Abstract
The molecular function of occludin, an integral membrane component of tight junctions, remains unclear. VEGF-induced phosphorylation sites were mapped on occludin by combining MS data analysis with bioinformatics. In vivo phosphorylation of Ser490 was validated and protein interaction studies combined with crystal structure analysis suggest that Ser490 phosphorylation attenuates the interaction between occludin and ZO-1. This study demonstrates that combining MS data and bioinformatics can successfully identify novel phosphorylation sites from limiting samples.
Keywords: tight junction, occludin, phosphorylation, VEGF, phosphosite mapping
Introduction
The capacity of multicellular organisms to define and regulate the composition of tissue compartments depends upon the junctional complex, which affords intercellular interactions within the epithelium and endothelium. The tight junctions create high resistance barriers in a variety of tissues, such as the intestinal epithelium and the vascular endothelium of the blood–brain and blood–retinal barrier, which is necessary to control the flux of water, ions and solutes. The importance of paracellular barrier properties provided by tight junctions is highlighted by the number of pathophysiological states involving alterations in permeability including inflammatory bowl disease, brain tumors and diabetic retinopathy.1–7
Occludin is an integral membrane protein component of tight junctions with tetraspan topology that orients both the amine and carboxy termini in the cytoplasm. Occludin contains four different functional domains.8 The intracellular N-terminal cytoplasmic tail (aa 1–60) regulates proteosome mediated occludin degradation9 and neutrophil transmigration across epithelial sheets.8,10 The tetraspan helices and corresponding loops possess sequence homology to the MARVEL domain (aa 61–265), a protein motif involved in vesicle trafficking and membrane apposition events.11 The proximal C-terminus (aa 266–412) has not been associated with a functional motif, while the distal C-terminus (aa 413–522) forms a coiled-coil domain. Crystal structure and mutational analysis of this coiled-coil domain demonstrates a positively charged face required for binding to the tight junction organizing protein ZO-1.12
Several lines of evidence suggest that occludin modulates barrier permeability (as reviewed in refs 13 and 14). Most recently, stable expression of siRNA directed to occludin resulted in a selective increase in paracellular permeability of small organic cations and altered the cells response to cholesterol depletion.15 The use of siRNA to occludin also increased permeability to a fluorescent marker after application of transmural pressure (Manuscript submitted). These data are consistent with current models whereby the type and amount of claudins and/or occludin located at tight junctions determine the selectivity of the paracellular pathway.16 Although data from occludin gene deletion mice suggest that occludin is not required for the formation of tight junction strands, the numerous phenotypic alterations together with the siRNA studies suggest a role for occludin in appropriately regulating the barrier properties of tight junctions.17
Recent studies suggest that occludin may also contribute to the regulation of cell growth and differentiation.18,19 Occludin gene deletion led to increased gut epithelial hyperplasia.17,19 Induction of epithelial-mesenchymal transition is associated with decreased expression of occludin20 and decreased occludin expression has been associated with enhanced cancer invasiveness.21,22 Moreover, overexpression of occludin in raf1-transformed cells returned cells to an epithelial phenotype19 and expression of occludin in various cancer cell lines attenuated several metastatic indices.23
Post-translational modification (PTM) of occludin may regulate occludin function. Changes in occludin phosphorylation state have been associated with increased endothelial permeability in response to vascular endothelial growth factor (VEGF).24,25 However, studies relating site-specific occludin phosphorylation to occludin function are lacking. A complex pattern of occludin phosphorylation has been suggested through the use of two-dimensional gel electrophoresis. These studies have revealed that in primary bovine retinal endothelial cells (BREC), occludin is basally phosphorylated on two resides and growth factor stimulation leads to the phosphorylation of at least three additional sites.25 Importantly, similar results have been observed by others.26 Moreover, phosphoamino acid analysis of brain extracts,27 as well as endothelial26 and MDCK cell28 lysates, suggest that occludin is primarily phosphorylated on serine and threonine residues and that the slower migrating, hyperphosphorylated form of occludin is associated with phosphorylation on multiple serine residues.28 Previously, an occludin phosphorylation site had been identified from occludin phosphorylated in vitro with a cocktail of protein kinase C isoforms.29 However, no known phosphosites have been determined in vivo.
In the adjoining paper, we have demonstrated the effectiveness of identifying in vivo phosphorylation sites through a combined MS and bioinformatics approach using the well-studied phosphoprotein AKT1. In this study, we have applied this approach to identify five putative in vivo phosphosites on occludin in VEGF-treated primary retinal endothelial cells. Phosphorylation of occludin on one of these sites, Ser490, was subsequently verified with a phosphosite-specific antibody and shown to be VEGF responsive. To the best of our knowledge, this is the first validation of an occludin phosphosite in vivo. Protein interaction studies and crystal structure analysis of occludin mutated at Ser 490 to aspartate (S490D) suggest that phosphorylation at Ser490 attenuates the interaction between occludin and ZO-1 by altering the charge distribution within the coiled-coil domain of occludin. Taken together, these data suggest that growth factor stimulation of Ser490 phosphorylation alters binding to ZO-1 and may act as a molecular switch to disassemble occludin from tight junctions. Furthermore, these studies confirm the utility of our combined MS and bioinformatics approach for identifying novel phosphorylation sites from biological systems where only limited sample is available.
Materials and Methods
Materials
Recombinant human VEGF165 was purchased from R&D Systems (Minneapolis, MN). ZO-1 rat monoclonal antibody was produced from a clone (R40–76) obtained from Dr. Bruce Stevenson. For immunoblot detection, anti-rabbit IgG-alkaline phosphatase, anti-mouse IgG-alkaline phosphatase, anti-mouse IgG-horseradish peroxidase, and anti-rabbit IgG-horseradish peroxidase were obtained from Amersham Pharmacia Biotech (Piscataway, NJ). Goat serum was obtained from Jackson Immunoresearch Laboratories (West Grove, PA) and highly cross-adsorbed goat anti-mouse Alexa Flour-488, goat anti-rabbit Alexa Flour-555, and goat anti-rat Alexa Flour-647 were from Invitrogen.
Cell Culture and Immunoblotting
Primary BREC were isolated and cultured as described previously.30 For all experiments, 2 days after confluence, the BREC were treated with VEGF (50 ng/mL) for 15 min or the time indicated. Cell lysates were harvested and Western blots were perfermed as described previously.25 Cell lysis buffer is described below. Primary antibodies were used at the following dilutions: α-Occ-pS490 (1:250), α-Occ-486-494 (1:250), rabbit α-Occ (1:1000, Zymed), mouse α-Occ (1:1000, Zymed), rabbit α-phospho-p42/44 MAPK (1:1000, Cell Signaling Technology). Primary antibodies were detected by either ECF, ECL Plus, or ECL Advance. Images detected with ECF were quantified by ImageQuant 5.0 software (Molecular Dynamics, Sunnyvale, CA), while images detected with ECL plus and ECL advance were quantified with GeneSnap software (SynGene, Frederick MD).
Mass Spectrometry. 1. Crude Membrane Preparation and Immunoprecipitation
For each condition, BREC on 4 × 100 mm polystyrene dishes were harvested by washing two times with ice-cold PBS and subjected to a crude membrane preparation. BREC were scraped in Buffer A (250 mM sucrose, 10 mM Tris, pH 7.4, 1 mM EGTA, 4 mM EDTA, 1 mM benzamidine, 1 mM NaV04, 10 mM NaF, 10 mM sodium pyrophosphate, 1 mM microcystin, and a cømplete (EDTA free) protease inhibitor cocktail tablet), homogenized by dounce, and centrifuged at 38 200 rpm for 20 min at 4 °C. The pellet was resuspended in Buffer B (10 mM Tris, pH 7.4, 1 mM EGTA, and 4 mM EDTA), homogenized by dounce, and centrifuged at 38 200 rpm for 20 min at 4 °C. The pellet was resuspended in lysis buffer (1% Trition X100, 0.5% sodium deoxycholate, 0.2% SDS, 100 mM NaCl, 2 mM EDTA, 10 mM Hepes, pH 7.5, 1 mM benzamidine, 1 mM NaV04, 10 mM NaF, 10 mM sodium pyrophosphate, 1 mM microcystin and a Complete (EDTA free) protease inhibitor cocktail tablet) and sonicated. A portion of this crude membrane preparation was used for Western blot.
2. Immunoprecipitation
The crude membrane preparation was precleared with protein G beads (Amersham) for 1 h at 4 °C. Following microcentrifugation, the supernatant was moved to a fresh tube and 10 μg of rabbit antioccludin was added to each reaction which proceeded overnight at 4 °C. To complete the reaction, protein G beads (Amersham) were added and the samples were rocked for 2 h at 4 °C. Each reaction was washed three times with lysis buffer and the immunoprecipitated occludin was eluted with boiling Laemmli sample buffer.
3. SDS-PAGE and Tryptic Digestion
Occludin IP samples were subjected to SDS-PAGE and stained with Sypro Ruby (Molecular Probes). The location of occludin containing bands was determined by running a fraction of each IP reaction in parallel and immunoblotting with rabbit α-occludin (1:1000, Zymed). Control and VEGF lanes were excised as a ladder of three bands denoted low, medium or high which represent differentially phosphorylated forms of occludin. Samples were destained twice (50% acetonitrile (AcN) containing 200 mM NH4CO3, pH 8) for 45 min at 37 °C and dried completely. Each sample was reduced (2 mM TCEP, 25 mM NH4CO3, pH 8) for 15 min at 37 °C with agitation and then alkylated (20 mM iodoacetamide, 25 mM NH4CO3, pH 8) for 30 min at 37 °C in the dark. All samples were washed three times with 25 mM NH4CO3, pH 8, and dried in a SpeedVac. Each sample was rehydrated with 1.5× original gel slice volume of 0.02 μg/μL of trypsin (sequencing grade modified trypsin, Promega) in 50% AcN, 40 mM NH4CO3, pH 8, and 0.1% (w/v) n-octylglucoside for 60 min at room temperature to allow for the concentrated trypsin to diffuse into the gel slice. An additional 50 μL of 50% AcN, 40 mM NH4CO3, pH 8, and 0.1% n-octylglucoside was added and the samples were incubated for 16–18 h at 37 °C with agitation. The supernatant, containing occludin fragments, was removed, dried and resuspended in 200 μL of water three times. After the last wash, the samples were resuspended in 20 μL of water and cleaned (10 × 3 min binding time) with strong cation exchange ZipTip (ZipTipSCX; Eppindorf) columns according to the manufacturer’s instructions.
4. Data Acquisition
The ZipTipSCX eluant was spotted on MALDI target plates with α-cyano-4-hydroxycinnamic acid (CHCA). The samples were analyzed by peptide mass fingerprinting analysis on the MS instrument (Applied Biosystems 4700 Proteomics Analyzer MALDI TOF-TOF). Mass spectra were acquired in positive ion mode.
5. Data Analysis
FindMod (www.expasy.org) was used to compare observed MS data to predicted tryptic masses of bovine occludin in order to identify potential phosphopeptides which were identified by an observed mass difference of 80 Da. Specific parameters used in FindMod include a mass tolerance of 100 ppm and an allowance of 2 missed tryptic cuts. Phosphorylation site prediction of bovine occludin protein sequence was obtained using Scansite (www.Scansite.mit.edu; medium stringency), ScanProsite (www.expasy.org), and NetPhos 2.0 (www.expasy.org). Scansite prediction results include the kinase predicted to phosphorylate a given site and the associated likelihood percentile. ScanProsite results include the kinase associated with the predicted site, while NetPhos results include the predictions greater than 0.5 where closer to 1.0 is associated with more accurate predictions.
Phosphospecific Antibody Production and Purification
A rabbit anti-phosphopeptide antibody was created in conjunction with Quality Controlled Biochemicals (Hopkington, MA), by immunizing rabbits with Ac-CLKQVKGpSADYK, a phosphopeptide covering human occludin residues 486–494. Serum was purified three times over the corresponding nonphosphopeptide column (Ac-CLKQVKGSADYK) to remove antibodies binding to the peptide in the absence of phosphorylation. The flow-through from the nonphosphopeptide column was purified over the phosphopeptide column and denoted anti-occludin phospho-Ser 490 (α-Occ-pS490). Specificity to the phosphorylated form of the peptide antigen was tested by conducting peptide blocking experiments. Prior to incubation with transferred BREC lysates, α-Occ-pS490 was combined with 50× molar excess of either Ac-CLKQVKGpSADYK or Ac-CLKQVKGSADYK and incubated at 37 °C for 20 min. The mixture was subsequently diluted in TBST with 5% milk and applied to blots of transferred BREC lysates. A nonphospho antibody was obtained from the fraction of serum bound to the nonphosphopeptide column which was eluted and denoted α-486–494-Occ. Peptide blocking studies demonstrated that this antibody is blocked by both the nonphosphopeptide and phosphopeptide, suggesting this antibody detects occludin independent of Ser490 phosphorylation (data not shown).
Immunoprecipitation
BRECs were treated with 50 ng/mL VEGF for 15 min and lysed with 200 μL of lysis buffer (1% Triton X-100, 50 mM HEPES, 10 mM EDTA, 10 mM sodium pyrophosphate, 100 mM sodium fluoride, 1 mM sodium orthovanadate, 1 μg/mL aprotinin, 1 μg/mL leupeptin, and 2 mM phenylmethylsulfonyl fluoride). After centrifugation at 12 000 rpm for 10 min, 1.0 mg of protein was subjected to immunoprecipitation. To clear protein extracts, protein G-Sepharose (20 μL of a 50% suspension) was added to the cell lysates, after which they were incubated for 1 h, followed by centrifugation and collection of the supernatant. Rabbit α-Occ (Zymed) or α-Occ-pS490 was added followed by incubation at 4 °C for 2 h with rocking; 20 μL of protein G-Sepharose was then added, and the sample was incubated for another 2 h at 4 °C with rocking. Protein G-Sepharose antigen–antibody conjugates were separated by centrifugation, washed five times, and boiled for 3 min in Laemmli sample buffer. Immunoblotting was performed with rabbit α-Occ (Zymed) or α-Occ-pS490 and immunoreactivity was detected by ECL plus or ECL advance. The membranes were stripped according to manufacturer’s instruction (Re-Blot Western Blot Recycling Kit; Chemicon) and the reciprocal antibody was applied and immunoblotting conducted.
Cloning and Purification of hOcc-wt 413-522 and hOcc-S490D413-522
Full-length human occludin cDNA (hOcc-wt-pENTR221) was obtained (Invitrogen) and used as a template to create the S490D mutant using Quick Change Site-Directed Mutagenisis Kit (Stratagene). These constructs were used as templates to clone hOcc-wt413–522 and hOcc-S490D413–522 into pET151 (Invitrogen), a bacterial expression plasmid endoding an N-terminal his-tag and TEV cleavage site. Cloning was achieved by a PCR reaction followed by subcloning with topoisomerase. The hOcc-wt413–522 and hOcc-S490D413–522 were used for PCR reaction with the following primers to introduce TOPO cloning sites: FP, 5′-CACCGAGGAGGACTGGATCAGGGAA-3′, and RP, 5′-CTATGTTTTCTGTCTATCATAGTCCCT-3′. The PCR products were purified using QIAquick PCR purification kit, and subjected to a TOPO cloning reaction as per the manufacturer’s instructions. TOP10 competent cells were then transformed with the TOPO cloning reaction product, plasmids were purified using Invitrogen’s mini prep kit, and sequence was verified. Protein purification of human occludin fragments encoding amino acids 413–522 was achieved by nickel column chromatography. Plasmids containing hOcc-wt413–522 and hOcc-S490D413–522 in pET151 were expressed transformed into Escherichia coli BL21 (DE3) and protein was expressed by autoinduction in ZYP-5052 medium.31 Cells were lysed with a solution of Bugbuster detergent (Novagen), 30 U rLysozyme (Novagen), and 75 U benzonase (Novagen) and spun at 21 000g for 30 min. For crystallization studies, purification to near homogeneity was achieved by applying the supernatant to a Nickel-sepharose matrix, washing with Buffer A (25 mm phosphate, pH 7.5, 500 nM NaCl, 20 mM iimidazole) and eluting with a linear gradient with Buffer B (25 mM phosphate, pH 7.5, 500 mM NaCl, 500 mM iimidazole). This was followed by cleavage of the His tag by TEV protease and reapplication to the Nickel-sepharose matrix to remove the cleaved His tag fragment. The flow-through was subsequently applied to a Superdex 75 size-exclusion chromatography column (Amersham Biosciences). Fractions enriched in protein were collected and concentrated to 2 mg/mL in 50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.1 mM EDTA, and 1 mM DTT for crystallization. For pull-down assays, purification was similar to that described above with the exception that the His tag was not cleaved by TEV protease, and after elution from the Nickel column, fractions were directly applied to the S75 gel filtration column. Fractions were pooled, concentrated to 1 mg/mL and stored at −80 °C until needed.
His-Tag Pull Down Assay
At 2 days postconfluence, BREC were lysed (buffer: 1% Triton X100, 100 mM NaCl, 10 mM Hepes, pH 7.5, 1 mM benzamidine, 1 mM NaV04, 10 mM NaF, 10 mM sodium pyrophosphate, 1 mM microcystin and a complete (EDTA free) protease inhibitor cocktail tablet) and frozen. For each reaction, 1 mg of total BREC lysate was incubated with 0.2 μM hOcc-wt413–522 or hOcc-S490D413–522 peptide overnight at 4 °C. Probond Resin (50 μL) was equilibrated with binding buffer (50 mM sodium phosphate, 300 mM NaCl, 50 mM imidazole, pH 7.8) for 1 h, rinsed with PBS, and added to the reaction. Following a 1 h incubation at 4 °C, the resin was washed five times with wash buffer (1% Triton X100, 300 mM NaCl, 10 mM Hepes, and 50 mM imidazole, pH 7.5) and eluted (50 mM Sodium Phosphate, 300 mM NaCl, and 500 mM imidazole, pH 7.8). Samples were subjected to SDS-PAGE and immunoblotted with rabbit α-Occ (Zymed), rabbit α-ZO-1 (Zymed), and rat α-ZO-1 monoclonal antibodies.
Statistical Methods
Freidman test and one-way analysis of variance (ANOVA) with Tukey multiple comparison tests were performed using Prism 2.0, with statistical significance set at p < 0.05.
Crystallography
S490D was crystallized by vapor diffusion using the sitting drop method from a reservoir containing 2.2 M ammonium sulfate and 7% glycerol in 100 mM sodium citrate at pH 5.6. Drops were formed with 4 μL of reservoir and 4 μL of protein. Crystals appeared over a period of 1 week and were harvested by drawing through a solution containing 100% N-paratone as a cryoprotectant. They were plunged into liquid nitrogen for storage and subsequent transport to the NSLS. Data were collected on X-ray beamline X-12C at the NSLS using a wavelength of 1 Å. Crystals grew in space group P21 with unit cell dimensions a = 33.05 Å, b = 35.59 Å, c = 107.40 Å, α = β = γ = 90°. Data were indexed, integrated and scaled (Table 1) using the HKL2000 program suite.32 Crystals were isomorphous to the wild-type protein. Rigid body refinement in CNS33 was performed using the protein atoms of wild-type occludin (PDB code 1WPA), as a template. Serine 490 and residues in 5 Å radius were removed prior to initial simulated annealing at 3000 K which was followed by individual B factor refinement. Initial 2Fo – Fc and Fo – Fc electron density maps were calculated using CNS. The map and molecule was visualized using the program TURBO34 and atoms corresponding to aspartic acid at position 490, atoms within a 5 Å radius, and water molecules, added as appropriate. This protocol of refinement followed by manual rebuilding was repeated until no improvement could be made to the model as judged by a reduction in Rfree. The current model contains residues 414–522 and 99 water molecules, and has R = 23.5% and Rfree = 25.1% with good geometry for all residues between 20 and 2.0 Å (Table 1). Ninety-eight percent of residues are in most favored regions and the remaining 2% are in allowed regions of a Ramachandran map.
Table 1.
Data Collection and Refinement Statistics for hOcc S490D413–522
| human Occludin S490D413–522 | |
|---|---|
| Data Collection | |
| Wavelength (Å) | 1.0 |
| Resolution limit (Å) | 30–2.0 |
| No. Reflections | 9,081 |
| Redundancy | 6.7 (6.8) |
| I/σI | 8.9 (3.0) |
| Rmerge (%) | 8.1 (44.1) |
| Completeness (%) | 100 (100) |
| Refinement | |
| Resolution Range (Å) | 30–2.0 |
| No. Reflections (working/test) | 8185/896 |
| Rwork/Rfree | 23.58/25.13 |
| No. Protein atoms | 925 |
| No. water molecules | 99 |
| rmsd from ideality | |
| bonds length (Å) | 0.011 |
| bond angles (deg) | 1.3 |
Results and Discussion
Identification of Putative Occludin Phosphorylation Sites
Previous studies have demonstrated a phosphorylation response of the tight junction protein occludin after VEGF treatment in retinal endothelial cells that is closely associated with a change in permeability. However, no in vivo occludin phosphosites have been determined.To identify potential in vivo phosphorylation sites, mass spectrometry (MS) was combined with bioinformatics to analyze occludin immunoprecipitates from primary retinal endothelial cells. Occludin was isolated by crude membrane fractionation of BREC (+/− VEGF) and immunoprecipitation followed by SDS-PAGE (Figure 1a). Gels were stained with Sypro Ruby (Molecular Probes), but occludin content was below the limit of detection (data not shown). Multiple bands of occludin, which represent the various states of occludin phosphorylation,24 were detected using Western blotting in separate gels run in parallel (figure 1a). These Western blots were used to locate the position of occludin on the preparative gel. Occludin was excised in 3 gel fragments (alpha, beta and a section above beta, Figure 1a), digested with trypsin and analyzed by MALDI-TOF mass spectrometry. Tryptic peptides observed in MS analysis of three independent experiments are underlined within the Bos taurus occludin sequence (Figure 1b). Across all experiments, MS peptide coverage of the internal loop and carboxy terminus was 89%, while coverage of the amino terminus was 12%. In addition, several extracellular and transmembrane peptides were identified, and as expected, no phosphopeptides within either of the extracellular loops or transmembrane regions were found. Coverage across experiments was consistent with approximately 80% of the peptides being observed between any two experiments. For these experiments, bovine occludin was cloned by RT-PCR and sequenced. The resulting partial cDNA (aa 16–522) was submitted to GenBank (Accession: AY589500) and the remaining residues (aa 1–15) were later obtained from www.ensemble.org. It should be noted that while we were unable to detect our sample by Sypro Ruby staining, our sample was in sufficient quantity to be detectable by Western blotting (picogram quantities) and MS.
Figure 1.
VEGF-induced occludin phosphorylation sites. (a) Following crude membrane preparation of BREC, occludin was immunoprecipitated, and separated by SDS-PAGE. A portion of the lysate was transferred and probed with rabbit polyclonal anti-Occludin antibody (α-Occ-r) (Zymed) to localize occludin. The remainder of the lysate was separated by SDS-PAGE and three regions of the gel, corresponding to different occludin isoforms, were excised and analyzed by MALDI-TOF MS. C, Control; V, VEGF 15 min; NP, no primary antibody; OP, other primary antibody; NC, no cell (no cell lysate control). (b) Occludin phosphosites identified from VEGF-stimulated endothelial cells. Tryptic peptides of occludin were analyzed by FindMod and data were filtered by inclusion criteria defined in Materials and Methods. Occludin sequence was aligned with the ClustalV algorithm. MS coverage of occludin is indicated by a solid line under the B. taurus sequence. Green shading represents transmembrane domains and blue represents the coiled-coil domain, while other domains are indicated above the aligned residues. Putative phosphosites, red; conserved S/T, yellow.
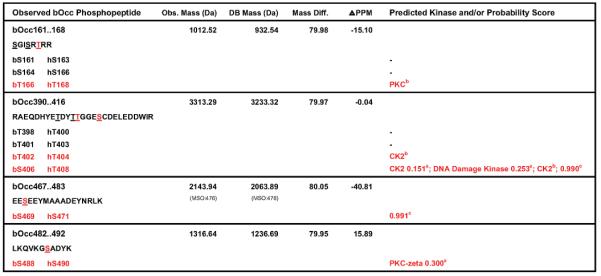
Several attempts to identify phosphosites using tandem MS failed to yield reliable results. On the basis of our work identifying phosphosites on AKT1 (see adjoining paper), we applied a series of inclusion/exclusion criteria to identify putative phosphopeptides within the MALDI-TOF profiles. We confined our analysis to phosphorylation of serine and threonine residues as immunoprecipitation and subsequent phosphotyrosine blotting did not provide evidence for tyrosine phosphorylation in this experimental system (data not shown). Each potential phosphopeptide was required to contain at least one conserved serine and/or threonine residue (Figure 1b) between mammalian and Gallus gallus occludin as this evolutionary distance is appropriate for the resolution required for post-translational modification mapping.35 In addition, each phosphopeptide must have been observed in multiple experiments, in samples from gel-shifted bands (associated with increased phosphorylation), in VEGF-stimulated cells, in overlapping fragments, and with high mass accuracy (within 100 ppm). Four putative phosphopeptides (Table 2) were identified by this analysis and each was scanned with multiple phosphosite prediction algorithms (Scansite, ScanProsite, or NetPhos 2.0). Application of these criteria yielded a total of five putative phosphosites (Figure 1b). This data is consistent with 2D gel analysis whereby occludin is basally phosphorylated on up to two sites and growth factor stimulation of cells lead to the phosphorylation of three additional residues.25 This method identified known AKT1 phosphosites with high specificity and sensitivity (see adjoining paper).
Table 2.
Occludin phosphopeptidesa
|
Tryptic peptides of occludin obtained from VEGF-stimulated BREC were analyzed by FindMod and data were filtered by inclusion criteria defind in Materials and Methods. The putative phosphopeptide observed mass (Obs. Mass), mass predicted from the data base for unphsphorylated peptide (DB mass), mass differential (Mass Diff.) and difference in parts per million (PPM) form predicted phosphopeptides are shown. Putative phosphoamino acids are shown in red and underlined. Other conserved serine and threonine residues are underlined. Bovine occludin was scanned for predicted phosphorylation sites and the predicted kinase and/or associated percentiles/probability score are shown. Predictions for each scanner are noted: Scansitea, Scanprositeb, or Netphosc.
Analysis of the domain distribution of putative occludin phosphorylation sites reveals that bovine occludin Thr166 (bOccThr166) is located within the MARVEL domain, while the remaining sites are located within the cytoplasmic C-terminus (Figure 2a). Both bOccThr402 and bOccSer406 are located within the proximal C-terminus and both sites are predicted to be phosphorylated by casein kinase II (Table 2). These results are consistent with the in vitro identification of occludin phosphosites from casein kinase II and brain extract kinase reactions.27,36,37 The remaining two putative sites, bOccSer469 and bOccSer488, are located within the coiled-coil domain of occludin (Figure 2a). Notably, both bOccSer469 and bOccSer488 are located in turns between helices (Figure 2b), and as such, they are available as substrates for protein kinases. Moreover, bOccSer488 is predicted to be phosphorylated by PKC-ζ (Table 2), an isoform of PKC associated with tight junctions38 that has also been shown to interact with occludin in vitro.39 These two modifications are of particular interest since the coiled-coil domain is known to function in protein interactions39 as well as in the targeting of occludin to tight junctions.
Figure 2.
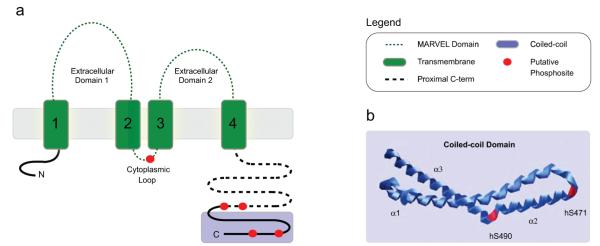
Location of occludin phosphorylation sites. (a) Schematic of occludin domains and phosphosite distribution. Putative phosphosites are depicted in red. Human Thr168 (hThr168) is located within the cytoplasmic loop of the MARVEL domain. The hThr404 and hSer408 residues are located within the proximal C-terminus, while hSer471 and hSer490 are located within the coiled-coil domain. (b) Structural location of putative phosphosites hSer471 and hSer490. Phosphosites, red.
Additionally, a mass consistent with a phosphopeptide containing bOccSer338 was observed. This site has been previously identified as a PKC phosphorylation site in vitro.29 However, this phosphopeptide failed to meet our inclusion criteria as Ser338 is not conserved from mammalian to G. gallus occludin.
Taken together, we have identified five potential in vivo occludin phosphorylation sites in VEGF-stimulated primary endothelial cells by MS analysis combined with bioinformatics inclusion criteria. Two of these sites, bOccThr402 and bOccSer406, were previously identified in vitro and this data confirms that one or both of these sites are likely to be phosphorylated in vivo. As discussed below, a third site, bOccSer488 matching human occludin Ser 490 (hOccSer490), was validated using phosphospecific antibodies. As such, a site-specific map of occludin phosphorylation in endothelial cells has emerged from this analysis. (Human occludin residue numbers are used for subsequent designation of phosphospecific antibodies and crystal structure analysis. For reference, both bovine and human residues are given in Table 2.)
Confirmation of Occludin Phosphorylation at Ser490
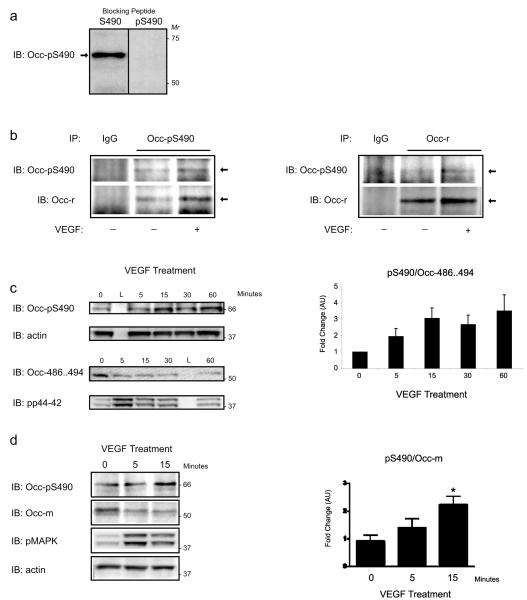
To confirm Ser490 phosphorylation in vivo, a phosphosite-specific antibody was created. Rabbit anti-phosphopeptide antibodies (Quality Controlled Biochemicals; Hopkinton, MA) were generated against hOcc486–494 phosphorylated at Ser490 (AcCLKQVKGpSADYK). Following purification, antibodies were screened for reactivity with BREC lysates. Dilution optimization experiments demonstrated a prominent band at 66 kDa that corresponds to the hyperphosphorylated form of occludin observed previously. Peptide blocking experiments demonstrated that this band was specifically blocked by preincubation of α-Occ-pS490 antibody with the phosphorylated peptide (AcCLKQVKGpSADYK). Importantly, preincubation with the non-phosphorylated form of the peptide failed to block reactivity to this band (Figure 3a). These data demonstrate that α-Occ-pS490 antibody specifically binds occludin when Ser490 is phosphorylated.
Figure 3.
Characterization of ROcc-pS490. (a) BREC lysates (50 μg) were subjected to SDS-PAGE and immunoblotted for α-Occ-pS490 after preincubation of antibody with a 50× molar excess of Ser490 peptide (hOcc: 486–494, CLKQVKGSADYK), left panel, or pSer490 peptide (hOcc: 486–494, CLKQVKGpSADYK), right panel. (b) BREC lysates were treated with VEGF for 15 min, lysed, and incubated with either α-Occ-pS490, left panel, or anti-occludin rabbit polyclonal antibody (α-Occ-r), right panel. Protein G-Sepharose, antigen–antibody conjugates were separated by centrifugation and washed, and the immunoprecipitated proteins were resolved by SDS-PAGE and subjected to immunoblot analysis with either α-Occ-pS490 or α-Occ-r. (c) Time course of Ser490 phosphorylation. VEGF (50 ng/mL) treated BREC were harvested at different time points and immunoblotted for α-Occ-pS490, α-actin, α-Occ-486–494, and α-pErk. The graph depicts the results from six replicates at each time point and reveals that α-Occ-pS490/α-Occ-486–494 is significantly elevated after VEGF treatment in BREC, p < 0.05, Freidman test. As a positive control for VEGF stimulation, pErk is elevated 5 min after VEGF treatment. (c) Analysis of VEGF-stimulated α-Occ-pS490. BREC were treated with VEGF (50 ng/mL) for 0, 5, or 15 min. A representative immunoblot is shown for α-Occ-pS490 and occludin monoclonal antibody (Occ-m). The graph depicts the results from four replicates at each time point and reveals that α-Occ-pS490/Occ-m is significantly elevated compared to controls, 15 min after VEGF treatment in BREC, *p < 0.05, ANOVA with Tukey multiple comparison test.
To confirm that α-Occ-pS490 antibody is specific for endogenous occludin, BREC lysates were treated with VEGF for 15 min, lysed, and immunoprecipitated with either α-Occ-pS490 or anti-occludin rabbit polyclonal antibody (α-Occ-r) (Zymed). The immunoprecipitated proteins were resolved by SDS-PAGE and subjected to immunoblot analysis with either α-Occ-pS490 or α-Occ-r. As seen in Figure 3b, immunoprecipitation with α-Occ-pS490 leads to a 66 kDa band with positive immunoreactivity to the α-Occ-r antibody. In addition, immunoprecipitation with α-Occ-r leads to 66 kDa band with positive immunoreactivity to α-Occ-pS490 antibody. Taken together with the peptide blocking studies, these data represent the first demonstration of a phosphosite-specific antibody to occludin and confirm that occludin is phosphorylated at Ser490 in vivo.
VEGF Stimulates Occludin Phosphorylation at Ser490
To determine if Ser490 phosphorylation was dynamically regulated, BREC were treated with VEGF (50 ng/mL) and harvested at various time points. Immunoblot analysis demonstrated that occludin was basally phosphorylated at Ser490 and that VEGF significantly stimulated Ser490 phosphorylation in a time dependent manner that was maximal by 15 min (Figure 3c; p < 0.05 Friedman test). Further experimentation showed that Ser490 is significantly elevated relative to controls following 15 min of VEGF stimulation (Figure 3d; p < 0.05, ANOVA with Tukey multiple comparison test). The time and magnitude of this effect is consistent with previous studies from our laboratory that assessed global changes in occludin phosphorylation in response to VEGF.24
S490D Mutation Attenuates the Interaction between Occludin413–522 and ZO-1
To determine whether Ser490 phosphorylation affects the interaction between occludin and ZO-1, a phosphomimic of occludin was created by substituting Ser490 with Asp490 (S490D). Constructs expressing his-tagged wt-Occ413–522 and S490D-Occ413–522 fragments were used in capture assays with BREC lysates. Eluted protein complexes were separated by SDS-PAGE and analyzed by immunoblot. Wt-Occ413–522 captured endogenous ZO-1 as detected by either rabbit α-ZO-1 (α-ZO-1-rab) (Figure 4a) or rat α-ZO-1 (α-ZO-1-rat) (Figure 4b). In contrast, the S490D-Occ413–522 phosphomimic significantly attenuated ZO-1 binding. (Figure 4 is a representative blot from 4 separate experiments.) These data confirm that the coiled-coil domain of occludin is sufficient for occludin/ZO-1 interaction and demonstrate that the phosphomimic point mutation at residue 490 attenuates the interaction between occludin and ZO-1 in vitro. These results suggest that the interaction between endogenous occludin and ZO-1 may be regulated by Ser490 phosphorylation of occludin in vivo. However, the potential contribution from other sites of post-translational modification cannot be ruled out.
Figure 4.
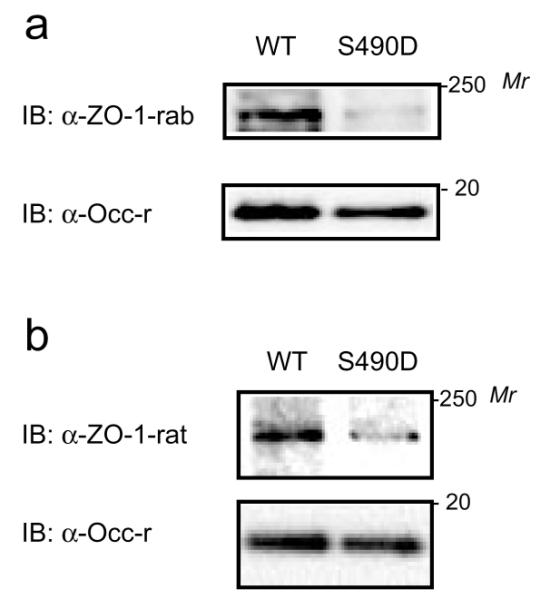
S490D mutation attenuates the occludin/ZO-1 interaction. (a) BREC lysates (1 mg) were incubated with his-tagged wt-hOcc413–522 or S490D-hOcc413–522. The occludin fragment was captured with IDA-Ni2+ ProBond Resin and complexes were washed followed by elution with imidazole. Protein was resolved by SDS-PAGE, and subjected to immunoblot analysis for the presence of ZO-1 using anti-ZO-1 rabbit polyclonal (α-ZO-1-rab) antibody. (b) As in (a) but the blots were analyzed with rat α-ZO-1. Shown are representative blots of 4 separate experiments.
Phosphorylation Mimic at Ser490 Alters the Surface Charge Distribution
To gain insight into the molecular mechanisms of Ser490-regulated ZO-1 binding, we determined the structure of S490D-Occ413–522 at 2 Å resolution and compared this to the previously solved wt-Occ413–522. Crystals of the phosphomimic were isomorphous to those of the wild-type construct previously solved by Li et al.12 Rigid body refinement was performed using protein coordinates of wt-Occ413–522 as a template and atoms within 5 Å of Cα490 were removed. The overall structure of both S490D-Occ413–522 and wt-Occ413–522 are highly similar (rmsd = 0.25 Å for all backbone atoms). Specifically, S490D-Occ413–522 and wt-Occ413–522 both fold into a coiled-coil approximately 70 Å long and 20 Å wide and are comprised of three α-helices preceded by an N-terminal loop12 (Figure 5).
Figure 5.
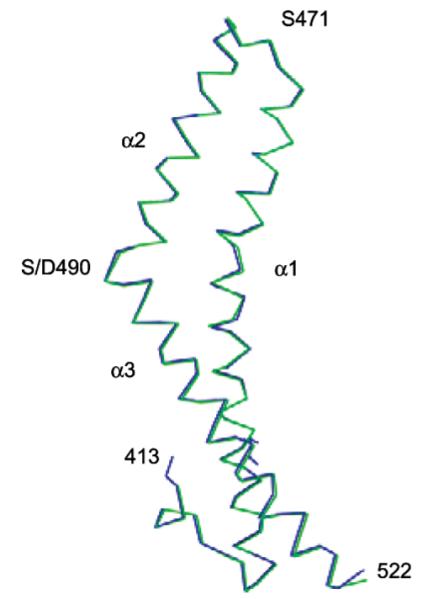
Structure of S490D-hOcc413–522. (a) Main chain Cα carbon atoms of S490D-hOcc413–522 superimposed onto wt-hOcc413–522. The fold of this domain of occludin is preserved after mutation of Ser490 to Asp490 and consists of 3 α-helices which form 2 antiparallel coiled-coils.
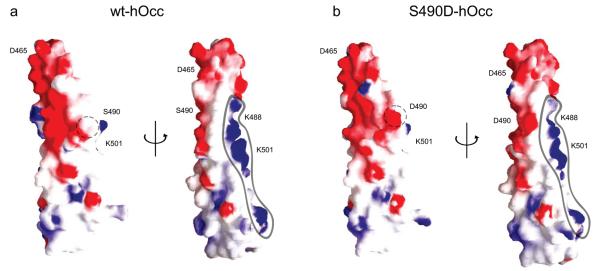
Although the structure of the S490D variant and the wild-type are nearly identical, the surface charge potential of S490D-Occ413–522 is significantly altered relative to wt-Occ413–522. Specifically, within the phosphomimic structure, the positively charged ZO-1 binding surface is disrupted at both Lys485 and Lys488 (Figure 6). Importantly, the contribution of positive charge provided by these residues to the local surface potential has recently been shown to be required for ZO-1 binding.12 Moreover, it is expected that phosphorylation of Ser490 would provide an even larger disruption of this positively charged ZO-1 binding surface. As such, an introduction of a phosphate group onto Ser490 would not only contribute negative charge in the midst of this positively charged binding surface, but also potentially neutralize the charge contribution of Lys485 and Lys488 necessary for ZO-1 binding. Taken together, these data suggest that Ser490 phosphorylation may function as a molecular switch to regulate the interaction between occludin and ZO-1.
Figure 6.
S490D disrupts occludin binding domain for ZO-1. (a). Comparison of surface charge potentials of wt-hOcc413–522 and S490D-hOcc413–522. The two images on the left were rotated 180° to obtain the corresponding image on the right. The positively charged ZO-1 interaction surface potential is outlined to highlight the partial disruption induced by the phosphomimic. S/D490 is circled; red, negative charge; blue, positive charge.
Conclusions
The studies herein have validated a combined MS and bioinformatics approach to identify novel phosphosites. The results suggest that occludin is phosphorylated on five residues in vascular endothelial growth factor stimulated endothelial cells and that these sites are distributed across three different functional domains. In addition, regulated phosphorylation within the coiled-coiled domain may function to dynamically regulate interaction with ZO-1. Previously, it was demonstrated that diabetes induces internalization of occludin in retinal capillaries and arterioles40 and this can be recapitulated in endothelial cells with VEGF treatment.41 Here, it is shown that Ser490 phosphorylation of occludin is stimulated in response to growth factor addition. As such, these studies suggest that dynamic movement and redistribution of occludin in response to growth factors may be associated with reduced ZO-1 binding following Ser490 phosphorylation. Mutational analysis studies will be required to determine the potential role of Ser490 phosphorylation in the regulation of occludin localization, endothelial barrier properties, and cell growth and differentiation.
Finally, data from a number of laboratories have demonstrated that a variety of TJ proteins (e.g., occludin, tricellulin, ZO-1, ZO-2, and claudin-5) are phosphoproteins, and future experiments directed at understanding the relationship between dynamic phosphorylation of these proteins and tight junction function are required. Currently, the major obstacles in identifying post-translational modifications are the modest amount of material available for analysis and the generally low stoichiometry of phosphorylation. To circumvent this problem, substrates are often phosphorylated in vitro. Although this frequently results in reliable tandem MS data and phosphosite identification, these reactions are generally promiscuous and may lead to a solution set with false positives. Overexpression of transgenes may also lead to false positives in a similar fashion. In an effort to obtain the most physiologically relevant solution set, occludin phosphorylation sites were identified from endogenously expressed occludin derived from primary cells. Although several strategies were utilized to obtain tandem MS spectra, including alternative matrices and phosphopeptide enrichment, each method failed to generate spectra capable of unambiguously identifying phosphosites. As such, the use of bioinformatics inclusion criteria proved to be a key element in the identification of putative in vivo phosphosites from MALDI-TOF profiles.
Acknowledgment
This research was supported by NEI-EY012021 (D.A.A.), from funding by the Juvenile Diabetes Research Foundation (D.A.A.) and the Pennsylvania Lions Sight Conservation and Eye Research Foundation (D.A.A., J.M.F.). We thank Dr. Thomas Gardner for his valued advice and direction. We also thank Mike Guilford of the Proteomics/Mass Spectrometry Core Facility of the Section of Research Resources, Penn State College of Medicine.
Abbreviations
- BREC
bovine retinal endothelial cells
- CKI
casein kinase I.
- CKII
casein kinase II
- PKC
protein kinase C
- MARVEL
MAL-related proteins for vesicle trafficking and membrane link domain
- MS
mass spectrometry
- ppm
parts per million VEGF, vascular endothelial growth factor
- PTM
post-translational modifications
References
- (1).Chiba H, Kojima T, Osanai M, Sawada N. The significance of interferon-gamma-triggered internalization of tight-junction proteins in inflammatory bowel disease. Sci. STKE. 2006;2006(316):pe1. doi: 10.1126/stke.3162006pe1. [DOI] [PubMed] [Google Scholar]
- (2).Ciccocioppo R, Finamore A, Ara C, Di Sabatino A, Mengheri E, Corazza GR. Altered expression, localization, and phosphorylation of epithelial junctional proteins in celiac disease. Am. J. Clin. Pathol. 2006;125(4):502–11. doi: 10.1309/DTYR-A91G-8R0K-TM8M. [DOI] [PubMed] [Google Scholar]
- (3).Harhaj NS, Antonetti DA. Regulation of tight junctions and loss of barrier function in pathophysiology. Int. J. Biochem. Cell Biol. 2004;36(7):1206–37. doi: 10.1016/j.biocel.2003.08.007. [DOI] [PubMed] [Google Scholar]
- (4).Hawkins BT, Davis TP. The blood-brain barrier/neurovascular unit in health and disease. Pharmacol. Rev. 2005;57(2):173–85. doi: 10.1124/pr.57.2.4. [DOI] [PubMed] [Google Scholar]
- (5).Lee DB, Huang E, Ward HJ. Tight junction biology and kidney dysfunction. Am. J. Physiol.: Renal Physiol. 2006;290(1):F20–34. doi: 10.1152/ajprenal.00052.2005. [DOI] [PubMed] [Google Scholar]
- (6).Papadopoulos MC, Saadoun S, Binder DK, Manley GT, Krishna S, Verkman AS. Molecular mechanisms of brain tumor edema. Neuroscience. 2004;129(4):1011–20. doi: 10.1016/j.neuroscience.2004.05.044. [DOI] [PubMed] [Google Scholar]
- (7).Simonovic I, Rosenberg J, Koutsouris A, Hecht G. Enteropathogenic Escherichia coli dephosphorylates and dissociates occludin from intestinal epithelial tight junctions. Cell Microbiol. 2000;2(4):305–15. doi: 10.1046/j.1462-5822.2000.00055.x. [DOI] [PubMed] [Google Scholar]
- (8).Erickson KK, Sundstrom JM, Antonetti DA. Vascular permeability in ocular disease and the role of tight junctions. Angiogenesis. 2007;10(2):103–17. doi: 10.1007/s10456-007-9067-z. [DOI] [PubMed] [Google Scholar]
- (9).Traweger A, Fang D, Liu YC, Stelzhammer W, Krizbai IA, Fresser F, Bauer HC, Bauer H. The tight junction-specific protein occludin is a functional target of the E3 ubiquitin protein ligase itch. J. Biol. Chem. 2002;277(12):10201–8. doi: 10.1074/jbc.M111384200. [DOI] [PubMed] [Google Scholar]
- (10).Huber D, Balda MS, Matter K. Occludin modulates transepithelial migration of neutrophils. J. Biol. Chem. 2000;275(8):5773–8. doi: 10.1074/jbc.275.8.5773. [DOI] [PubMed] [Google Scholar]
- (11).Sanchez-Pulido L, Martin-Belmonte F, Valencia A, Alonso MA. MARVEL: a conserved domain involved in membrane apposition events. Trends Biochem. Sci. 2002;27(12):599–601. doi: 10.1016/s0968-0004(02)02229-6. [DOI] [PubMed] [Google Scholar]
- (12).Li Y, Fanning AS, Anderson JM, Lavie A. Structure of the conserved cytoplasmic C-terminal domain of occludin: identification of the ZO-1 binding surface. J. Mol. Biol. 2005;352(1):151–64. doi: 10.1016/j.jmb.2005.07.017. [DOI] [PubMed] [Google Scholar]
- (13).Feldman GJ, Mullin JM, Ryan MP. Occludin: structure, function and regulation. Adv. Drug Delivery Rev. 2005;57(6):883–917. doi: 10.1016/j.addr.2005.01.009. [DOI] [PubMed] [Google Scholar]
- (14).Matter K, Balda MS. Occludin and the functions of tight junctions. Int. Rev. Cytol. 1999;186:117–46. doi: 10.1016/s0074-7696(08)61052-9. [DOI] [PubMed] [Google Scholar]
- (15).Yu AS, McCarthy KM, Francis SA, McCormack JM, Lai J, Rogers RA, Lynch RD, Schneeberger EE. Knockdown of occludin expression leads to diverse phenotypic alterations in epithelial cells. Am. J. Physiol.: Cell Physiol. 2005;288(6):C1231–41. doi: 10.1152/ajpcell.00581.2004. [DOI] [PubMed] [Google Scholar]
- (16).Matter K, Balda MS. Signalling to and from tight junctions. Nat. Rev. Mol. Cell Biol. 2003;4(3):225–36. doi: 10.1038/nrm1055. [DOI] [PubMed] [Google Scholar]
- (17).Saitou M, Furuse M, Sasaki H, Schulzke JD, Fromm M, Takano H, Noda T, Tsukita S. Complex phenotype of mice lacking occludin, a component of tight junction strands. Mol. Biol. Cell. 2000;11(12):4131–42. doi: 10.1091/mbc.11.12.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Wang Z, Mandell KJ, Parkos CA, Mrsny RJ, Nusrat A. The second loop of occludin is required for suppression of Raf1-induced tumor growth. Oncogene. 2005;24(27):4412–20. doi: 10.1038/sj.onc.1208634. [DOI] [PubMed] [Google Scholar]
- (19).Murata M, Kojima T, Yamamoto T, Go M, Takano K, Osanai M, Chiba H, Sawada N. Down-regulation of survival signaling through MAPK and Akt in occludin-deficient mouse hepatocytes in vitro. Exp. Cell Res. 2005;310(1):140–51. doi: 10.1016/j.yexcr.2005.07.017. [DOI] [PubMed] [Google Scholar]
- (20).Medici D, Hay ED, Goodenough DA. Cooperation between snail and LEF-1 transcription factors is essential for TGF-beta1-induced epithelial-mesenchymal transition. Mol. Biol. Cell. 2006;17(4):1871–9. doi: 10.1091/mbc.E05-08-0767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Marzioni D, Banita M, Felici A, Paradinas FJ, Newlands E, De Nictolis M, Muhlhauser J, Castellucci M. Expression of ZO-1 and occludin in normal human placenta and in hydatidiform moles. Mol. Hum. Reprod. 2001;7(3):279–85. doi: 10.1093/molehr/7.3.279. [DOI] [PubMed] [Google Scholar]
- (22).Tobioka H, Isomura H, Kokai Y, Tokunaga Y, Yamaguchi J, Sawada N. Occludin expression decreases with the progression of human endometrial carcinoma. Hum. Pathol. 2004;35(2):159–64. doi: 10.1016/j.humpath.2003.09.013. [DOI] [PubMed] [Google Scholar]
- (23).Osanai M, Murata M, Nishikiori N, Chiba H, Kojima T, Sawada N. Epigenetic silencing of occludin promotes tumorigenic and metastatic properties of cancer cells via modulations of unique sets of apoptosis-associated genes. Cancer Res. 2006;66(18):9125–33. doi: 10.1158/0008-5472.CAN-06-1864. [DOI] [PubMed] [Google Scholar]
- (24).Antonetti DA, Barber AJ, Hollinger LA, Wolpert EB, Gardner TW. Vascular endothelial growth factor induces rapid phosphorylation of tight junction proteins occludin and zonula occluden 1. A potential mechanism for vascular permeability in diabetic retinopathy and tumors. J. Biol. Chem. 1999;274(33):23463–7. doi: 10.1074/jbc.274.33.23463. [DOI] [PubMed] [Google Scholar]
- (25).Harhaj NS, Felinski EA, Wolpert EB, Sundstrom JM, Gardner TW, Antonetti DA. VEGF activation of protein kinase C stimulates occludin phosphorylation and contributes to endothelial permeability. Invest. Ophthalmol. Visual Sci. 2006;47(11):5106–15. doi: 10.1167/iovs.06-0322. [DOI] [PubMed] [Google Scholar]
- (26).Hirase T, Kawashima S, Wong EY, Ueyama T, Rikitake Y, Tsukita S, Yokoyama M, Staddon JM. Regulation of tight junction permeability and occludin phosphorylation by Rhoa-p160ROCK-dependent and -independent mechanisms. J. Biol. Chem. 2001;276(13):10423–31. doi: 10.1074/jbc.M007136200. [DOI] [PubMed] [Google Scholar]
- (27).Smales C, Ellis M, Baumber R, Hussain N, Desmond H, Staddon JM. Occludin phosphorylation: identification of an occludin kinase in brain and cell extracts as CK2. FEBS Lett. 2003;545(2–3):161–6. doi: 10.1016/s0014-5793(03)00525-8. [DOI] [PubMed] [Google Scholar]
- (28).Sakakibara A, Furuse M, Saitou M, Ando-Akatsuka Y, Tsukita S. Possible involvement of phosphorylation of occludin in tight junction formation. J. Cell Biol. 1997;137(6):1393–401. doi: 10.1083/jcb.137.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Andreeva AY, Krause E, Muller EC, Blasig IE, Utepbergenov DI. Protein kinase C regulates the phosphorylation and cellular localization of occludin. J. Biol. Chem. 2001;276(42):38480–6. doi: 10.1074/jbc.M104923200. [DOI] [PubMed] [Google Scholar]
- (30).Antonetti DA, Wolpert EB. Isolation and Characterization of Retinal Endothelial Cells. In: Nag S, editor. The Blood-Brain Barrier, Biology and Research Protocols. Numana Press; Totowa, NJ: 2003. pp. 365–374. [DOI] [PubMed] [Google Scholar]
- (31).Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expression Purif. 2005;41(1):207–34. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- (32).Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–26. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- (33).Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1998;54(Pt 5):905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- (34).Jones T. A graphics model building and refinement system for macromolecules. J. Appl. Crystallogr. 1978;11:268–272. [Google Scholar]
- (35).Cooper GM, Brudno M, Green ED, Batzoglou S, Sidow A. Quantitative estimates of sequence divergence for comparative analyses of mammalian genomes. Genome Res. 2003;13(5):813–20. doi: 10.1101/gr.1064503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).McKenzie JA, Riento K, Ridley AJ. Casein kinase I epsilon associates with and phosphorylates the tight junction protein occludin. FEBS Lett. 2006;580(9):2388–94. doi: 10.1016/j.febslet.2006.03.048. [DOI] [PubMed] [Google Scholar]
- (37).Cordenonsi M, Turco F, D’Atri F, Hammar E, Martinucci G, Meggio F, Citi S. Xenopus laevis occludin. Identification of in vitro phosphorylation sites by protein kinase CK2 and association with cingulin. Eur. J. Biochem. 1999;264(2):374–84. doi: 10.1046/j.1432-1327.1999.00616.x. [DOI] [PubMed] [Google Scholar]
- (38).Dodane V, Kachar B. Identification of isoforms of G proteins and PKC that colocalize with tight junctions. J. Membr. Biol. 1996;149(3):199–209. doi: 10.1007/s002329900020. [DOI] [PubMed] [Google Scholar]
- (39).Nusrat A, Chen JA, Foley CS, Liang TW, Tom J, Cromwell M, Quan C, Mrsny RJ. The coiled-coil domain of occludin can act to organize structural and functional elements of the epithelial tight junction. J. Biol. Chem. 2000;275(38):29816–22. doi: 10.1074/jbc.M002450200. [DOI] [PubMed] [Google Scholar]
- (40).Barber AJ, Antonetti DA. Mapping the blood vessels with paracellular permeability in the retinas of diabetic rats. Invest. Ophthalmol. Visual Sci. 2003;44(12):5410–6. doi: 10.1167/iovs.03-0244. [DOI] [PubMed] [Google Scholar]
- (41).Kevil CG, Payne DK, Mire E, Alexander JS. Vascular permeability factor/vascular endothelial cell growth factor-mediated permeability occurs through disorganization of endothelial junctional proteins. J. Biol. Chem. 1998;273(24):15099–103. doi: 10.1074/jbc.273.24.15099. [DOI] [PubMed] [Google Scholar]