Abstract
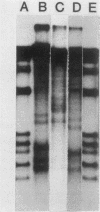
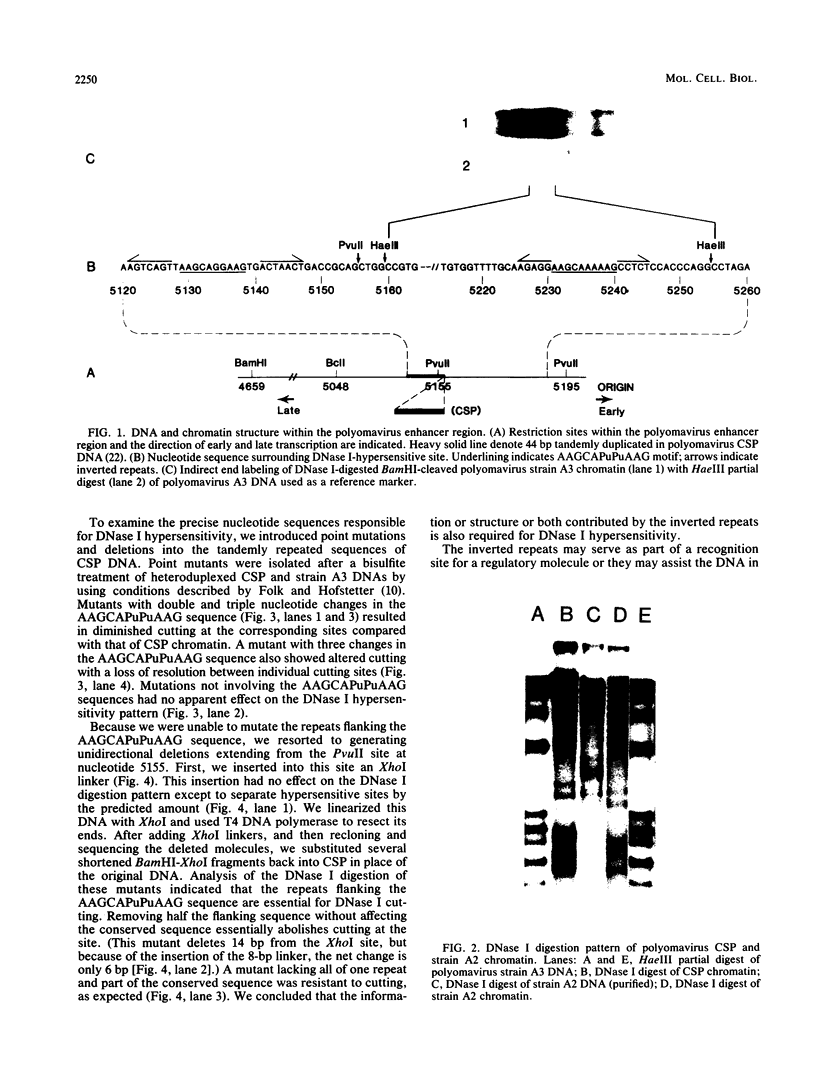
DNase I preferentially cleaves polyomavirus minichromosomes at two sites in the enhancer, each of which comprises the sequence AAGCAPuPuAAG flanked by short inverted repeats. A tandem duplication of this sequence generates an additional hypersensitive locus. Mutations which alter either the AAGCAPuPuAAG or flanking repeats diminish hypersensitivity. This region must determine the chromatin conformation recognized by DNase I.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerji J., Olson L., Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983 Jul;33(3):729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Bryan P. N., Destree O. H. Activation and function of chromatin. Horiz Biochem Biophys. 1983;7:43–70. [PubMed] [Google Scholar]
- Cereghini S., Herbomel P., Jouanneau J., Saragosti S., Katinka M., Bourachot B., de Crombrugghe B., Yaniv M. Structure and function of the promoter-enhancer region of polyoma and SV40. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):935–944. doi: 10.1101/sqb.1983.047.01.107. [DOI] [PubMed] [Google Scholar]
- Costlow N. A., Simon J. A., Lis J. T. A hypersensitive site in hsp70 chromatin requires adjacent not internal DNA sequence. Nature. 1985 Jan 10;313(5998):147–149. doi: 10.1038/313147a0. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. DNAase I-hypersensitive sites of chromatin. Cell. 1981 Dec;27(3 Pt 2):413–415. doi: 10.1016/0092-8674(81)90381-0. [DOI] [PubMed] [Google Scholar]
- Emerson B. M., Felsenfeld G. Specific factor conferring nuclease hypersensitivity at the 5' end of the chicken adult beta-globin gene. Proc Natl Acad Sci U S A. 1984 Jan;81(1):95–99. doi: 10.1073/pnas.81.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson B. M., Lewis C. D., Felsenfeld G. Interaction of specific nuclear factors with the nuclease-hypersensitive region of the chicken adult beta-globin gene: nature of the binding domain. Cell. 1985 May;41(1):21–30. doi: 10.1016/0092-8674(85)90057-1. [DOI] [PubMed] [Google Scholar]
- Folk W. R., Hofstetter H. A detailed mutational analysis of the eucaryotic tRNAmet1 gene promoter. Cell. 1983 Jun;33(2):585–593. doi: 10.1016/0092-8674(83)90439-7. [DOI] [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Hearing P., Shenk T. The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell. 1983 Jul;33(3):695–703. doi: 10.1016/0092-8674(83)90012-0. [DOI] [PubMed] [Google Scholar]
- Herbomel P., Bourachot B., Yaniv M. Two distinct enhancers with different cell specificities coexist in the regulatory region of polyoma. Cell. 1984 Dec;39(3 Pt 2):653–662. doi: 10.1016/0092-8674(84)90472-0. [DOI] [PubMed] [Google Scholar]
- Herbomel P., Saragosti S., Blangy D., Yaniv M. Fine structure of the origin-proximal DNAase I-hypersensitive region in wild-type and EC mutant polyoma. Cell. 1981 Sep;25(3):651–658. doi: 10.1016/0092-8674(81)90172-0. [DOI] [PubMed] [Google Scholar]
- Linney E., Donerly S. DNA fragments from F9 PyEC mutants increase expression of heterologous genes in transfected F9 cells. Cell. 1983 Dec;35(3 Pt 2):693–699. doi: 10.1016/0092-8674(83)90102-2. [DOI] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- Muller W. J., Mueller C. R., Mes A. M., Hassell J. A. Polyomavirus origin for DNA replication comprises multiple genetic elements. J Virol. 1983 Sep;47(3):586–599. doi: 10.1128/jvi.47.3.586-599.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussinov R., Shapiro B., Lipkin L. E., Maizel J. V., Jr DNAase I hypersensitive sites may be correlated with genomic regions of large structural variation. J Mol Biol. 1984 Aug 25;177(4):591–607. doi: 10.1016/0022-2836(84)90039-1. [DOI] [PubMed] [Google Scholar]
- Piette J., Kryszke M. H., Yaniv M. Specific interaction of cellular factors with the B enhancer of polyoma virus. EMBO J. 1985 Oct;4(10):2675–2685. doi: 10.1002/j.1460-2075.1985.tb03987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen C., Baltimore D. Immunoglobulin gene transcription is activated by downstream sequence elements. Cell. 1983 Jul;33(3):741–748. doi: 10.1016/0092-8674(83)90016-8. [DOI] [PubMed] [Google Scholar]
- Rothwell V. M., Folk W. R. Comparison of the DNA sequence of the Crawford small-plaque variant of polyomavirus with those of polyomaviruses A2 and strain 3. J Virol. 1983 Nov;48(2):472–480. doi: 10.1128/jvi.48.2.472-480.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassone-Corsi P., Wildeman A., Chambon P. A trans-acting factor is responsible for the simian virus 40 enhancer activity in vitro. Nature. 1985 Feb 7;313(6002):458–463. doi: 10.1038/313458a0. [DOI] [PubMed] [Google Scholar]
- Schöler H. R., Gruss P. Specific interaction between enhancer-containing molecules and cellular components. Cell. 1984 Feb;36(2):403–411. doi: 10.1016/0092-8674(84)90233-2. [DOI] [PubMed] [Google Scholar]
- Tyndall C., La Mantia G., Thacker C. M., Favaloro J., Kamen R. A region of the polyoma virus genome between the replication origin and late protein coding sequences is required in cis for both early gene expression and viral DNA replication. Nucleic Acids Res. 1981 Dec 11;9(23):6231–6250. doi: 10.1093/nar/9.23.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Lupton S., Kamen R. Polyomavirus enhancer contains multiple redundant sequence elements that activate both DNA replication and gene expression. Mol Cell Biol. 1985 Apr;5(4):649–658. doi: 10.1128/mcb.5.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- de Villiers J., Olson L., Tyndall C., Schaffner W. Transcriptional 'enhancers' from SV40 and polyoma virus show a cell type preference. Nucleic Acids Res. 1982 Dec 20;10(24):7965–7976. doi: 10.1093/nar/10.24.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers J., Schaffner W., Tyndall C., Lupton S., Kamen R. Polyoma virus DNA replication requires an enhancer. Nature. 1984 Nov 15;312(5991):242–246. doi: 10.1038/312242a0. [DOI] [PubMed] [Google Scholar]