Abstract
Background
Experimental and epidemiological evidence have suggested that chronic inflammation may play a critical role in endometrial carcinogenesis.
Methods
To investigate this hypothesis, a two-stage study was carried out to evaluate single nucleotide polymorphisms (SNPs) in inflammatory pathway genes in association with endometrial cancer risk. In stage 1, 64 candidate pathway genes were identified and 4,542 directly genotyped or imputed SNPs were analyzed among 832 endometrial cancer cases and 2,049 controls, using data from the Shanghai Endometrial Cancer Genetics Study. Linkage disequilibrium of stage 1 SNPs significantly associated with endometrial cancer (P<0.05) indicated that the majority of associations could be linked to one of 24 distinct loci. One SNP from each of the 24 loci was then selected for follow-up genotyping. Of these, 21 SNPs were successfully designed and genotyped in stage 2, which consisted of ten additional studies including 6,604 endometrial cancer cases and 8,511 controls.
Results
Five of the 21 SNPs had significant allelic odds ratios and 95% confidence intervals as follows: FABP1, 0.92 (0.85-0.99); CXCL3, 1.16 (1.05-1.29); IL6, 1.08 (1.00-1.17); MSR1, 0.90 (0.82-0.98); and MMP9, 0.91 (0.87-0.97). Two of these polymorphisms were independently significant in the replication sample (rs352038 in CXCL3 and rs3918249 in MMP9). The association for the MMP9 polymorphism remained significant after Bonferroni correction and showed a significant association with endometrial cancer in both Asian- and European-ancestry samples.
Conclusions
These findings lend support to the hypothesis that genetic polymorphisms in genes involved in the inflammatory pathway may contribute to genetic susceptibility to endometrial cancer.
Impact Statement
This study adds to the growing evidence that inflammation plays an important role in endometrial carcinogenesis.
Keywords: endometrial cancer, inflammation, genetic risk variants, meta-analysis
INTRODUCTION
Endometrial cancer is the most common gynecological malignancy in developed countries and the second most common in the world (1, 2). In China, the incidence of endometrial cancer has increased 90% over the past two decades to 7.62/100,000 in 2007 (3), although it is still substantially lower than the incidence seen in developed countries (US 22.0/100,000; Europe 11.8-12.5/100,000) (2). Obesity, early age at menarche, late age at menopause, nulliparity, and use of estrogen hormone replacement therapy are established risk factors for endometrial cancer (4).
Although the genetics of endometrial cancer are poorly understood, its heritability of approximately 0.5 indicates that there is a strong genetic component for disease risk (5, 6). A number of lines of experimental and epidemiological evidence have indicated that inflammation may play an important role in the transition from normal endometrium to malignancy. Of the many risk factors associated with endometrial cancer, several -- including use of unopposed estrogen (7), anovulation (8), endometriosis (9), early age at menarche (10), late age at menopause (11), nulliparity (12, 13), polycystic ovary syndrome (PCOS) (14), and obesity (15) -- may contribute to a state of prolonged exposure to inflammation (16). Chronic inflammation can result in derangement of cellular processes, leading to excessive mitosis, decreased apoptosis, the accumulation of DNA damage, and thus initiate and promote neoplastic transformation (12, 17). Given that inflammatory process are influenced by inflammation-related genes, we hypothesized that common genetic polymorphisms in inflammatory pathway genes may also influence the risk of endometrial cancer.
To investigate this hypothesis, a two-stage study was carried out to determine whether common variants in genes involved in the inflammatory response were associated with endometrial cancer risk using the resources of the Shanghai Endometrial Cancer Genetics Study and ten additional studies of endometrial cancer conducted among women in the US, Australia, Europe, and China.
MATERIALS AND METHODS
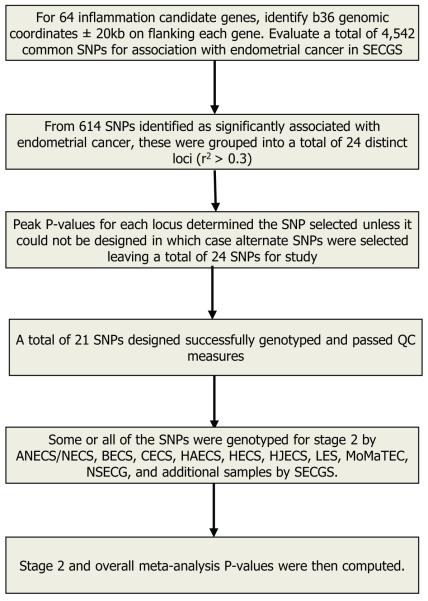
This study involved two stages, as shown in Table 1. Study populations are described below and the overall study design and SNP selection procedure are depicted in Figure 1.
Table 1.
Study populations included.
| Study | Abbreviation | General Setting | Cases | Controls | Genotyping platform |
|---|---|---|---|---|---|
| Stage 1 Sample Sets | Stage 1 | ||||
| Shanghai Endometrial Cancer Genetic Study |
SECGS-I | Shanghai, China; Population-based, case-control studies |
832 | 2,682 | Affymetrix 6.0 |
| Stage2 Sample Sets | Stage 2 | ||||
| Australian National Endometrial Cancer Study/Newcastle Endometrial Cancer Study |
ANECS/NECS | Australia; Population- based, case-control study/Hospital-based study |
1,436 | 1,175 | Sequenom |
| Bavarian Endometrial Cancer Study |
BECS | Germany; Population- based, case-control study |
202 | 387 | Sequenom |
| Connecticut Endometrial Cancer Study |
CECS | Connecticut, USA; Population-based, case-control study |
534 | 621 | Sequenom |
| Hannover-Almaty Endometrial Cancer Study |
HAECS | Kazakhstan; Hospital- based, case-control study |
218 | 232 | Taqman |
| Hawaii Endometrial Cancer Study |
HECS | Hawaii, USA; Population-based, case-control study |
168 | 574 | Sequenom |
| Hannover-Jena Endometrial Cancer Study |
HJECS | Germany; Hospital- based, case-control study |
229 | 554 | Taqman |
| Leuven Endometrial Cancer Study |
LES | Belgium; Hospital- based, case-control study |
264 | 591 | Sequenom |
| Molecular Markers in Treatment of Endometrial Cancer |
MoMaTEC | Norway; Population- based, case-control study |
411 | 210 | Sequenom |
| National Study of the Genetics of Endometrial Cancer |
NSECG | United Kingdom; Population-based, case-control study |
1,514 | 507 | Illumina 550K / Sequenom |
| Shanghai Endometrial Cancer Genetic Study |
SECGS-II | Shanghai, China; Population-based, case-control studies |
796 | 978 | Sequenom |
| Total | 6,604 | 8,511 |
Figure 1.
Selection and prioritization of inflammation-related SNPs for meta-analysis.
Study population
Stage 1 was conducted among the participants of the Shanghai Endometrial Cancer Genetics Study (SECGS), which included 832 cases from the Shanghai Endometrial Cancer Study (SECS) and 2,049 controls from the Shanghai Breast Cancer Study (SBCS) and the Shanghai Women’s Health Study (SWHS). Details of these studies have been described previously (18). Briefly, among 1,199 endometrial cancer cases included in the SECS, 832 women who donated a blood sample to the study and were successfully genotyped by Affymetrix 6.0 array were included in the stage 1 study. Genome-wide scan data from 2,049 women from the SBCS served as controls. The mean age of cases was 54.7 and for controls was 51.7; 45% of cases and 30% of controls were post-menopausal. Data for stage 2 included 6,604 cases and 8,511 controls from a total of 10 studies (Table 1). IRB approval was obtained for all of the parent studies from all contributing institutions, and informed consent was obtained from all participants.
Candidate SNP selection
The SNP selection scheme is shown in Figure 1. Sixty-four candidate genes involved in inflammatory pathways were identified based on literature review and bioinformatics searches. In order to cast a comprehensive net, we did literature review of genes involved in inflammatory pathways, searched Vanderbilt’s Gene List Automatically Derived for You (19), and String-DB (20) for related inflammatory network genes (Supplementary Table 1). A total of 4,542 SNPs with minor allele frequencies of 0.05 or greater were located in or near (± 20kb) RefSeq transcripts of these genes were identified for the stage 1 study. Genotyping of these SNPs was carried out as part of a larger genome-wide association study previously described (18). Only SNPs that passed quality control (QC) from the Affymetrix 6.0 array (Affymetrix, Santa Clara, CA, USA) or that could be imputed were eligible for selection. SNPs for stage 2 were selected, using data from HapMap, release 28, after evaluation of linkage disequilibrium (LD) between the associated SNPs. From this, it was determined that the majority of associations could be linked to one of 24 distinct loci as determined by LD to other SNPs (see Supplementary Figure 1 for an example in MMP9 and CXCL3). The SNP with the lowest P value from each of the 24 loci was selected for follow-up genotyping in stage 2 unless assay design parameters indicated it would fail genotyping. In the latter case, the next most significant SNP was chosen for validation.
Genotyping, quality control, and imputation
Stage 1 genotyping and QC procedures have been described in detail in previous publications (18, 21). Briefly, genotyping was performed using the Affymetrix 6.0 array, which includes 906,602 SNPs. The Birdseed v2 algorithm was used to call genotypes (22). QC samples from Coriell Cell Repositories (Camden, NJ) were included on each 96-well plate, and the average concordance percentage among QC samples was 99.85%. Female sex was confirmed for all samples. Multidimensional scaling analysis of the genotypes with 210 unrelated HapMap samples indicated that all participants clustered with HapMap Asian samples (CHB+JPT). All potential relatives with pairwise identity by descent (IBD) of PI_HAT>0.25 were removed. SNPs that failed the Hardy-Weinberg equilibrium test (P<0.0001) and SNPs that had significantly different missing genotyping rates for cases and controls (P<0.0001) were excluded. After QC was completed, the Hidden Markov Model as implemented in Mach 1.0 was used to impute the genotype for variants of interest that were not directly genotyped using Asian genotyping data from HapMap phase 2 for reference genotypes (23).
In stage 2, 21 of the 24 SNPs selected for replication genotyping as described above, were successfully genotyped. Some stage 2 studies (e.g. HAECS and HJECS) genotyped fewer than 21 SNPs. Only SNPs which met QC criteria similar to that applied for stage 1 were included in the stage 2 analysis. Imputed genotypes were used for some SNPs in ANECS/NECS, NSECG, and control samples derived from the WTCCC when direct genotyping data were not available (24).
Statistical analysis
Unconditional logistic regression was used to calculate odds ratios (ORs) and 95% confidence intervals (CIs) for associations between genotypes and endometrial cancer risk in stage 1. Covariates adjusted for included age, income, and education. Directly genotyped or imputed information for 4,542 SNPs was evaluated for associations with endometrial cancer and 614 SNPs showed a nominal association with endometrial cancer (P<0.05).
Unconditional logistic regression was used to analyze the 21 SNPs selected for stage 2. These analyses were adjusted for age only, because a unifying set of common demographic or anthropometric covariates was not available across all studies. Using the ORs derived from individual studies, a meta-analysis was conducted derive summary statistics (25). An overall Z-statistic and P value based on the weighted average of the individual statistics was calculated. The resulting ORs and 95CIs are based on the fixed effect model, unless heterogeneity across studies was evident (P<0.05 for homogeneity test). In the latter case, ORs, 95 CIs, and P values derived from the random effect model are presented. All P values presented are based on two-tailed tests.
SNP functional annotation
The relationship between P values LD measures relative to two sample SNPs selected for stage 2 genotyping are shown in Supplementary Figure 1 and was done using LocusZoom plotting P values for stage 1 data (26). Functional annotation of the SNPs of interest was carried out using the NIEHS SNP Info Webserver’s SNP function prediction module (27).
RESULTS
Stage 1, Stage 2, and combined results for the 21 SNPs promoted to Stage 2 study along with the number of studies and samples contributing to the analysis are presented in Table 2. In total, five of the 21 SNPs had significant allelic ORs (95%CIs) in the overall dataset: FABP1, 0.92 (0.85-0.99); CXCL3, 1.16 (1.05-1.29); IL6, 1.08 (1.00-1.17); MSR1, 0.90 (0.82-0.98); and MMP9, 0.91 (0.87-0.97). The directions of association in the discovery and replication samples were consistent for all five SNPs. Of these SNPs, only the polymorphisms near CXCL3 and in MMP9 were significantly associated with endometrial cancer risk in the replication stage. No heterogeneity across studies was found for these five SNPs.
Table 2.
Associations with endometrial cancer for the 21 SNPs included in each stage and overall.
| Discovery |
Replication |
Overall |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| rsID | Reference allelea |
Adjacent Genes |
OR (95% CI)b | P c | Studiesd | OR (95%CI)e | P f | OR meta (95%CI)g |
P h | Pj | Heterogeneity P-value |
|
|
|
|
|
||||||||
| rs3918249 | C | MMP9 | 0.81 (0.70-0.92) | 0.002 | 10 | 0.94(0.88-1.00) | 0.042 | 0.91(0.87-0.97) | 0.001 | 0.021 | 0.153 |
| rs352038 | G | CXCL3 | 1.26 (1.06-1.50) | 0.008 | 10 | 1.14(1.00-1.29) | 0.050 | 1.16(1.05-1.29) | 0.003 | 0.063 | 0.498 |
| rs10503574 | C | MSR1 | 0.81 (0.70-0.94) | 0.006 | 6 | 0.97(0.86-1.08) | 0.547 | 0.90(0.82-0.98) | 0.016 | 0.336 | 0.088 |
| rs2970924 | T | FABP1 | 0.80 (0.68-0.96) | 0.013 | 7 | 0.95(0.87-1.03) | 0.214 | 0.92(0.85-0.99) | 0.024 | 0.504 | 0.244 |
| rs2069852 | A | IL6 | 1.19 (1.04-1.36) | 0.013 | 7 | 1.05(0.96-1.16) | 0.284 | 1.08(1.00-1.17) | 0.049 | 1.000 | 0.154 |
| rs1472095 | T | PPARGC1A | 1.41 (1.13-1.77) | 0.003 | 8 | 1.09(0.97- 1.24) i |
0.152 | 1.13(1.00-1.28) i | 0.054 | 1.000 | 0.006 |
| rs4149319 | A | ABCA1 | 0.76 (0.63-0.91) | 0.003 | 8 | 0.99(0.87-1.13) | 0.937 | 0.91(0.82-1.01) | 0.074 | 1.000 | 0.194 |
| rs7709864 | C | LOC729123 | 1.25 (1.07-1.46) | 0.006 | 8 | 1.20(0.94- 1.52) i |
0.137 | 1.17(0.98-1.41) i | 0.084 | 1.000 | 0.001 |
| rs12368672 | G | STAT6 | 1.32 (1.15-1.53) | 1.05E-04 | 8 | 1.00(0.94-1.06) | 0.987 | 1.04(0.99-1.10) | 0.139 | 1.000 | 0.096 |
| rs2239349 | A | IL4R | 1.17 (1.00-1.36) | 0.046 | 8 | 1.14(0.92- 1.40) i |
0.235 | 1.13(0.96-1.34) i | 0.15 | 1.000 | 0.001 |
| rs2780815 | G | JAK1 | 0.74 (0.63-0.88) | 3.72E-04 | 8 | 0.98(0.92-1.04) | 0.471 | 0.94(0.86-1.03) i | 0.193 | 1.000 | 0.032 |
| rs6914211 | A | ESR1 | 1.40 (1.15-1.70) | 0.001 | 8 | 0.99(0.90-1.09) | 0.905 | 1.05(0.97-1.15) | 0.237 | 1.000 | 0.341 |
| rs9839934 | G | THRB | 0.80 (0.69-0.94) | 0.006 | 8 | 1.00(0.94-1.07) | 0.951 | 0.97(0.91-1.02) | 0.253 | 1.000 | 0.282 |
| rs933360 | C | GRB10 | 0.75 (0.65-0.87) | 8.40E-05 | 8 | 1.02(0.96-1.09) | 0.542 | 0.97(0.91-1.03) | 0.269 | 1.000 | 0.067 |
| rs12757165 | G | ESRRG | 0.78 (0.68-0.89) | 2.49E-04 | 8 | 0.99(0.93-1.05) | 0.638 | 0.95(0.87-1.04) i | 0.299 | 1.000 | 0.023 |
| rs2735188 | C | HDAC3 | 1.38 (1.09-1.75) | 0.007 | 8 | 1.00(0.90-1.10) | 0.939 | 1.05(0.96-1.14) | 0.311 | 1.000 | 0.099 |
| rs310247 | A | JAK1 | 0.81 (0.71-0.91) | 0.001 | 8 | 0.99(0.90- 1.08) i |
0.769 | 0.96(0.88-1.06) i | 0.412 | 1.000 | 0.006 |
| rs3781619 | A | DDB2 | 1.18 (1.04-1.35) | 0.013 | 8 | 0.93(0.87-1.00) | 0.062 | 0.96(0.87-1.06) i | 0.457 | 1.000 | 0.041 |
| rs17627111 | G | ESRRG | 0.72 (0.62-0.85) | 0.0000493 | 8 | 0.99(0.93-1.05) | 0.782 | 0.98(0.89-1.08) i | 0.681 | 1.000 | 0.01 |
| rs1421894 | T | CENTD3 | 0.86 (0.75-0.98) | 0.028 | 8 | 1.04(0.97-1.12) | 0.225 | 0.98(0.89-1.09) i | 0.767 | 1.000 | 0.021 |
| rs9896401 | C | SAMD14 | 1.43 (1.14-1.80) | 0.002 | 8 | 0.96(0.90-1.03) | 0.286 | 1.00(0.93-1.07) | 0.944 | 1.000 | 0.061 |
Allele associated with the ORs specified in the table.
OR in discovery stage of inflammation study.
P-value for discovery stage (SECGS-I data)
Number of studies contributing data to replication stage.
OR meta based on some or all of the following studies ANECS, BECS, CECS, HAECS, HECS, HJECS, LES, MoMaTEC, NSECG, and SECGS-II.
Meta-analysis P-value for replication stage including ANECS, BECS, CECS, HAECS, HECS, HJECS, LES, MoMaTEC, NSECG, and SECGS-II
OR for all studies combined.
P-value for overall meta-analysis including replication and discovery stages.
Random effects model used
P-value Bonferroni corrected
Table 3 presents the heterozygous, homozygous, and per allele associations with type 1 endometrial (endometroid) cancer for the five significant SNPs among all women combined, among women of Asian ancestry, and among women of European ancestry. SNP rs3918249 in MMP9 was associated with endometrial cancer risk in women of both Asian and European ancestry. Other SNPs were not significantly associated with endometrial cancer in European-ancestry women. SNP rs10503574 in MSR1 was more significant in Asian-ancestry women than in the overall sample. When restricting analyses to women with type 1 endometrial cancer, the results were largely unchanged.
Table 3.
A ssociation with endometrial cancer risk for selected variants by ethnicity and histological type.
| N |
Allele Freq |
OR (95% CI) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Population | SNP | Cases | Controls | Cases | Controls | Heterozygous | Homozygous | Allelic | P |
|
|
|
|
|||||||
| All women, endometrial cancer cases vs. controls | |||||||||
| rs2970924 | 5832 | 7037 | 0.15 | 0.16 | 0.90(0.82-0.98) | 0.93(0.72-1.20) | 0.92(0.85-0.99) | 0.024 | |
| rs352038 | 6568 | 8405 | 0.06 | 0.08 | 1.16(1.03-1.30) | 1.30(0.90-1.86) | 1.16(1.05-1.29) | 0.003 | |
| rs2069852 | 5784 | 6922 | 0.21 | 0.38 | 0.98(0.86-1.13) | 1.08(0.90-1.29) | 1.08(1.00-1.17) | 0.049 | |
| rs10503574 | 3026 | 6685 | 0.16 | 0.17 | 0.93(0.84-1.04) | 0.76(0.59-0.98) | 0.90(0.82-0.98) | 0.016 | |
| rs3918249 | 6561 | 8273 | 0.44 | 0.53 | 0.95(0.87-1.04) | 0.83(0.73-0.93) | 0.91(0.87-0.97) | 0.001 | |
| All Asian-ancestry endometrial cancer cases vs. controls | |||||||||
| rs2970924 | 1714 | 3783 | 0.15 | 0.16 | 0.87(0.70-1.08) | 0.77(0.48-1.25) | 0.82(0.63-1.07) | 0.140 a | |
| rs352038 | 1693 | 3773 | 0.17 | 0.16 | 1.11(0.98-1.27) | 1.28(0.88-1.88) | 1.12(1.00-1.26) | 0.047 | |
| rs2069852 | 1635 | 3675 | 0.66 | 0.65 | 0.91(0.75-1.11) | 1.07(0.88-1.30) | 1.08(0.99-1.18) | 0.101 | |
| rs10503574 | 1685 | 3823 | 0.24 | 0.26 | 0.89(0.79-1.01) | 0.70(0.54-0.91) | 0.86(0.78-0.95) | 0.003 | |
| rs3918249 | 1700 | 3654 | 0.70 | 0.72 | 0.91(0.73-1.13) | 0.78(0.63-0.98) | 0.88(0.80-0.97) | 0.008 | |
| All European-ancestry endometrial cancer cases vs. controls |
|||||||||
| rs2970924 | 3856 | 2856 | 0.16 | 0.15 | 0.89(0.79-1.00) | 1.07(0.77-1.50) | 0.94(0.84-1.04) | 0.206 | |
| rs352038 | 4553 | 4111 | 0.02 | 0.01 | 1.16(0.87-1.54) | 1.23(0.99-1.54) | 1.18(0.89-1.57) | 0.250 | |
| rs2069852 | 3889 | 2850 | 0.03 | 0.03 | 1.00(0.80-1.26) | 0.31(0.06-1.49) | 1.00(0.80-1.25) | 0.997 | |
| rs10503574 | 1214 | 2450 | 0.05 | 0.04 | 1.10(0.82-1.49) | 0.31(0.06-1.49) | 1.07(0.80-1.43) | 0.653 | |
| rs3918249 | 4539 | 4098 | 0.35 | 0.36 | 0.97(0.87-1.08) | 0.82(0.70-0.96) | 0.92(0.86-0.99) | 0.024 | |
| All women, type I endometrial cancer cases vs. controls | |||||||||
| rs2970924 | 4703 | 7037 | 0.15 | 0.16 | 0.89(0.81-0.98) | 0.94(0.72-1.22) | 0.91(0.84-0.99) | 0.027 | |
| rs352038 | 5285 | 8405 | 0.06 | 0.08 | 1.17(1.03-1.32) | 1.28(0.87-1.88) | 1.17(1.05-1.30) | 0.004 | |
| rs2069852 | 4653 | 6922 | 0.22 | 0.38 | 0.98(0.85-1.13) | 1.08(0.89-1.31) | 1.10(1.01-1.19) | 0.030 | |
| rs10503574 | 2605 | 6685 | 0.16 | 0.17 | 0.93(0.83-1.04) | 0.70(0.53-0.92) | 0.88(0.80-0.96) | 0.007 | |
| rs3918249 | 5484 | 8273 | 0.45 | 0.53 | 0.97(0.88-1.07) | 0.82(0.72-0.93) | 0.91(0.86-0.97) | 0.002 | |
| Asian-ancestry women, type I endometrial cancer cases vs. controls | |||||||||
| rs2970924 | 1464 | 3783 | 0.15 | 0.16 | 0.90(0.78-1.04) | 0.79(0.52-1.20) | 0.89(0.79-1.00) | 0.055 | |
| rs352038 | 1448 | 3773 | 0.17 | 0.16 | 1.14(0.99-1.31) | 1.24(0.83-1.87) | 1.13(1.00-1.28) | 0.041 | |
| rs2069852 | 1393 | 3675 | 0.67 | 0.65 | 0.88(0.71-1.07) | 1.07(0.88-1.32) | 1.09(0.99-1.20) | 0.075 | |
| rs10503574 | 1439 | 3823 | 0.23 | 0.26 | 0.88(0.78-1.01) | 0.68(0.51-0.90) | 0.85(0.77-0.94) | 0.002 | |
| rs3918249 | 1453 | 3654 | 0.70 | 0.72 | 0.93(0.74-1.18) | 0.80(0.63-1.01) | 0.89(0.80-0.98) | 0.015 | |
| European-ancestry women, type I endometrial cancer cases vs. controls | |||||||||
| rs2970924 | 3037 | 2856 | 0.15 | 0.15 | 0.86(0.76-0.98) | 1.05(0.74-1.49) | 0.91(0.82-1.02) | 0.099 | |
| rs352038 | 3580 | 4111 | 0.02 | 0.01 | 1.16(0.86-1.57) | 1.26(1.00-1.59) | 1.19(0.88-1.60) | 0.255 | |
| rs2069852 | 3060 | 2850 | 0.03 | 0.03 | 1.07(0.72-1.60) | 0.54(0.05-5.40) | 1.08(0.72-1.62) | 0.703 a | |
| rs10503574 | 1061 | 2450 | 0.05 | 0.04 | 1.12(0.82-1.53) | 0.54(0.05-5.40) | 1.07(0.80-1.45) | 0.644 | |
| rs3918249 | 3574 | 4098 | 0.35 | 0.36 | 0.98(0.88-1.10) | 0.79(0.67-0.93) | 0.91(0.84-0.98) | 0.017 | |
Random effects model used
DISCUSSION
The link between inflammation and endometrial cancer is supported by a great deal of experimental and epidemiological evidence; conditions related to chronic inflammation, such as prolonged menstruation, obesity, unopposed menopausal estrogen use, and other factors, have all been linked to and increased risk of endometrial cancer (28, 29). Menstruation itself, during which the endometrium goes through proliferative, secretory, and menstrual phases, mimics an inflammatory process and is associated with the activation of inflammatory cytokines that results in the shedding of the endometrium (29). Estrogen directly regulates the production of a number of inflammatory cytokines, growth factors, and corresponding receptors (30). Inflammation increases mitotic activity in endometrial epithelial cells, which in turn results in increased DNA replication and repair errors, subsequently leading to somatic mutations that may ultimately give rise to hyperplasia and endometrial cancer (12).
In this large two-stage study, including samples from both Asian- and European-ancestry populations, we found that genetic variants in five candidate genes, FABP1, CXCL3, IL6, MSR1, and MMP9, were associated with endometrial cancer in combined analyses. Of these, the CXCL3 and MMP9 polymorphisms had significant associations in the stage 2 analysis. Only rs3918249, the MMP9 variant, was associated with endometrial cancer in both Asian- and European-ancestry samples and remained statistically significant after adjustment for multiple comparisons.
MMP9 encodes a matrix metalloproteinase, involved in the breakdown of the extracellular matrix, a process which has been well studied for its relationship with cancer. MMP9 is secreted from endometrial stromal cells in response to induction by growth factors, such as HGF, in endometrial cancer cell lines, which, in turn, increases cancer cell invasiveness (31). Expression of MMP9 is known to be up-regulated through pro-inflammatory cytokines, including nuclear factor kappa B, IL8, and TNF-alpha, leading to increased tumor cell proliferation (32-34). MMP9 expression level has been correlated to the grade and stage of endometrial cancer (35). The MMP9 protein has been shown to be frequently expressed in endometriosis, a benign disease, in which MMP9 expression level is higher in aggressive lesions than in normal endometrium (36, 37). MMP9 transgenic mice show significantly increased susceptibility to chemically induced cancer (38). The significant SNP we found, rs3918249, resides in a promoter region of MMP9, and is predicted to be in a transcription factor binding site and has modestly strong LD with some sites predicted to act as miRNA binding sites or splice enhancers. Further, it is in LD with two non-synonymous coding SNPs, rs17576 and rs2250889, in MMP9 (Supplementary Table 2). Further investigation of the role of this gene in endometrial carcinogenesis is warranted as is fine-mapping of this locus for other possible causal alleles.
SNP rs352038 near the CXCL3 gene was our second most significant finding overall and, like MMP9, independently significant in the replication sample. CXCL3 is an attractive candidate gene, although rs352038 is not located in the CXCL3 gene, but 14.2kb downstream. However, it is in LD with SNPs in other CXC chemokine genes in the 4q21 region, including CXCL2 and CXCL5. CXCL3 is upregulated in breast cancer, is present at higher levels in metastases, and is associated with shorter relapse-free survival in patients treated with tamoxifen (39). Consistent with the hormonal etiology of endometrial cancer, gonadotropin releasing hormone (GnRH) I and II may regulate the expression of CXCL3 (40). CXCL3 has shown to be up-regulated in uterine smooth muscle. Inhibition of CXCL3 and IL6 has been shown in cancer cell lines to reduce Stat3 activation (41). It is worth noting that the genotyped SNP rs352038 is predicted to act as an eQTL for another inflammatory gene, IL8 (P = 0.007), though this gene is over 300kb distant from rs352038 (42). This SNP is in LD with two other SNPs predicted to be potential transcription factor binding sites (Supplementary Table 2).
Three other SNPs in or near FABP1 (rs2970294), IL6 (rs2069852), and MSR1 (rs10503574) with significant associations in stage 1 data were also significant in the overall dataset, although they were not replicated in stage 2.
The present study has a number of strengths and weaknesses. The study benefits from its collection of a relatively large number of case and control samples from a number of study sites. The increased sample size and consistent directions of association across a number of study sites strengthens the evidence that these findings — particularly for the CXCL3 and MMP9 SNPs — are much more likely to represent true associations. Limitations include that stage 1 was carried out in an Asian population, and only one SNP per region was selected for the replication study. Some association findings may not extend to non-Asian populations, because of LD structure differences resulting in false negative results, as may be the case for rs10503574 in MSR1, where LD blocks as defined by D-prime are quite different between HapMap samples for CEU and CHB+JPT. False positive findings resulting from multiple testing is another concern. Minor allele frequencies in European populations were quite low for three of the five SNPs significant in stage I (in CXCL3, IL6, and MSR1), suggesting low statistical power for validating these associations. Furthermore, we did not have information on most of the non-genetic risk factors for stage 2 data, which limited our ability to evaluate the potential confounding effects of these factors. However, within stage 1 data, adjusting for known non-genetic factors, including age BMI, age at menarche, age at menopause, nulliparity, and HRT use did not materially alter point estimates for SNPs selected for stage 2 replication genotyping, Last, this analysis was restricted to SNPs in or near (within 20kb) the 64 candidate inflammation genes. Future studies may wish to expand investigations to SNPs known to be eQTLs for inflammatory genes, some of which may be more distant or even in trans to the genes they regulate. Such variations may offer more potent explanations of the expression levels of inflammatory genes. As new resources such as The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression project (GTEx) are developed, the tools to determine the SNPs controlling the expression of these genes in relevant tissue types will allow more specific tests to be carried out.
In summary, this study found evidence for the involvement of MMP9 and CXCL3 in endometrial carcinogenesis in both Asian- and European-ancestry populations. These findings may warrant additional and functional studies to determine the mechanisms by which these common variants increase disease risk. Future studies may focus on specific eQTL SNPs in the tissues of interest and seek to better explore the link between these inflammatory pathway genes and endometrial carcinogenesis.
Supplementary Material
ACKNOWLEDGEMENTS
The SECS GWAS thanks Drs. Wang-Hong Xu, Fan Jin, and other research staff for their contributions to the field operation, Ms. Regina Courtney for DNA preparation, and Ms. Bethanie Rammer and Mrs. Jacqueline Stern for editorial support in the preparation of this manuscript.
The HJECS thanks Dr. Wen Zheng, Prof. Peter Hillemanns, and Prof. Ingo Runnebaum for their support in patient recruitment. The HAECS gratefully acknowledges Prof. Tjoung-Won Park-Simon for her support.3
ANECS gratefully acknowledges the contributions of Study Investigator Penelope Webb and support of recruitment by project grants from the National Health and Medical Research Council of Australia (ID#339435), The Cancer Council Queensland (ID#4196615), and Cancer Council Tasmania (ID#403031 and ID#457636).
The cooperation of 28 Connecticut hospitals, including Charlotte Hungerford Hospital, Bridgeport Hospital, Danbury Hospital, Hartford Hospital, Middlesex Hospital, New Britain General Hospital, Bradley Memorial Hospital, Yale/New Haven Hospital, St. Francis Hospital and Medical Center, St. Mary’s Hospital, Hospital of St. Raphael, St. Vincent’s Medical Center, Stamford Hospital, William W. Backus Hospital, Windham Hospital, Eastern Connecticut Health Network, Griffin Hospital, Bristol Hospital, Johnson Memorial Hospital, Day Kimball Hospital, Greenwich Hospital, Lawrence and Memorial Hospital, Milford Hospital, New Milford Hospital, Norwalk Hospital, MidState Medical Center, John Dempsey Hospital and Waterbury Hospital, in allowing patient access, is gratefully acknowledged.
The authors take sole responsibility for the content of this article.
Grant Support: The SECS was supported by a US PHS grant R01 CA098285 (PI: X.-O. Shu) from the National Institutes of Health, National Cancer Institute (NIH/NCI). Other studies that contributed to the SECS GWAS were funded by NIH/NCI US PHS grants, R01 CA064277, R01 CA090899, and R37 CA070869 (PI: W. Zheng). The Stage 2 ANECS research was supported by the National Health and Medical Research Council (ID#552402), The Wellcome Trust and by Cancer Research UK grants C1287/A10118, C490/A1021, C8197/A10865 & C8197/A10123. A.B.S. is an NHMRC Senior Research Fellow. T.O’M. is supported by an Australian Postgraduate Award, an Institute of Health and Biomedical Innovation PhD Top-Up, and a Smart State PhD Award. D.F.E. is a Principal Research Fellow of Cancer Research UK. A.M.D is supported by the Joseph Mitchell Trust. I.T. is supported by Cancer Research UK and the Oxford Comprehensive Biomedical Research Centre. We acknowledge use of DNA from the British 1958 Birth Cohort collection, funded by the Medical Research Council grant G0000934 and the Wellcome Trust grant 068545/Z/02. Funding for this project was provided by the Wellcome Trust under award 085475. P.A.F. was partly funded by the Dr. Mildred Scheel Stiftung of the Deutsche Krebshilfe (German Cancer Aid). A.B. Spurdle, F. Lose, and T. O’Mara represent the ANECS. ANECS gratefully acknowledges the contributions of Study Investigator Penelope Webb and support of recruitment by project grants from the National Health and Medical Research Council of Australia (ID#339435), The Cancer Council Queensland (ID#4196615), and Cancer Council Tasmania (ID#403031 and ID#457636). The Bavarian Endometrial Cancer Study (BECS) was partly funded by the ELAN fund of the University of Erlangen. This study was supported by NCI-NIH grant 5R01CA098346. This study was approved by the State of Connecticut Department of Public Health Human Investigation Committee. Certain data used in this study were obtained from the Connecticut Tumor Registry in the Connecticut Department of Public Health. The Leuven Endometrium Study (LES) was supported by the Verelst Foundation for endometrial cancer. MoMaTEC received financial support from a Helse Vest Grant, the University of Bergen, Melzer Foundation, The Norwegian Cancer Society (Harald Andersens legat), The Research Council of Norway and Haukeland University Hospital. The Shanghai Endometrial Cancer Genetics Study (SECGS) was supported by grants from the National Cancer Institute of United States Public Health Service (R01 CA092585, R01 CA90899, R01 CA064277). SEARCH is funded by a programme grant from Cancer Research UK [C490/A10124].
Footnotes
Conflict of interest: The authors have no conflicts of interest to declare.
Reference List
- (1).Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. International Journal of Cancer. 2010;127(12):2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- (2).Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- (3).Shanghai Municipal Center for Disease Control and Prevention Shanghai Cancer Report of 2009. Jan 1, 2009.
- (4).Mueck AO, Seeger H, Rabe T. Hormonal contraception and risk of endometrial cancer: a systematic review. Endocr Relat Cancer. 2010;17(4):R263–R271. doi: 10.1677/ERC-10-0076. [DOI] [PubMed] [Google Scholar]
- (5).Schildkraut JM, Risch N, Thompson WD. Evaluating genetic association among ovarian, breast, and endometrial cancer: evidence for a breast/ovarian cancer relationship. Am J Hum Genet. 1989;45(4):521–9. [PMC free article] [PubMed] [Google Scholar]
- (6).Sangrajrang S, Sato Y, Sakamoto H, Ohnami S, Laird NM, Khuhaprema T, et al. Genetic polymorphisms of estrogen metabolizing enzyme and breast cancer risk in Thai women. International Journal of Cancer. 2009;125(4):837–43. doi: 10.1002/ijc.24434. [DOI] [PubMed] [Google Scholar]
- (7).Grady D, Gebretsadik T, Kerlikowske K, Ernster V, Petitti D. Hormone replacement therapy and endometrial cancer risk: a meta-analysis. Obstetrics & Gynecology. 1995;85(2):304–13. doi: 10.1016/0029-7844(94)00383-O. [DOI] [PubMed] [Google Scholar]
- (8).McPherson CP, Sellers TA, Potter JD, Bostick RM, Folsom AR. Reproductive Factors and Risk of Endometrial Cancer The Iowa Women’s Health Study. American journal of Epidemiology. 1996;143(12):1195–202. doi: 10.1093/oxfordjournals.aje.a008707. [DOI] [PubMed] [Google Scholar]
- (9).Brinton LA, Sakoda LC, Sherman ME, Frederiksen K, Kjaer SK, Graubard BI, et al. Relationship of benign gynecologic diseases to subsequent risk of ovarian and uterine tumors. Cancer Epidemiology Biomarkers & Prevention. 2005;14(12):2929–35. doi: 10.1158/1055-9965.EPI-05-0394. [DOI] [PubMed] [Google Scholar]
- (10).Brinton LA, Berman ML, Mortel R, Twiggs LB, Barrett RJ, Wilbanks GD, et al. Reproductive, menstrual, and medical risk factors for endometrial cancer: results from a case-control study. American journal of obstetrics and gynecology. 1992;167(5):1317. doi: 10.1016/s0002-9378(11)91709-8. [DOI] [PubMed] [Google Scholar]
- (11).Kalandidi A, Tzonou A, Lipworth L, Gamatsi I, Filippa D, Trichopoulos D. A case-control study of endometrial cancer in relation to reproductive, somatometric, and life-style variables. Oncology. 1996;53(5):354–9. doi: 10.1159/000227587. [DOI] [PubMed] [Google Scholar]
- (12).Key TJ, Pike MC. The dose-effect relationship between’unopposed’oestrogens and endometrial mitotic rate: its central role in explaining and predicting endometrial cancer risk. British journal of cancer. 1988;57(2):205. doi: 10.1038/bjc.1988.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Xu WH, Xiang YB, Ruan ZX, Zheng W, Cheng JR, Dai Q, et al. Menstrual and reproductive factors and endometrial cancer risk: Results from a population–based case control study in urban Shanghai. International Journal of Cancer. 2004;108(4):613–9. doi: 10.1002/ijc.11598. [DOI] [PubMed] [Google Scholar]
- (14).Hardiman P, Pillay OS, Atiomo W. Polycystic ovary syndrome and endometrial carcinoma. The lancet. 2003;361(9371):1810–2. doi: 10.1016/s0140-6736(03)13409-5. [DOI] [PubMed] [Google Scholar]
- (15).Wisse BE. The inflammatory syndrome: the role of adipose tissue cytokines in metabolic disorders linked to obesity. Journal of the American Society of Nephrology. 2004;15(11):2792–800. doi: 10.1097/01.ASN.0000141966.69934.21. [DOI] [PubMed] [Google Scholar]
- (16).Modugno F, Ness RB, Chen C, Weiss NS. Inflammation and endometrial cancer: a hypothesis. Cancer Epidemiology Biomarkers & Prevention. 2005;14(12):2840–7. doi: 10.1158/1055-9965.EPI-05-0493. [DOI] [PubMed] [Google Scholar]
- (17).Preston-Martin S, Pike MC, Ross RK, Jones PA, Henderson BE. Increased cell division as a cause of human cancer. Cancer Research. 1990;50(23):7415. [PubMed] [Google Scholar]
- (18).Long J, Zheng W, Xiang YB, Lose FA, Thompson DJ, Tomlinson I, et al. Genome-wide association study identifies a possible susceptibility locus for endometrial cancer. Cancer Epidemiology Biomarkers & Prevention. 2012 doi: 10.1158/1055-9965.EPI-11-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Jourquin J. Gene List Automatically Derived For You (GLAD4U): deriving and prioritizing gene lists from PubMed literature. 2010. [DOI] [PMC free article] [PubMed]
- (20).Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic acids research. 2011;39(suppl 1):D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Zheng W, Long J, Gao YT, Li C, Zheng Y, Xiang YB, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41(3):324–8. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nature genetics. 2008;40(10):1253–60. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genetic epidemiology. 2010;34(8):816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Spurdle AB, Thompson DJ, Ahmed S, Ferguson K, Healey CS, O’Mara T, et al. Genome-wide association study identifies a common variant associated with risk of endometrial cancer. Nat Genet. 2011 doi: 10.1038/ng.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Normand SLT. Tutorial in biostatistics meta-analysis: formulating, evaluating, combining, and reporting. Statistics in medicine. 1999;18(3):321–59. doi: 10.1002/(sici)1097-0258(19990215)18:3<321::aid-sim28>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- (26).Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic acids research. 2009;37(suppl 2):W600–W605. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Gangemi M, Meneghetti G, Predebon O, Scappatura R, Rocco A. Obesity as a risk factor for endometrial cancer. Clinical and experimental obstetrics & gynecology. 1987;14(2):119. [PubMed] [Google Scholar]
- (29).Sugino N, Karube-Harada A, Taketani T, Sakata A, Nakamura Y. Withdrawal of Ovarian Steroids Stimulates Prostaglandin F2α Production Through Nuclear Factor-κB Activation via Oxygen Radicals in Human Endometrial Stromal Cells: Potential Relevance to Menstruation. Journal of Reproduction and Development. 2004;50(2):215–25. doi: 10.1262/jrd.50.215. [DOI] [PubMed] [Google Scholar]
- (30).Tabibzadeh S. Cytokines and the hypothalamic—pituitary—ovarian—endometrial axis. Human Reproduction. 1994;9(5):947–67. doi: 10.1093/oxfordjournals.humrep.a138621. [DOI] [PubMed] [Google Scholar]
- (31).Park Y, Ryu H, Choi D, Chang K, Park D, Min CK. Effects of hepatocyte growth factor on the expression of matrix metalloproteinases and their tissue inhibitors during the endometrial cancer invasion in a three-dimensional coculture. International Journal of Gynecological Cancer. 2003;13(1):53–60. doi: 10.1046/j.1525-1438.2003.13033.x. [DOI] [PubMed] [Google Scholar]
- (32).Oh JH, Kim JH, Ahn HJ, Yoon JH, Yoo SC, Choi DS, et al. Syndecan-1 enhances the endometrial cancer invasion by modulating matrix metalloproteinase-9 expression through nuclear factor κB. Gynecologic oncology. 2009;114(3):509–15. doi: 10.1016/j.ygyno.2009.05.027. [DOI] [PubMed] [Google Scholar]
- (33).Laterveer L, Lindley IJ, Heemskerk DP, Camps JA, Pauwels EK, Willemze R, et al. Rapid mobilization of hematopoietic progenitor cells in rhesus monkeys by a single intravenous injection of interleukin-8. Blood. 1996;87(2):781–8. [PubMed] [Google Scholar]
- (34).Zhu X, Liu Q, Wang M, Liang M, Yang X, Xu X, et al. Activation of Sirt1 by Resveratrol Inhibits TNF-α Induced Inflammation in Fibroblasts. PloS one. 2011;6(11):e27081. doi: 10.1371/journal.pone.0027081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Aglund K, Rauvala M, Puistola U, Angström T, Turpeenniemi-Hujanen T, Zackrisson B, et al. Gelatinases A and B (MMP-2 and MMP-9) in endometrial cancer-MMP-9 correlates to the grade and the stage. Gynecologic oncology. 2004;94(3):699–704. doi: 10.1016/j.ygyno.2004.06.028. [DOI] [PubMed] [Google Scholar]
- (36).Weigel MT, Krämer J, Schem C, Wenners A, Alkatout I, Jonat W, et al. Differential expression of MMP-2, MMP-9 and PCNA in endometriosis and endometrial carcinoma. European Journal of Obstetrics & Gynecology and Reproductive Biology. 2011 doi: 10.1016/j.ejogrb.2011.09.040. [DOI] [PubMed] [Google Scholar]
- (37).Ueda M, Yamashita Y, Takehara M, Terai Y, Kumagai K, Ueki K, et al. Survivin gene expression in endometriosis. Journal of Clinical Endocrinology & Metabolism. 2002;87(7):3452–9. doi: 10.1210/jcem.87.7.8682. [DOI] [PubMed] [Google Scholar]
- (38).Mohammed FF, Pennington CJ, Kassiri Z, Rubin JS, Soloway PD, Ruther U, et al. Metalloproteinase inhibitor TIMP̃1 affects hepatocyte cell cycle via HGF activation in murine liver regeneration. Hepatology. 2005;41(4):857–67. doi: 10.1002/hep.20618. [DOI] [PubMed] [Google Scholar]
- (39).Bièche I, Chavey C, Andrieu C, Busson M, Vacher S, Le Corre L, et al. CXC chemokines located in the 4q21 region are up-regulated in breast cancer. Endocrine-related cancer. 2007;14(4):1039–52. doi: 10.1677/erc.1.01301. [DOI] [PubMed] [Google Scholar]
- (40).Cavanagh PC, Dunk C, Pampillo M, Szereszewski JM, Taylor JE, Kahiri C, et al. Gonadotropin-releasing hormone-regulated chemokine expression in human placentation. American Journal of Physiology-Cell Physiology. 2009;297(1):C17–C27. doi: 10.1152/ajpcell.00013.2009. [DOI] [PubMed] [Google Scholar]
- (41).Marotta LLC, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, et al. The JAK2/STAT3 signaling pathway is required for growth of CD44+ CD24–stem cell–like breast cancer cells in human tumors. The Journal of Clinical Investigation. 2011;121(7):2723. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Veyrieras JB, Kudaravalli S, Kim SY, Dermitzakis ET, Gilad Y, Stephens M, et al. High-resolution mapping of expression-QTLs yields insight into human gene regulation. PLoS genetics. 2008;4(10):e1000214. doi: 10.1371/journal.pgen.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.