumor survival mechanisms.
1. CD20
CD20 is a transmembrane protein, originally identified as a B-cell surface marker involved in Ca++ channeling, B cell activation and proliferation (Somasundaram, Villanueva, & Herlyn, 2011; Tedder & Engel, 1994). Using gene expression profiling, CD20 has been identified as one of the top 22 genes in melanoma that defines the aggressive nature of the disease (Bittner, et al., 2000). Our group has shown that a small proportion of melanoma cells express CD20 when grown as tumor spheroids under in vitro culture conditions (Fang, et al., 2005). This CD20+ population was previously considered to be a cancer stem-like cell or-tumor initiating cell as it fulfilled some of the criteria of ‘tumor stemness’ by its ability to differentiate into multiple lineages including melanocytes, adipocytes or chondrocytes (Fang, et al., 2005). However, the concept of stem cells in melanoma has been challenged and remains controversial; along wth Morrison’s group, we have demonstrated that any melanoma cell can be a tumor-initiating cell. Our unpublished observations indicate that melanoma cells that are resistant to chemotherapeutic agents like cisplatin show higher expression of CD20. We and others have identified CD20+ melanoma cells in metastatic tumor lesions; the significance of melanoma cells expressing CD20 under in vivo conditions is not yet clear and is currently under investigation (Pinc, et al., 2012; Schmidt, et al., 2011).
Recently, Schmidt et al, were able to target a small population of melanoma cells expressing CD20 using chimeric antigen receptor (CAR) engineered T cells (Schmidt, et al., 2011) in a mouse xenograft model. They showed that by targeting a small subset of CD20+ tumor cells with engineered T cells with redirected specificity for CD20, complete inhibition of tumor growth in mice could be achieved. Inhibition of tumor growth was long-lasting; furthermore, no tumor relapse in mice was observed for more than 36 weeks. Moreover, in a recent study, we reported that when advanced melanoma patients were treated with anti-CD20 antibody in an adjuvant setting, the majority of patients remain disease free during the three year period of observation (Pinc, et al., 2012). Similarly staged patients in historical controls showed less than 1 year of survival. In a single case study, Abken’s group has confirmed the regression of metastatic melanoma lesions in a patient treated with anti-CD20 in a non-adjuvant setting (Schlaak, et al., 2012). Overall, the above studies strongly suggest that a CD20+ melanoma subpopulation could be a major driver of tumor progression and elimination of this subset could result in disease free survival.
2. ABCB5/ABCG2/ABCB8
ABC transporters such as ABCB5, ABCB8 and ABCG2 are frequently reported to be present in various cancers including melanoma (Dean, et al., 2005; Szakacs, et al., 2004). Recently, Schatton et al reported a subpopulation of melanoma cells that have high expression of ABCB5 with tumor initiating properties (Schatton, et al., 2008). These cells were highly chemoresistant and targeting of the ABCB5 subpopulation resulted in inhibition of tumor growth in immunodeficient nude mice. This group also reported that the expression of ABCB5 was higher in metastatic melanomas when compared to primary or melanocytic nevi tissues. Melanoma cells obtained from nodal metastatic lesions had higher expression of ABCB5 as compared to cells obtained from visceral metastasis. Using immunodeficient SCID mice, the authors showed that ABCB5+ cells were more tumorigenic than ABCB5 negative melanoma cells. The CD133+ melanoma subpopulation that is chemoresistant is known to co-expresses ABCG2 (Monzani, et al., 2007; Taghizadeh, et al., 2011). Given the selective expression of ABCG2 in a minor subpopulation of CD133+ cells, its expression in melanoma tissue sections has not yet been confirmed. In vivo xenograft studies indicate the aggressive potential of cells that co-express CD133 and ABCG2 cells (Monzani, et al., 2007). In addition to ABCB5 and ABCG2, an in vitro study has shown the presence of an ABCB8+ melanoma subpopulation that is resistant to drugs such as doxorubicin (Elliott & Al-Hajj, 2009). However, melanoma tumor tissue staining of ABCB8 has not been confirmed thus far.
3. CD133
CD133, a transmembrane glycoprotein also known as prominin-1, is normally expressed on undifferentiated cells including endothelial progenitor cells, hematopoietic stem cells, fetal brainstem cells, and prostate epithelial cells (Neuzil, et al., 2007). CD133 has also been identified as a cancer stem cell marker with tumor initiating properties (Monzani, et al., 2007; Shmelkov, et al., 2008). Various solid tumors including brain, breast, colon, liver, lung, pancreatic and prostate cancers show expression of CD133 (Dembinski & Krauss, 2009; Liu, et al., 2006; Ricci-Vitiani, et al., 2007; Salmaggi, et al., 2006; Shmelkov, et al., 2008). A small proportion of melanomas and primary human melanocytes are known to express CD133 (Klein, et al., 2007; Rappa, Fodstad, & Lorico, 2008). Klein et al observed a significant increase in the expression of stem-cell markers CD133, CD166, and nestin in primary and metastatic melanomas compared with benign nevi (Klein, et al., 2007). Aggressive melanomas were usually associated with greater expression of these markers. However, there are some discrepancies regarding immune-detection of CD133 likely due to differences in the binding affinity of different antibody clones to the glycosylation sites of CD133 that vary between tumor and normal cells (Kemper, et al., 2010). Some reports indicate that only CD133+ melanoma cells are capable of forming tumors in immunodeficient NOD/SCID IL2Rγc (NSG) null mice, whereas CD133- cells failed to form tumors; these data imply that CD133+ cells are key drivers of tumor cell repopulation under the experimental conditions (Monzani, et al., 2007). However, we find that both CD133+ and CD133- melanoma cells are equally capable of forming tumors (unpublished). Drug resistant tumor subpopulations that were obtained from breast, glioma and lung tumors after chemotherapy frequently express CD133 (Levina, Marrangoni, DeMarco, Gorelik, & Lokshin, 2008; Liu, et al., 2006; Visvader & Lindeman, 2008). Higher expression of CD133 has been associated with up regulation of anti-apoptotic proteins and increased survival mechanisms. CD133+ drug resistant tumor subpopulations usually express increased levels of Nestin (NES) presence, which has been associated with de-differentiation and more aggressive behavior of the disease (Grichnik, et al., 2006; Klein, et al., 2007). NES co-expression is frequently observed in CD133+ and CD271+ tumor initiating subpopulations of melanomas (Grichnik, et al., 2006; Klein, et al., 2007). As melanocytes share common lineage with neural crest cells, co-expression of nestin, CD133, CD271 (nerve growth factor receptor [NGFR]) and other embryonic markers in melanoma subpopulations is expected.
4. CD271 (NGFR, also referred as p75 neurotrophin receptor)
CD271 or NGFR, a transmembrane protein, is found in a number of human neural-crest-derived tissues and in cancers from breast, colon, pancreas, prostate, ovaries and melanomas. Recently, Boiko et al have shown that CD271+ melanoma subpopulations derived from patient tissues are more tumorigenic and aggressive than CD271- subpopulations when transplanted in immunodeficient Rag2−/−γc−/− mice (Boiko, et al., 2010). Many of the melanoma-associated antigens such as MART1, MAGE, and tyrosinase were lost or down modulated in CD271+ cells (Boiko, et al., 2010). These antigen losses in subpopulation variants are mostly likely linked to the selection of immunologically resistant melanoma cells in vivo. Civenni et al found that the expression of CD271 correlated with higher metastatic potential and poor prognosis in an analysis performed in many biopsy specimens from melanoma patients (Civenni, et al., 2011). The authors have observed that CD271+ subpopulations of melanoma cells frequently show higher expression of ABCB5 transport proteins and lower expression of MAA, indicating that these cells may have survived drug therapy and anti-melanoma reactive immune T cells.
5. JARID1B
We have identified a slow cycling subpopulation of melanoma cells representing ~1–5% of all cells in tumor lesions that have stem-like or cancer initiating properties (Roesch, et al., 2010). These cells show high expression of histone demethylases jumonji ARID (JARID, also referred as lysine demethylase 5 [KDM5]) 1B, known to be critically involved in regulating gene expression and transcriptional activities. Preliminary data indicate that JARID1B expression is influenced by the TME. In prostate cancer, JARID1B up regulation is usually associated with increased androgen receptor expression; activation of androgen receptors is known to confer resistance to therapies. The expression of JARID1B in breast cancer cells is associated with increased proliferation due to specific repression of an anti-oncogene such as BRCA1 and members of the let-7 family of microRNA tumor suppressors (Mitra, Das, Huynh, & Jones, 2011). We have shown that isolated JARID1B+ melanoma cells can give rise to a rapidly proliferating progeny that is again heterogeneous (JARID1B+ and JARID1B–) like the parental tumor cells (Roesch, et al., 2010). Additionally, stable knockdown of JARID1B led to an initial acceleration of tumor growth followed by exhaustion, as determined by serial xeno-transplantation experiments in NSG null mice, suggesting that JARID1B has an essential role in continuous melanoma growth (Roesch, et al., 2010). Notably, Settleman’s group has recently reported that JARID1A, a close homolog of JARID1B, is required for drug resistance in non small-cell lung cancer cells (Sharma, et al., 2010), suggesting that slow cycling cells can survive most conventional and targeted therapies and that this subpopulation needs to be selectively targeted.
VI. New approaches to therapy
Melanomas are heterogeneous tumors, comprised of many genotypic and phenotypic subtypes. Given the complexity of the tumor cells, earlier therapeutic approaches designed to treat melanomas as a single disease using chemotherapeutic agents, such as DTIC or temozolomide, resulted in dismal response rates of < 15%. Moreover, the majority of patients developed resistance to most available therapies very early during treatment. In the last decade, the identification of mutations in the genes involved in MAPK activation, including BRAF and NRAS, or alterations or mutations in cell-cycle regulatory genes/proteins such as CCND1/CDK4 or C-KIT, have led to the development of targeted therapy approaches using small molecule inhibitors that are either approved (e.g.vemurafenib) or in late stage clinical trials (e.g: the BRAF inhibitor dabrafenib, the MEK inhibitor tramatenib) (Flaherty, Hodi, et al., 2010; Vidwans, et al., 2011). BRAF-V600E+ melanoma patients treated with vemurafenib experience dramatic tumor regression and improved survival compared to patients treated with conventional therapies (Flaherty, Puzanov, et al., 2010). Despite the impressive regression of bulky tumor lesions in patients treated with BRAF inhibitors, many of them eventually develop resistance to treatment (Vidwans, et al., 2011). Resistance to targeted agents can be mediated by diverse mechanisms, including development of secondary mutations, epigenetic changes in the target gene and activation of compensatory signaling pathways that result in increased tumor survival (Vidwans, et al., 2011; J. Villanueva, et al., 2011; J. Villanueva, et al., 2010). Several biological and chemical inhibitors are available to target multiple pathways that support proliferation and cell survival (Vidwans, et al., 2011; J. Villanueva, et al., 2011). For example, MEK inhibitors, which can block reactivation of the MAPK pathway, are in advanced stages of clinical investigation as single agents or in combination with BRAF inhibitors. RTK inhibitors or inhibitors of the PI3K pathway could also be used to block compensatory survival mechanisms that usually become activated in drug resistant tumors. A multi-modal therapy approach that combines targeting multiple pathways that promote maintenance of the bulk of the tumor with targeting melanoma subpopulations with a panel of antibodies or inhibitors may be necessary to prolonged disease free survival of melanoma patients (see Figure3). For this approach, each melanoma patient’s tumor needs to be profiled before and after therapy to determine the best combination therapy approach to target each individual tumor. Drug resistant tumor subpopulations that are frequently selected after therapy need to be analyzed for epigenetic and phenotypic changes in order to design a personalized targeted approach. Potentially, antibodies or drugs that neutralize IGF-1, PDGF or other tyrosine kinase receptors could be used to target drug resistant subpopulations that are known to have enhanced IGF1or PDFGF receptor signaling (J. Villanueva, et al., 2011; J. Villanueva, et al., 2010). An alternative approach is to use antibodies such as anti-CD20 or anti-CD133 or anti-CD271 or anti-ABCB5 to deplete respective minor drug resistant subpopulations. JARID1B+ subpopulation can be depleted by use of inhibitors. This strategy will help prevent tumor recurrence and thus obtain long lasting responses. Additionally, a marked increase in CD8+ T cell responses in regressing tumors after vemurafenib treatment (Wilmott, et al., 2012) supports the recent strategy of combined use of immune check point reagents such as anti-CTLA4 or anti-PD1 antibodies with vemurafenib. Preliminary results from these combination approaches, barring some skin sensitivity issues (Harding, Pulitzer, & Chapman, 2012), are encouraging but it is still too early to know if this treatment modality will improve overall survival of melanoma patients.
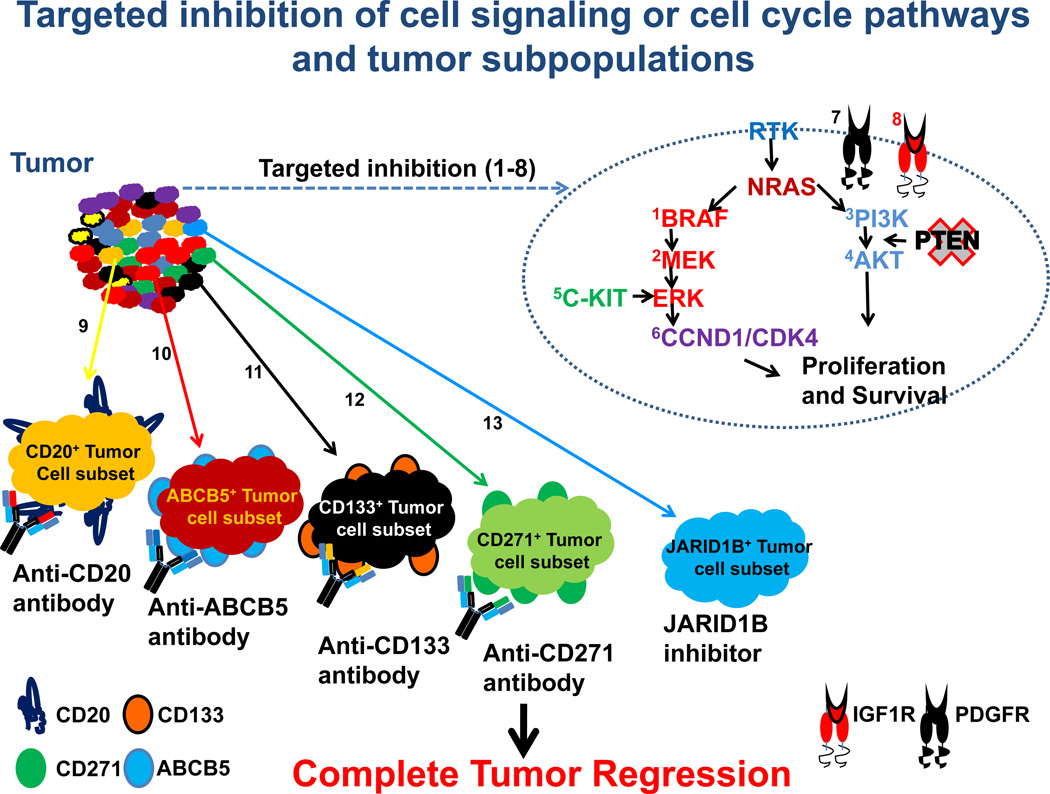
Figure 3. Potential new therapeutic approaches to target melanoma.
A heterogeneous tumor such as melanoma will require multi-targeted inhibition of signaling pathways (e.g. BRAF) or cell-cycle regulatory proteins (e.g. CDK inhibitors) (1–8) and depletion of minor subpopulations (e.g. CD20) that sustain the tumor using a combination of antibodies or inhibitors (9–13). This strategy will help prevent tumor recurrence and thus obtain long-lasting responses.
VII. Future directions
The recent development of advanced molecular techniques and their application to classify tumor subtypes based on gene signatures and protein expression profiles has revolutionized cancer treatment approaches. As described above, combination therapies targeting multiple signaling and cell-cycle pathways may be a useful approach to treat melanoma patients. This approach combined with immune checkpoint reagents using anti-CTLA4 and anti-PD1 antibodies will extend the expansion and retention of circulating anti-melanoma reactive cytotoxic T cells that are observed after targeted therapy. The presence of anti-melanoma reactive T cells could prevent recurrence of lesions after therapy withdrawal. Several recent reports suggest that primary or early stage lesions may have the genetic footprint for invasive potential of the neoplastic disease (Albini, Mirisola, & Pfeffer, 2008; Chin, et al., 2006; Ramaswamy, Ross, Lander, & Golub, 2003). The metastatic potential of these tumors is supported by the stromal-derived cells such as macrophages, fibroblasts or other leukocytes. In this context, a combination approach targeting the tumor stromal-derived cells and the tumor may be beneficial, providing long lasting responses and tumor regression.
VIII. Conclusion
Malignant melanoma, like other cancers, is a heterogeneous tumor comprised of many subpopulations with unique genotypic and phenotypic signatures. Single agent therapies such as DTIC or temozolomide resulted in low (<15%) response rates that were frequently followed by drug resistance. Molecular identification of mutant BRAFV600E and other gene mutations has led to the development of a number of targeted therapy drugs that have shown dramatic response rates in patients. Unfortunately, the responses to targeted therapy drugs are also transient and many patients develop resistance. A personalized therapy approach of treating patients based on the genotype and phenotype of their tumors with a combination of targeted therapy drugs that inhibit multiple signaling and cell-cycle pathways will be necessary for long lasting regression of melanoma lesions. Additionally, targeting tumor subpopulations that are generally drug resistant will be beneficial in preventing melanoma recurrence. Inclusion of immune checkpoint reagents such as anti-CTLA4 or anti-PD1 antibodies with targeted therapy drugs in the treatment regimen may provide additional benefits by expansion and retention of anti-melanoma reactive T cells that have a potential to prevent emergence of drug resistant tumor cells.
Acknowledgement
The authors thank Jessica Blodgett for editorial revisions and the NIH support (CA-114046, CA-025874 and CA-010815) for melanoma research.
M. Herlyn received support from GlaxoSmithKline (GSK).
Abbreviations
- ACS
American Cancer Society
- ATP
Adenosine-5'-triphosphate
- BCL
B-cell lymphoma
- BER
Base excision repair
- CAR
Chimeric antigen receptor
- CCDND1
Cyclin D1
- CDK
Cyclin-dependent kinase
- CDKN2a
Cyclin-dependent kinas inhibitor
- CTLA4
Cytotoxic T-lymphocyte antigen 4
- FDA
Food and Drug Administration
- HHR
Homologous recombinational repair
- IGF
insulin-dependent growth factor
- IFN
Interferon
- IL
Interleukin
- INPP4b
Inositol polyphosphate 4-phosphatase type II
- MAPK
Mitogen activated protein kinase
- MCL
Myeloid cell leukemia
- MDR
Multiple drug resistance
- MGMT
Methylguanine-DNA methyltransferase
- MITF
Microphthalmia associated transcription factor
- MMR
Mismatch repair
- NER
Nucleotide excision repair
- NES
Nestin
- NHEJ
Non-homologous end joining
- NOD
Non obese diabetic
- NSG
NOD SCID IL2 receptor gamma chain knockout
- PARP
Polyadenosine diphosphate-ribose polymerase
- PD
Programmed cell death
- PDGF
Platelet derived growth factor
- PI
Phosphoinositide
- PTEN
Phosphotase and tensin
- RGP
Radial growth phase
- RNI
Reactive nitrogen intermediate
- ROS
Reactive oxygen species
- RTK
Receptor tyrosine kinase
- SCID
Severe combined immunodeficiency mice
- TKi
Tyrosine kinase inhibitors
- TME
Tumor microenvironment
- VGP
Vertical growth phase
- WT
Wild type
Footnotes
Conflict of interest
The other authors disclosed no potential conflicts of interest.
References
- Abdullah C, Wang X, Becker D. Expression analysis and molecular targeting of cyclin-dependent kinases in advanced melanoma. Cell Cycle. 2011;10(6):977–988. doi: 10.4161/cc.10.6.15079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addla SK, Brown MD, Hart CA, Ramani VA, Clarke NW. Characterization of the Hoechst33342 side population from normal and malignant human renal epithelial cells. Am J Physiol Renal Physiol. 2008 doi: 10.1152/ajprenal.90286.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albini A, Mirisola V, Pfeffer U. Metastasis signatures: genes regulating tumormicroenvironment interactions predict metastatic behavior. Cancer Metastasis Rev. 2008;27(1):75–83. doi: 10.1007/s10555-007-9111-x. [DOI] [PubMed] [Google Scholar]
- Atefi M, von Euw E, Attar N, Ng C, Chu C, Guo D, et al. Reversing melanoma cross-resistance to BRAF and MEK inhibitors by co-targeting the AKT/mTOR pathway. PLoS One. 2011;6(12):e28973. doi: 10.1371/journal.pone.0028973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine CK, Yoo JS, Potti A, Yoshimoto Y, Zipfel PA, Friedman HS, et al. Genomic and molecular profiling predicts response to temozolomide in melanoma. Clin Cancer Res. 2009;15(2):502–510. doi: 10.1158/1078-0432.CCR-08-1916. [DOI] [PubMed] [Google Scholar]
- Bittner M, Meltzer P, Chen Y, Jiang Y, Seftor E, Hendrix M, et al. Molecular classification of cutaneous malignant melanoma by gene expression profiling. Nature. 2000;406(6795):536–540. doi: 10.1038/35020115. [DOI] [PubMed] [Google Scholar]
- Bloethner S, Snellman E, Bermejo JL, Hiripi E, Gast A, Thirumaran RK, et al. Differential gene expression in melanocytic nevi with the V600E BRAF mutation. Genes Chromosomes Cancer. 2007;46(11):1019–1027. doi: 10.1002/gcc.20488. [DOI] [PubMed] [Google Scholar]
- Boiko AD, Razorenova OV, van de Rijn M, Swetter SM, Johnson DL, Ly DP, et al. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature. 2010;466(7302):133–137. doi: 10.1038/nature09161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PA, Middleton MR. DNA repair pathways in drug resistance in melanoma. Anticancer Drugs. 2004;15(5):421–426. doi: 10.1097/01.cad.0000127665.74096.93. [DOI] [PubMed] [Google Scholar]
- Brahmer JR, Drake CG, Wollner I, Powderly JD, Picus J, Sharfman WH, et al. Phase I study of single-agent anti-programmed death-1 (MDX-1106) in refractory solid tumors: safety, clinical activity, pharmacodynamics, and immunologic correlates. J Clin Oncol. 2010;28(19):3167–3175. doi: 10.1200/JCO.2009.26.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennen WN, Isaacs JT, Denmeade SR. Rationale behind targeting fibroblast activation protein-expressing carcinoma-associated fibroblasts as a novel chemotherapeutic strategy. Mol Cancer Ther. 2012;11(2):257–266. doi: 10.1158/1535-7163.MCT-11-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campoli M, Ferris R, Ferrone S, Wang X. Immunotherapy of malignant disease with tumor antigen-specific monoclonal antibodies. Clin Cancer Res. 2009;16(1):11–20. doi: 10.1158/1078-0432.CCR-09-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364(26):2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Garraway LA, Fisher DE. Malignant melanoma: genetics and therapeutics in the genomic era. Genes Dev. 2006;20(16):2149–2182. doi: 10.1101/gad.1437206. [DOI] [PubMed] [Google Scholar]
- Chung C-Y, Khanna P, Gowda R, Sharma A, Neves R, Dong C, Robertson GP. Vemurafenib (PLX4032) promotes epigenetic changes in melanoma cells leading to development of more invasive metastatic disease. [Abstract] Pigment Cell & Melanoma Research. 2011;24(5):1000. [Google Scholar]
- Civenni G, Walter A, Kobert N, Mihic-Probst D, Zipser M, Belloni B, et al. Human CD271-positive melanoma stem cells associated with metastasis establish tumor heterogeneity and long-term growth. Cancer Res. 2011;71(8):3098–3109. doi: 10.1158/0008-5472.CAN-10-3997. [DOI] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- de Souza CF, Morais AS, Jasiulionis MG. Biomarkers as key contributors in treating malignant melanoma metastases. Dermatol Res Pract. 2012;2012:156068. doi: 10.1155/2012/156068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5(4):275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- Dembinski JL, Krauss S. Characterization and functional analysis of a slow cycling stem cell-like subpopulation in pancreas adenocarcinoma. Clin Exp Metastasis. 2009 doi: 10.1007/s10585-009-9260-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denardo DG, Brennan DJ, Rexhepaj E, Ruffell B, Shiao SL, Madden SF, et al. Leukocyte Complexity Predicts Breast Cancer Survival and Functionally Regulates Response to Chemotherapy. Cancer Discov. 2011;1:54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick JE. Looking ahead in cancer stem cell research. Nat Biotechnol. 2009;27(1):44–46. doi: 10.1038/nbt0109-44. [DOI] [PubMed] [Google Scholar]
- Dissanayake SK, Olkhanud PB, O'Connell MP, Carter A, French AD, Camilli TC, et al. Wnt5A regulates expression of tumor-associated antigens in melanoma via changes in signal transducers and activators of transcription 3 phosphorylation. Cancer Res. 2008;68(24):10205–10214. doi: 10.1158/0008-5472.CAN-08-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott AM, Al-Hajj MA. ABCB8 mediates doxorubicin resistance in melanoma cells by protecting the mitochondrial genome. Mol Cancer Res. 2009;7(1):79–87. doi: 10.1158/1541-7786.MCR-08-0235. [DOI] [PubMed] [Google Scholar]
- Fang D, Nguyen TK, Leishear K, Finko R, Kulp AN, Hotz S, et al. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65(20):9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- Fecher LA, Cummings SD, Keefe MJ, Alani RM. Toward a molecular classification of melanoma. J Clin Oncol. 2007;25(12):1606–1620. doi: 10.1200/JCO.2006.06.0442. [DOI] [PubMed] [Google Scholar]
- Flaherty KT, Hodi FS, Bastian BC. Mutation-driven drug development in melanoma. Curr Opin Oncol. 2010;22(3):178–183. doi: 10.1097/cco.0b013e32833888ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty KT, Puzanov I, Kim KB, Ribas A, McArthur GA, Sosman JA, et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med. 2010;363(9):809–819. doi: 10.1056/NEJMoa1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank NY, Margaryan A, Huang Y, Schatton T, Waaga-Gasser AM, Gasser M, et al. ABCB5-mediated doxorubicin transport and chemoresistance in human malignant melanoma. Cancer Res. 2005;65(10):4320–4333. doi: 10.1158/0008-5472.CAN-04-3327. [DOI] [PubMed] [Google Scholar]
- Friedlander P, Hodi FS. Advances in targeted therapy for melanoma. Clin Adv Hematol Oncol. 2010;8(9):619–627. [PubMed] [Google Scholar]
- Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemadmoghadam D, Bowtell D, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 2009;16(2):115–125. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grichnik JM, Burch JA, Schulteis RD, Shan S, Liu J, Darrow TL, et al. Melanoma, a tumor based on a mutant stem cell? J Invest Dermatol. 2006;126(1):142–153. doi: 10.1038/sj.jid.5700017. [DOI] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadnagy A, Gaboury L, Beaulieu R, Balicki D. SP analysis may be used to identify cancer stem cell populations. Exp Cell Res. 2006;312(19):3701–3710. doi: 10.1016/j.yexcr.2006.08.030. [DOI] [PubMed] [Google Scholar]
- Halaban R, Krauthammer M, Pelizzola M, Cheng E, Kovacs D, Sznol M, et al. Integrative analysis of epigenetic modulation in melanoma cell response to decitabine: clinical implications. PLoS One. 2009;4(2):e4563. doi: 10.1371/journal.pone.0004563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding JJ, Pulitzer M, Chapman PB. Vemurafenib sensitivity skin reaction after ipilimumab. N Engl J Med. 2012;366(9):866–868. doi: 10.1056/NEJMc1114329. [DOI] [PubMed] [Google Scholar]
- Ho MM, Ng AV, Lam S, Hung JY. Side population in human lung cancer cell lines and tumors is enriched with stem-like cancer cells. Cancer Res. 2007;67(10):4827–4833. doi: 10.1158/0008-5472.CAN-06-3557. [DOI] [PubMed] [Google Scholar]
- Hodi FS, O'Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363(8):711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell PM, Jr, Liu S, Ren S, Behlen C, Fodstad O, Riker AI. Epigenetics in human melanoma. Cancer Control. 2009;16(3):200–218. doi: 10.1177/107327480901600302. [DOI] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468(7326):968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C, Plummer ER. PARP inhibitors and cancer therapy - early results and potential applications. Br J Radiol. 2008;81(Spec No 1):S2–S5. doi: 10.1259/bjr/30872348. [DOI] [PubMed] [Google Scholar]
- Kauffmann A, Rosselli F, Lazar V, Winnepenninckx V, Mansuet-Lupo A, Dessen P, et al. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene. 2008;27(5):565–573. doi: 10.1038/sj.onc.1210700. [DOI] [PubMed] [Google Scholar]
- Kemper K, Sprick MR, de Bree M, Scopelliti A, Vermeulen L, Hoek M, et al. The AC133 epitope, but not the CD133 protein, is lost upon cancer stem cell differentiation. Cancer Res. 2010;70(2):719–729. doi: 10.1158/0008-5472.CAN-09-1820. [DOI] [PubMed] [Google Scholar]
- Klein WM, Wu BP, Zhao S, Wu H, Klein-Szanto AJ, Tahan SR. Increased expression of stem cell markers in malignant melanoma. Mod Pathol. 2007;20(1):102–107. doi: 10.1038/modpathol.3800720. [DOI] [PubMed] [Google Scholar]
- Kline J, Gajewski TF. Clinical development of mAbs to block the PD1 pathway as an immunotherapy for cancer. Curr Opin Investig Drugs. 2010;11(12):1354–1359. [PubMed] [Google Scholar]
- Koh HK. Cutaneous melanoma. N Engl J Med. 1991;325(3):171–182. doi: 10.1056/NEJM199107183250306. [DOI] [PubMed] [Google Scholar]
- Korkaya H, Liu S, Wicha MS. Breast cancer stem cells, cytokine networks, and the tumor microenvironment. J Clin Invest. 2011a;121(10):3804–3809. doi: 10.1172/JCI57099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkaya H, Liu S, Wicha MS. Regulation of cancer stem cells by cytokine networks: attacking cancer's inflammatory roots. Clin Cancer Res. 2011b;17(19):6125–6129. doi: 10.1158/1078-0432.CCR-10-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levina V, Marrangoni AM, DeMarco R, Gorelik E, Lokshin AE. Drugselected human lung cancer stem cells: cytokine network, tumorigenic and metastatic properties. PLoS One. 2008;3(8):e3077. doi: 10.1371/journal.pone.0003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AJ, Mihm MC., Jr Melanoma. N Engl J Med. 2006;355(1):51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- Mitra D, Das PM, Huynh FC, Jones FE. Jumonji/ARID1 B (JARID1B) protein promotes breast tumor cell cycle progression through epigenetic repression of microRNA let-7e. J Biol Chem. 2011;286(47):40531–40535. doi: 10.1074/jbc.M111.304865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagut C, Sharma SV, Shioda T, McDermott U, Ulman M, Ulkus LE, et al. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer Res. 2008;68(12):4853–4861. doi: 10.1158/0008-5472.CAN-07-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzani E, Facchetti F, Galmozzi E, Corsini E, Benetti A, Cavazzin C, et al. Melanoma contains CD133 and ABCG2 positive cells with enhanced tumourigenic potential. Eur J Cancer. 2007;43(5):935–946. doi: 10.1016/j.ejca.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468(7326):973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuzil J, Stantic M, Zobalova R, Chladova J, Wang X, Prochazka L, et al. Tumour-initiating cells vs. cancer 'stem' cells and CD133: what's in the name? Biochem Biophys Res Commun. 2007;355(4):855–859. doi: 10.1016/j.bbrc.2007.01.159. [DOI] [PubMed] [Google Scholar]
- Nishimura EK, Jordan SA, Oshima H, Yoshida H, Osawa M, Moriyama M, et al. Dominant role of the niche in melanocyte stem-cell fate determination. Nature. 2002;416(6883):854–860. doi: 10.1038/416854a. [DOI] [PubMed] [Google Scholar]
- Pinc A, Somasundaram R, Wagner C, Hormann M, Karanikas G, Jalili A, et al. Targeting CD20 in Melanoma Patients at High Risk of Disease Recurrence. Mol Ther. 2012 doi: 10.1038/mt.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer R, Jones C, Middleton M, Wilson R, Evans J, Olsen A, et al. Phase I study of the poly(ADP-ribose) polymerase inhibitor, AG014699, in combination with temozolomide in patients with advanced solid tumors. Clin Cancer Res. 2008;14(23):7917–7923. doi: 10.1158/1078-0432.CCR-08-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480(7377):387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahallad A, Sun C, Huang S, Di Nicolantonio F, Salazar R, Zecchin D, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483(7387):100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33(1):49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- Rappa G, Fodstad O, Lorico A. The stem cell-associated antigen CD133 (Prominin-1) is a molecular therapeutic target for metastatic melanoma. Stem Cells. 2008;26(12):3008–3017. doi: 10.1634/stemcells.2008-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastetter M, Schagdarsurengin U, Lahtz C, Fiedler E, Marsch W, Dammann R, et al. Frequent intra-tumoural heterogeneity of promoter hypermethylation in malignant melanoma. Histol Histopathol. 2007;22(9):1005–1015. doi: 10.14670/HH-22.1005. [DOI] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445(7123):111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141(4):583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runger TM, Emmert S, Schadendorf D, Diem C, Epe B, Hellfritsch D. Alterations of DNA repair in melanoma cell lines resistant to cisplatin, fotemustine, or etoposide. J Invest Dermatol. 2000;114(1):34–39. doi: 10.1046/j.1523-1747.2000.00844.x. [DOI] [PubMed] [Google Scholar]
- Salmaggi A, Boiardi A, Gelati M, Russo A, Calatozzolo C, Ciusani E, et al. Glioblastoma-derived tumorospheres identify a population of tumor stem-like cells with angiogenic potential and enhanced multidrug resistance phenotype. Glia. 2006;54(8):850–860. doi: 10.1002/glia.20414. [DOI] [PubMed] [Google Scholar]
- Sarasin A, Kauffmann A. Overexpression of DNA repair genes is associated with metastasis: a new hypothesis. Mutat Res. 2008;659(1–2):49–55. doi: 10.1016/j.mrrev.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Satyamoorthy K, Herlyn M. Cellular and molecular biology of human melanoma. Cancer Biol Ther. 2002;1(1):14–17. doi: 10.4161/cbt.1.1.32. [DOI] [PubMed] [Google Scholar]
- Scharenberg CW, Harkey MA, Torok-Storb B. The ABCG2 transporter is an efficient Hoechst 33342 efflux pump and is preferentially expressed by immature human hematopoietic progenitors. Blood. 2002;99(2):507–512. doi: 10.1182/blood.v99.2.507. [DOI] [PubMed] [Google Scholar]
- Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, et al. Identification of cells initiating human melanomas. Nature. 2008;451(7176):345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2009;31(1):37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaak M, Schmidt P, Bangard C, Kurschat P, Mauch C, Abken H. Regression of metastatic melanoma in a patient by antibody targeting of cancer stem cells. Oncotarget. 2012;3(1):22–30. doi: 10.18632/oncotarget.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt P, Kopecky C, Hombach A, Zigrino P, Mauch C, Abken H. Eradication of melanomas by targeted elimination of a minor subset of tumor cells. Proc Natl Acad Sci U S A. 2011;108(6):2474–2479. doi: 10.1073/pnas.1009069108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt P, Losch C, Manista K, Kopecky C, Hombach A, Loffek S, et al. Targeted elimination of cancer stem cells eradicates melanoma. Human Gene Therapy. 2009;20(11):1391–1391. [Google Scholar]
- Sensi M, Nicolini G, Petti C, Bersani I, Lozupone F, Molla A, et al. Mutually exclusive NRASQ61R and BRAFV600E mutations at the single-cell level in the same human melanoma. Oncogene. 2006;25(24):3357–3364. doi: 10.1038/sj.onc.1209379. [DOI] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141(1):69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shmelkov SV, Butler JM, Hooper AT, Hormigo A, Kushner J, Milde T, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133- metastatic colon cancer cells initiate tumors. J Clin Invest. 2008;118(6):2111–2120. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Somasundaram R, Villanueva J, Herlyn M. Will engineered T cells expressing CD20 scFv eradicate Melanoma? Mol Ther. 2011;19(4):638–640. doi: 10.1038/mt.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366(8):707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakacs G, Annereau JP, Lababidi S, Shankavaram U, Arciello A, Bussey KJ, et al. Predicting drug sensitivity and resistance: profiling ABC transporter genes in cancer cells. Cancer Cell. 2004;6(2):129–137. doi: 10.1016/j.ccr.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Taghizadeh R, Noh M, Huh YH, Ciusani E, Sigalotti L, Maio M, et al. CXCR6, a newly defined biomarker of tissue-specific stem cell asymmetric self-renewal, identifies more aggressive human melanoma cancer stem cells. PLoS One. 2011;5(12):e15183. doi: 10.1371/journal.pone.0015183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor ST, Hickman JA, Dive C. Epigenetic determinants of resistance to etoposide regulation of Bcl-X(L) and Bax by tumor microenvironmental factors. J Natl Cancer Inst. 2000;92(1):18–23. doi: 10.1093/jnci/92.1.18. [DOI] [PubMed] [Google Scholar]
- Tedder TF, Engel P. CD20: a regulator of cell-cycle progression of B lymphocytes. Immunol Today. 1994;15(9):450–454. doi: 10.1016/0167-5699(94)90276-3. [DOI] [PubMed] [Google Scholar]
- Tredan O, Galmarini CM, Patel K, Tannock IF. Drug resistance and the solid tumor microenvironment. J Natl Cancer Inst. 2007;99(19):1441–1454. doi: 10.1093/jnci/djm135. [DOI] [PubMed] [Google Scholar]
- van Kempen LC, Ruiter DJ, van Muijen GN, Coussens LM. The tumor microenvironment: a critical determinant of neoplastic evolution. Eur J Cell Biol. 2003;82(11):539–548. doi: 10.1078/0171-9335-00346. [DOI] [PubMed] [Google Scholar]
- Vidwans SJ, Flaherty KT, Fisher DE, Tenenbaum JM, Travers MD, Shrager J. A melanoma molecular disease model. PLoS One. 2011;6(3):e18257. doi: 10.1371/journal.pone.0018257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva J, Vultur A, Herlyn M. Resistance to BRAF inhibitors: unraveling mechanisms and future treatment options. Cancer Res. 2011;71(23):7137–7140. doi: 10.1158/0008-5472.CAN-11-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18(6):683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva MT. Targeted therapies: Smart tumor, smarter treatment. Nat Rev Clin Oncol. 2012;9(3):127. doi: 10.1038/nrclinonc.2012.4. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8(10):755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- Wilmott JS, Long GV, Howle JR, Haydu LE, Sharma RN, Thompson JF, et al. Selective BRAF Inhibitors Induce Marked T-cell Infiltration into Human Metastatic Melanoma. Clin Cancer Res. 2012 doi: 10.1158/1078-0432.CCR-11-2479. [DOI] [PubMed] [Google Scholar]
- Yancovitz M, Litterman A, Yoon J, Ng E, Shapiro RL, Berman RS, et al. Intraand inter-tumor heterogeneity of BRAF(V600E))mutations in primary and metastatic melanoma. PLoS One. 2012;7(1):e29336. doi: 10.1371/journal.pone.0029336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27(41):5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabierowski SE, Herlyn M. Melanoma stem cells: the dark seed of melanoma. J Clin Oncol. 2008;26(17):2890–2894. doi: 10.1200/JCO.2007.15.5465. [DOI] [PubMed] [Google Scholar]
- Zitvogel L, Apetoh L, Ghiringhelli F, Kroemer G. Immunological aspects of cancer chemotherapy. Nat Rev Immunol. 2008;8(1):59–73. doi: 10.1038/nri2216. [DOI] [PubMed] [Google Scholar]
- Zitvogel L, Kepp O, Kroemer G. Immune parameters affecting the efficacy of chemotherapeutic regimens. Nat Rev Clin Oncol. 2011;8(3):151–160. doi: 10.1038/nrclinonc.2010.223. [DOI] [PubMed] [Google Scholar]