Abstract
Objective
To test if the association between inflammation and pancreatic ductal adenocarcinoma (PC) is facilitated by host susceptibility, specifically by genetic polymorphisms in inflammation-related genes.
Summary Background Data
Inflammation has been linked to PC. Reports have cited an increased expression of proinflammatory mediators, such as NF-κB and COX, in PC but not in normal adjacent tissue, suggesting a possible role in carcinogenesis. We sought to further understand the role that genetic variants in the NF-κB inflammatory pathway play in the development and progression of PC.
Methods
We genotyped 1536 tag single nucleotide polymorphisms (SNPs) in 102 candidate genes of multiple inflammatory pathways in 1308 Caucasian patients with PC who were divided into three groups based on extent of disease: resected for cure (n = 400), locally advanced/unresected (n = 443), and metastatic (n = 465). Survival analysis was performed using Kaplan-Meier curves and Cox proportional hazards regression models. Statistical significance was set at <0.001, to control for multiple testing.
Results
Median age was 67 (28.0–91.0) years, and 57% were male. Median survival for each of the three groups (resected, locally advanced, and metastatic) was 23.7, 9.4, 6.6 months, respectively (p<0.0001). In the resected group, carriers of a minor allele for either rs3824872 (MAPK8IP1) or rs8064821 (SOCS3) were associated with a 10 - and 6-month survival advantage compared to non-carriers in patients with resected disease, with an additional 2-year survival if both minor alleles were present. With locally advanced disease, SNP rs1124736 (IGF1R) was associated with improved survival if they had a copy of the G allele, hazard ratio (HR) 0.57 (0.42– 0.77) p = 0.0002. Additionally, four SNPs in patients with metastatic disease were found to be associated with worse survival and two associated with improved overall survival but the differences in survival were deemed not clinically significant.
Conclusion
SNPs in the inflammatory pathway genes MAPK8IP1 and SOCS3 were associated with increased overall survival in patients undergoing potentially curative resection and may be used in the future as markers to predict survival. Future research is needed to determine the functional relevance of these loci.
Keywords: survival, SOCS3, NFKB, MAPKI8P1, pancreatic cancer, single nucleotide polymorphisms, inflammation
Introduction
The lethality of pancreatic ductal adenocarcinoma (PC) is remarkable. In the period from 1999–2006, the 5-year relative survival in the United States was only 5.4%.1 Operative resection is the single most important treatment modality that has a positive impact on survival. The overall 5-year survival after resection improves to approximately 20%;2, 3 however, in Stage IA disease the 5-year overall survival (OS) can be as high as 32%.4
Other clinicopathologic features that appear to have a positive impact on survival are the lack of lymph node metastases, a negative resection margin (R0), lack of need for blood transfusion, postoperative chemoradiation therapy and the deoxyribonucleic acid (DNA) content of tumor.5–10 Furthermore, predisposing disease processes, such as chronic pancreatitis,11 and environmental factors, such as smoking12 increase the risk of PC. A pathway shared in common between these risk factors is inflammation.
The genetic basis of the association between inflammation and PC is of considerable interest. While clinicopathologic factors allow us to relay favorable or unfavorable prognostic information to a patient with PC, they are imperfect in identifying with certainty the individuals that will develop PC. An ideal marker would be a germline marker, because it is measurable, not subject to change, and robust to storage and technical conditions. There is increasing interest in identifying a genetic marker to serve as a prognostic indicator for patients with PC. Most reports have focused on response to drug treatment. 13 In an effort to identify such a marker, we evaluated single nucleotide polymorphisms (SNPs) in candidate genes in the inflammatory pathway genes to assess their impact on survival in surgically treated patients with PC. Our hypothesis was that SNPs in certain genes in the inflammation pathway would affect survival after operative resection of adenocarcinoma in PC patients.
Methods
Subjects
After obtaining Institutional Review Board approval, 1328 patients with PC were recruited using an ultra-rapid, case-finding process during the years 2000–2008 at Mayo Clinic as described previously.14, 15 Informed consent was obtained on all patients. Each patient filled out a questionnaire regarding dietary history, medical and family history, and medication use, and each donated blood. DNA from lymphocyte samples were extracted and stored in a −80°C freezer until the time of testing. Analyses were restricted to Caucasians only who comprised 98% of the population (N=1308).
The patients were divided into three groups based on review of the medical record review: “resected” if they had an operative procedure to remove a portion or all of the pancreas as potentially curative treatment for pancreatic cancer, “locally advanced” if they were not an operative candidate or had unresectable disease but did not have metastatic disease (American Joint Committee on Cancer (AJCC) 6th Edition, Stage III Disease), and “metastatic” if they had evidence on imaging of distant disease (AJCC 6th Edition, Stage IV Disease).
Candidate Gene and Single Nucleotide Polymorphism Selection
Candidate genes were selected using a combination of literature review and bioinformatics to identify previously reported genes involved in the pathogenesis of PC that participate in the inflammatory pathways involving NF-κB. A list of preselected genes was used to generate a list of >10,000 candidate genes utilizing bioinformatics tools of Ingenuity® Systems (Redwood, CA), and MetaCore™ from GeneGo, Inc. (St Joseph, MI) based on pathway analysis and understanding the PC disease process from this list one hundred and two genes were selected for genotyping.
To create a panel of linkage disequilibrium (LD) tag SNPs for the 102 candidate genes, we used genotypes that were made available from the genome-wide genotyping project, HapMap Phase II,16 and two gene resequencing initiatives: SeattleSNPs17 and National Institute of Environmental Health Sciences (NIEHS) SNPs18. To determine the HapMap SNPs for each of the candidate genes, we picked SNPs 5 kb upstream and downstream of each gene. Our gene and SNP coordinates were based on RefSeq release 29 (NCBI build 36) and dbSNP build 129 (http://HapMap.ncbi.nlm.nih.gov/). If the gene was resequenced in SeattleSNPs or NIEHS SNPs, we used genotypes from those sources as well. At the time we picked these SNPs, NIEHS SNPs had re-sequenced four of our candidate genes, and the SeattleSNPs had re-sequenced 26.
To pick LD tag SNPs, we ran LD Select19 on each gene for each genotype source (HapMap, Seattle, NIEHS) for the Caucasian samples in those public sources. We used a r2 of 0.9 to select tagSNPs with high LD and a minor allele frequency (MAF) cutoff of 0.05. To determine the best source of genotypes for each gene where a gene had been re-sequenced, we selected the source with the greater number of LD bins for the Caucasian samples after bins were removed and that did not have a tag SNP with an assay score of ≥0.4. If each source (e.g. HapMap, SeattleSNPs) had the same number of bins, we used HapMap as the best source because of its greater number of samples (60 unrelated Caucasian samples). If there were >10 SNPs in a LD bin, two tag SNPs were selected from that bin; and if there were >30 SNPs in a bin, three tag SNPs were selected from that bin. HapMap was chosen as best source for 79 genes, SeattleSNPs for 19 genes, and NIEHS SNPs for 3 genes. After selecting a total of 1536 LD-based tag SNPs19 and prioritizing for inclusion nonsynonymous-tag SNPs, genotyping occurred in two phases form the patients described above, genotyping 768 SNPs each time for a total of 1536 SNPs genotyped.
Genotyping
High-throughput genotyping was performed using BeadLab™ technology from Illumina®, Inc. (San Diego, CA) at Mayo Clinic’s Advanced Genomics Technology Center (AGTC) Genotyping Shared Resource 20. The quality control results have been reported elsewhere.14, 15 Each plate had trios from the Centre d’Etude du Polymorphisme Humain (CEPH) in duplicates and two random duplicates of subject samples per plate.
Phase 1
During the first phase of genotyping, 61 genes were genotyped on a 1536 SNP panel using Illumina® GoldenGate (San Diego, CA) chemistry; of these 1536 SNPs, 768 were in the genes of interest herein. Coverage of the SNPs of any given gene was greater than 90% in 51 (83.6%) of the 61 genes. The ten (16.4%) remaining genes had 50–85% SNPs, largely because tag SNPs were not used if they were within 60 base pairs, within areas of insertions or deletions, had an Illumina® (San Diego, CA) design score of <0.6 (see below), or were obtained from complementary DNA (cDNA) sequences.
Phase 2
In the second phase of genotyping, 41 genes were genotyped on a 1536 SNP panel using Illumina® GoldenGate (San Diego, CA) chemistry; of these 1536 SNPs, 768 were in the genes of interest herein. Coverage of the SNPs of any given gene was >90% in 35 (85.4%) of the 41 genes. The six (14.6%) remaining genes had 35–89% SNP coverage for the same reasons as described above.
Overall Quality Control
Single nucleotide polymorphisms were considered to be genotyped successfully if the call rate was >95% for each subject; seven cases and six controls fell below this threshold, for a genotyping pass rate of 99.99%. If a SNP deviated from Hardy Weinberg Equilibrium (HWE) (p<0.00001), the SNP was excluded from further analysis. Of the 1536 SNPs that were genotyped, the following quality control (QC) concerns were identified: 30 SNPs were monomorphic, the lab failed 110 SNPs, and 12 SNPs had HWE p<0.00001. All duplicates had concordance >90%. Only one subject sample failed genotyping and as such was not included in the Kaplan Meier or Cox regression survival analyses.
Operative and Pathologic Data Collection
The medical record was abstract by a surgeon (KRL). Abstracted data include operative procedure, pathologic resection margin (R status), lymph node status (positive versus negative), perioperative mortality, date of death, and treatment with radiation and/or chemotherapy. In addition, operative details, such as vein resection and pathologic assessments of neural and vascular invasion, were included. For descriptive purposes, the presence of extensive fibrosis and inflammation was recorded if it was mentioned in the pathology report.
Statistical Analysis
The association between genotyped SNPs using the dominant model (0=non-carrier, 1=carrier of one or more copies of the minor allele) and survival was of primary interest. Kaplan Meier curves were used to visualize survival with estimated median survival presented in the tables. Multivariable Cox Proportional Hazards regression analysis was used to further investigate these associations between SNP and survival. Models were adjusted for age, sex, stage of disease, and categorized body mass index (BMI) (< 30 not obese or ≥30 obese). In addition, the following surgical variables were also included as adjusters in the analysis of the resected patients: margin status (R0 – no neoplastic cells at the surgical margin, R1 – margin positive microscopically for neoplastic cells, R2 – margin positive grossly for neoplastic cells), grade, lymph node status and maximum tumor dimension. The SNP-SNP interaction analyses using a dominant model coding for the minor allele (i.e. carrier status yes/no) were investigated for SNPs with a p<0.001.
Results
Clinical Characteristics
There were 1,328 patients eligible for inclusion and we limited to the current analysis to 1308 Caucasians. Patients were divided into three groups, resected PC (n=400), locally advanced and unresected disease (n=443), and those with metastatic disease (465). Overall, 57.4% were male, and the median overall age at initial diagnosis of PC was 67 years (range = 28–91). Overall there were 88 patients with a reported family history of PC, 28 (7%) in the resected group, 24 (5.4%) in the locally advanced group and 31 (6.7%) in the metastatic group. Fourteen patients (6, 4, and 4 in each group respectively) reported a history of chronic pancreatitis (CP) diagnosed three years or more prior to the diagnosis of PC and 150 (68, 42, 40 in each group respectively) reported a history of chronic pancreatitis diagnosed within a year of the diagnosis of PC.
Of the 400 patients with resected disease, 16.7% were stage I, and 83.3% stage II, (Table 1). In the locally advanced group 158 (35.7%) were Stage II, and the remaining 285 (64.3%) were Stage III. The types of resections in the resected group were pancreatoduodenectomy in 322 (80.5%), distal pancreatectomy in 67 (16.8%), and total pancreatectomy in 11 (2.8%). The majority of patients (83%) undergoing resection had a R0 resection, defined as no neoplastic cells at the margins of the operative specimen.
Table 1.
Clinical Characteristics
| Resected (N=400) | Locally Advanced (N=443) | Metastatic (N=465) | Total (N=1308) | p-value | |
|---|---|---|---|---|---|
| Sex | 0.3428 | ||||
| Female | 179 (44.8%) | 192 (43.3%) | 186 (40.0%) | 557 (42.6%) | |
| Male | 221 (55.3%) | 251 (56.7%) | 279 (60.0%) | 751 (57.4%) | |
| Age at Initial Diagnosis | 0.0004 | ||||
| Median | 66.0 (32–88) | 69.0 (28 –91) | 66.0 (32–91) | 67.0 (28–91) | |
| Body Mass Index | 0.2997 | ||||
| Median | 27.0 (15–50) | 28.0 (17–53) | 28.0 (17–59) | 28.0 (15–59) | |
| BMI < 30 | 283 (70.8%) | 298 (67.3%) | 315 (67.7%) | 896 (68.5%) | |
| BMI ≥ 30 | 117 (29.3%) | 145 (32.7%) | 150 (32.3%) | 412 (31.5%) | |
| Site of pancreatic mass | <0.0001 | ||||
| Head | 299 (74.8%) | 264 (59.9%) | 213 (46.1%) | 776 (59.6%) | |
| Body | 22 (5.5%) | 79 (17.9%) | 76 (16.5%) | 177 (13.6%) | |
| Head / Body | 10 (2.5%) | 42 (9.5%) | 30 (6.5%) | 82 (6.3%) | |
| Tail | 36 (9.0%) | 10 (2.3%) | 72 (15.6%) | 118 (9.1%) | |
| Body/ Tail | 11 (2.8%) | 23 (5.2%) | 44 (9.5%) | 78 (6.0%) | |
| Uncinate Process | 16 (4.0%) | 17 (3.9%) | 20 (4.3%) | 53 (4.1%) | |
| NOS | 6 (1.5%) | 4 (0.9%) | 7 (1.5%) | 17 (1.3%) | |
| Stage at Initial Diagnosis* | <0.0001 | ||||
| Resected: IA | 20 (5.0%) | - | - | 20 (1.5%) | |
| Resected: IB | 47 (11.7%) | - | - | 47 (3.6%) | |
| Resected: IIA | 110 (27.5%) | - | - | 110 (8.4%) | |
| Resected: IIB | 223 (55.8%) | - | - | 223 (17.0%) | |
| Locally Advanced: Unresected | - | 443 (100.0%) | - | 443 (33.9%) | |
| Metastatic: Unresected | - | - | 465 (100.0%) | 465 (35.6%) | |
| Type of Surgery | |||||
| Pancreaticoduodenectomy | 322 (80.5%) | - | - | 324 (80.5%) | |
| Distal Pancreatectomy | 67 (16.8%) | - | - | 67 (16.8%) | |
| Total Pancreatectomy | 11 (2.8%) | - | - | 11 (2.8%) | |
| Maximum Tumor Dimension (cm) | |||||
| Median | 3.0 (0.0–16.0) | 3.0 (0.0–16.0) | |||
| Number Lymph Nodes Sampled | |||||
| Median | 10.0 (0.0–74.0) | 10.0 (0.0–74.0) | |||
| Lymph Node Positive | |||||
| No | 176 (44.4%) | - | - | 176 (44.4%) | |
| Yes | 221 (55.6%) | - | - | 221 (55.6%) | |
| Surgery Margin Status | |||||
| R0:Negative Margins | 331 (83.0%) | - | - | 331 (83.0%) | |
| R1:Microscopically Positive Margins | 62 (15.5%) | - | - | 62 (15.5%) | |
| R2:Grossly Positive Margins | 6 (1.5%) | - | - | 6 (1.5%) | |
| Tumor Grade | |||||
| 1 | 3 (0.8%) | - | - | 3 (0.8%) | |
| 2 | 65 (16.7%) | - | - | 65 (16.7%) | |
| 3 | 264 (67.9%) | - | - | 264 (67.9%) | |
| 4 | 57 (14.7%) | - | - | 57 (14.7%) | |
| Chemotherapy (Adjuvant) | 0.1374 | ||||
| Yes | 325 (92.6%) | 241 (88.5%) | 242 (88.6%) | 808 (90.2%) | |
| No | 26 (7.4%) | 31 (11.4%) | 31 (11.4%) | 88 (9.8%) | |
| Missing | 49 | 171 | 192 | 412 | |
| Radiation (Adjuvant) | <0.0001 | ||||
| Yes | 265 (66.3%) | 212 (47.9%) | 34 (7.3%) | 511 (39.1%) | |
| No | 135 (33.6%) | 231 (52.1%) | 431 (92.7%) | 797 (60.9%) | |
| Survival from Initial Diagnosis (months) | <0.0001 | ||||
| Median Survival Time, months | 23.7 (20.4–25.9) | 9.4 (8.7 –10.3) | 6.6 (6.1–7.3) | 10.7 (10.0–11.3) | |
| 5-year survival | 20.3% | 0.9% | 0% | 6.5% |
American Joint Committee on Cancer, 6th Edition, 2002.
Chemoradiotherapy
The vast majority of all patients underwent chemotherapy, 90.2%, with no significant difference observed across stage (p=0.1374); chemotherapy was delivered to 92.6%, 88.5%, and 88.6%, in the resected, locally advanced, and metastatic group, respectively. Gemcitabine was the chemotherapy agent used most frequently in all groups: 79.8% for patients with resected disease, 71.7% in locally advanced and 87.0% in patients with metastatic disease (p=0.0001). The frequency of radiotherapy in the resected, locally advanced, and metastatic groups was 66.3%, 47.9%, and 7.3%, p < 0.0001 respectively, with an overall frequency of 39.1% for all patients.
Overall Survival
The median survival from time of diagnosis for patients in the resected group was 23.7 (20.4–25.9) months with a 5-year survival rate of 20.3% versus 9.4 (8.7–10.3) months in the locally advanced group and 6.6 (6.1–7.3) months in patients diagnosed with metastatic disease, (Table 1).
Resected Disease
Genetic Predictors of Improved Overall Survival
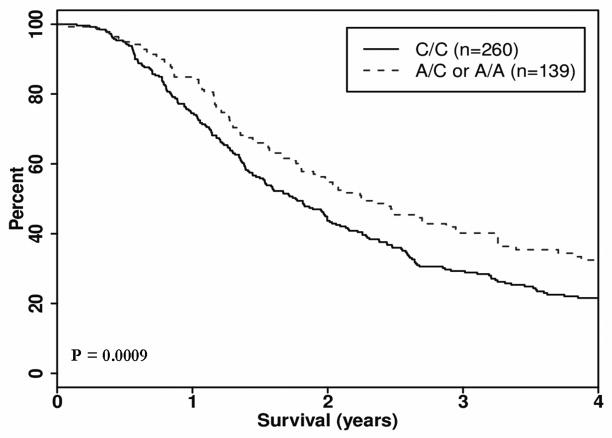
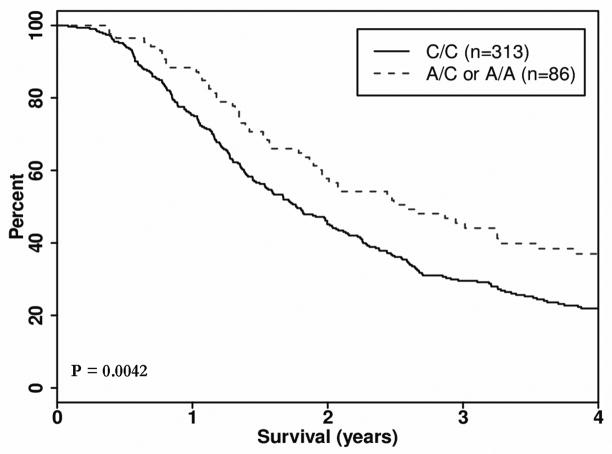
We evaluated 1,536 tag SNPs in 102 candidate genes and identified three genes harboring SNPS with a significant clinical impact on OS in the resected group (Table 2). In the MAPK8IP1 gene, SNP rs3824872 was associated with an approximate 6-month median survival advantage (27.5 months versus 21.5 months) in patients with at least one copy of the minor allele (A), HR 0.66 (95% confidence interval (CI): 0.52, 0.84) p = 0.0009, (Figure 1). The other clinically interesting SNP in the resected group, SOCS3 gene SNP rs8064821 was associated with a HR of 0.65 (0.49, 0.87), p = 0.0042 and a median survival advantage of approximately 10 months (31.4 versus 21.5 months) between carriers of the minor allele (C) and homozygote major allele carriers (Figure 2). For patients who carry a copy of a minor alleles (C/C) at both rs8064821 (SOCS3) and rs3824872 (MAPK8IP1), the observed median OS was 3.8 years compared to a median of 1.7 years for non-carriers (Table 3) (HR 0.372, 95% CI: (0.23, 0.60), p <0.0001 in the resected group.
Table 2.
SNPs with Clinical Potential: Dominant Genetic Model
| Chromosome | Gene | SNPs rs ID | Nucleotide Location* | Minor Allele Frequency | Major Allele/ Minor Allele | Total (N) ** | Deaths (N) ** | Median Survival (days) ** | Hazard Ratio *** (95% CI) | p-value *** |
|---|---|---|---|---|---|---|---|---|---|---|
| Resected | ||||||||||
| 11 | MAPK8IP1 | rs3824872 | 45862181 | 0.197 | A/C | 260/139 | 215/103 | 644/826 | 0.66 (0.52–0.84) | 0.0009 |
| 17 | SOCS3 | rs8064821 | 73868986 | 0.109 | A/C | 313/86 | 258/60 | 645/941 | 0.65 (0.49–0.87) | 0.0042 |
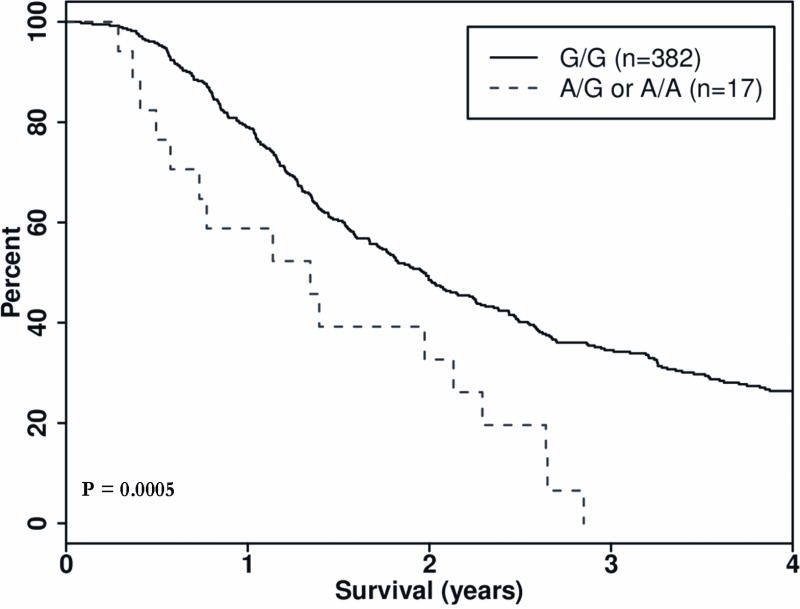
| 1 | PTGFRN | rs6688746 | 117284796 | 0.021 | A/G | 382/17 | 302/16 | 719/491 | 2.53 (1.50–4.26) | 0.0005 |
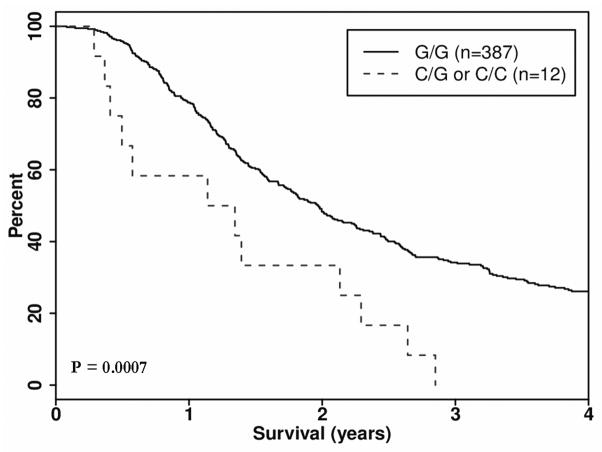
| 1 | PTGFRN | rs11583422 | 117272368 | 0.015 | C/G | 387/12 | 306/12 | 719/454 | 2.76 (1.53–4.97) | 0.0007 |
| Locally Advanced | ||||||||||
| 15 | IGF1R | rs11247367 | 97053799 | 0.070 | A/G | 378/59 | 369/51 | 269/339 | 0.57 (0.42–0.77) | 0.0002 |
| 14 | NFKB IA | rs7157810 | 34945522 | 0.202 | A/C | 278/161 | 268/155 | 305/268 | 1.43 (1.16–1.75) | 0.0008 |
| 7 | NOS3 | rs1799983 | 150327044 | 0.328 | A/C | 193/239 | 185/232 | 320/261 | 1.39 (1.14 – 1.70) | 0.0011 |
| Metastatic | ||||||||||
| 6 | RIPK1 | rs2326173 | 3020552 | 0.307 | A/G | 219/242 | 214/242 | 227/184 | 1.44 (1.20–1.74) | 0.0001 |
| 2 | STAT1 | rs2066795 | 191560142 | 0.111 | A/G | 361/96 | 358/94 | 188/266 | 0.67 (0.53–0.84) | 0.0006 |
| 2 | STAT1 | rs12693591 | 191568747 | 0.118 | A/C | 354/103 | 351/101 | 191/260 | 0.68 (0.55–0.86) | 0.0010 |
| 10 | PTPRE | rs7079639 | 129751782 | 0.254 | G/A | 260/201 | 257/199 | 226/176 | 1.39 (1.15–1.68) | 0.0007 |
| 10 | PTPRE | rs7093446 | 129750803 | 0.197 | A/G | 297/164 | 294/162 | 208/176 | 1.40 (1.16–1.71) | 0.0007 |
| 10 | GATA3 | rs10905278 | 8139829 | 0.046 | A/G | 421/40 | 416/40 | 205/149 | 1.82 (1.31–2.53) | 0.0004 |
Genome Build 36
Presented as Minor Allele non-carriers / Minor Allele carriers
Adjusted for age, sex, BMI class, Stage, margin status (R0, R1, R2), grade, tumor size and lymph node status
Figure 1.
Kaplan Meier survival curve, Dominant Model: Gene: MAPK8IP1, SNP rs3824872, Group: Resected
Figure 2.
Kaplan Meier survival curve, Dominant Model: Gene: SOCS3, SNP rs8064821, Group: Resected
Table 3.
Minor Allele Carrier Status of SNPs in MAPK8IP1 and SOCS3 Genes and Impact on Survival in Patients with Resected Pancreas Cancer
| rs3824872 (MAPK8IP1) | rs8064821 (SOCS3) | Total (N) | Deaths (N) | Median Survival (Days) | p-value |
|---|---|---|---|---|---|
| Non-carrier | Non-carrier | 210 | 176 | 625 | 0.0014 |
| Carrier | Non-carrier | 103 | 82 | 698 | |
| Non-carrier | Carrier | 50 | 39 | 712 | |
| Carrier | Carrier | 36 | 21 | 1395 |
American Joint Committee on Cancer 6 Edition, 2002
Genetic Predictors of Poor Overall Survival
In the resected group, one gene, PTGFRN, contained two SNPs that predicted worse OS. If a patient carried minor alleles rs6688746 and rs11583422 in the PTGFRN gene, there was an observed decrease in OS of 8 and 9 months respectively, with corresponding HRs of 2.53 95% CI 1.50, 4.26), p = 0.0005) and 2.76 (1.53, 4.97), p = 0.0007) (Table 2, Figure 3 and 4).
Figure 3.
Kaplan Meier survival curve, Dominant Model: Gene: PTGFRN, SNP rs6688746, Group: Resected
Figure 4.
Kaplan Meier survival curve, Dominant Model: Gene: PTGFRN, SNP rs1153422, Group: Resected
Impact of Chemotherapy and Radiation – Resected Group
While survival was improved for carriers of the minor alleles of SNPs rs3824872 (MAPK8IP1) and SNP rs8064821 (SOCS3) in resected patients, however this did not seem to be driven by the delivery of chemo or radiation therapy as survival was highest amongst the group of patients with the minor allele in each group evaluated (chemotherapy yes or no and radiation therapy yes or no), (Supplemental Table 1). The overall survival was highest among patients that carried at least one copy of the minor alleles and who were treated with chemoradiation, however even if a patient was not treated with chemoradiation therapy the survival was higher among those who were carriers of the minor allele for each SNP.
Similarly, patients that were carriers of the minor alleles associated with poor survival (rs6688746 and rs11583422) in the PTGFRN gene had observed worse overall survival even when treated with and chemotherapy and radiation therapy when compared to those who homozygote major allele carriers. (Supplemental Table 1).
Locally Advanced Disease
Genetic Predictors of Improved Overall Survival
In locally advanced disease, rs11247367 in the IGFIR gene was associated with improved survival if a patient carried a copy of the G allele, HR 0.57, (95% CI 0.42–0.77) p =0.0002. The median OS of carriers of the minor allele was two months longer than patients who were major allele carriers (11.3 months versus 9.0 months). If a patient carried two copies of the minor allele (G/G) there was an observed worse survival than carriers of only one copy of the minor allele or those who were homozygote carriers of the major allele (A). This SNP however has a low MAF of only 7% (Table 2).
Genetic Predictors of Poor Overall Survival
In the NFKBIA gene, SNP rs7157810 and rs150327044 in NOS3 were associated with worse OS by 1–2 months if patients were carriers of one of more copies of the variant (C) allele. The minor allele frequencies were 20.2% and 32.8%, (Table 2)
Metastatic disease
Genetic Predictors of Improved Overall Survival
In patients with metastatic disease, SNPs rs2066795 and rs12693591 in STAT1 were associated with an improvement of 2–2.5 months in OS among carriers of the variant (G and C respectively) allele, HR= 0.67 (95% CI 0.53–0.84), p = 0.0006 and HR=0.68 (95%CI 0.55–0.86), p=0.0010, respectively.
Genetic Predictors of Poor Overall Survival
SNP rs2326173 in RIPK1 was associated with decreased OS (by a median of 43 days) for carriers of the variant (G) allele, HR = 1.44 (1.20 –1.74), p-value = 0.0001. Similarly SNP rs10905278 in GATA3 had a decreased OS of 2 months for carriers of the minor allele (G), HR = 1.82 (95% CI 1.31–2.53) p = 0.0004. SNPs rs7079639 and rs7093446 in the PTPRE gene had a decreased OS by 2 and 1 months among A and G, allele carriers, p = 0.0007 and 0.0007, respectively (Table 2).
Discussion
In this candidate inflammation pathway gene study, SNPs in inflammatory-related genes of the NF-κB pathway were genotyped to assess their impact on survival in patients with PC. We have identified four potential markers that may be used to predict OS after resection of PC. The observed improvement in OS among these SNPs was from 6–10 months, an observation that was consistent across treatment with radiation and/or chemotherapy. The most interesting of the SNPs, rs8064821 in the SOCS3 gene, had an observed 10-month increase in OS among the resected patients. This association was observed even when patients did and did not receive radiation or chemotherapy in the resected group. Patients who carried the minor alleles of both rs3824872 (MAPK8IP1) and rs8064821 (SOCS3) had an observed doubling of their OS from a median of 1.7 years to a median survival of up to 3.8 years.
Resected Group
No prior studies have reported on the role of variants in MAPK8IP1 and SOCS3 as a possible predictor of OS. However, a recent report suggested a role for SOCS3 in the progression of precursor lesions of the pancreas, pancreatic intraepithelial neoplasm (PanIN) to PC via the IL6 signaling pathway in a mouse model.21 They found that SOCS3 and KRAS mutations were associated with PanIN progression to PC. The authors discussed that STAT3 is important in that it regulates KRAS dependent apoptosis and leads to tumor growth via a modulation of the stroma and the immune system. SOCS3 normally serves to inhibit STAT3 however if a homozygous deletion occur in the SOCS3 gene then its normally inhibitory function is reversed leading to increased levels of SOCS3 an observed progression to PC is observed.
STAT3 induction is achieved by phosphorylation of STAT3 via interleukin 1L6. In a previously reported study, IL6 levels >5.2 pg/mL in the serum was associated with worse OS in patients with PC.22 This observation may be due to the contribution of IL6 to cachexia and hypermetabolism 23 in PC patients or perhaps by contributing to the progression of disease. While genetic variants in STAT3 were not associated with improved OS using our cut off values, with improved survival (p=0.004–0.008), three SNPS were found to interact with SOCS3 SNP rs8064821 and were found to have improved OS if the subject carried copies of both the variant SOCS3 SNP and the variant STAT3 SNPs suggesting a potentially biological interaction. Our study, however, the study was not powered for this analysis and follow-up is needed.
In addition to genetic variants in SOCS3, SNP rs3824872 in the MAPK8IP1 was associated with improved OS. The MAPK8IP1 gene is thought to play an important role in the development of diabetes. MAPK8IP1 is a transcript of the IBI (insulin-brain-1) gene, which is a DNA-binding transactivator of the glucose transporter (GLUT2).24 IB1 is predominately found in pancreatic islet beta cells protein related to JIP-1, which is highly expressed in pancreatic beta-cells where it functions as a transactivator of the GLUT2 gene.25 The exact role of MAPK8IP1 in PC is unclear.
Both the minor alleles identified in the SOCS3 and MAPK8IP1 genes are located in the promotor region of the gene and has a medium risk of a functional effect as predicted by FastSNP. 26 Further exploration using the Alibaba2.1 net-based transcription factor binding site search tool ((http://www.gene-regulation.com/pub/databases.html) 27, 28 predicted in silico that the presence of the minor allele rs8064821 in the SOCS3 lead to an introduction of a new transcription binding site for the transcription factor nuclear-factor 1 (NF-1). There was no predicted functional effect for the minor allele, rs3824872 in the MAPK8IP1 gene. The SNPS in the PTFGRN genes are intronic with no predicted function effect.
Locally Advanced Group
In the locally advanced tumor group, we identified three genes with genetic variants that were associated with changes in OS. The first IGF1R was associated with improved survival while the other two NFKB1A and NOS3 were associated with worse overall survival. The clinical significance of the differences in the latter two genes was small. In the IGF1R gene, SNP rs11247367 was associated with improved OS by 2 months with improved survival modified by chemoradiation therapy. Dong, et al. recently reported on genetic variants in insulin-like growth factor pathways and identified two SNPs in the IGF1R gene with improved OS and HR, similar to our findings with HR ranging from 0.60 to 0.87, depending on genotype.29 IGF1R overexpression inhibits apoptosis of PC cells and allows for motility in the host via the PI3K/AKT/mTOR and RAS/MAPK/ERK pathways.30, 31 Similar observations were seen between our studies warrants further investigation to assess the functional implications of this intronic SNP as there were no in silico functional effect.
Metastatic Group
Six SNPs in four genes were associated with survival, two with improved survival and four with worse survival however the survival differences on average were only two months. These differences lack clinical significance as the clinical impact would be minor. The most important of these SNPs, rs2326173 of the RIPK1 gene had a predicted loss of three transcription factor binding sites, IRF9, LMOD1, IRF8, in silico. 27, 60 Recently Chae et al, published that the GA+AA genotype of the RIPK1 rs2272990 was associated with a worse disease free and disease specific survival in colorectal cancer after complete resection. The minor allele of RIPK1 rs2326173 in our study was associated with worse overall survival in PC with metastatic disease.32
The obvious strength of the study is the large sample of patients with surgically resected PC. Results were rigidly adjusted for known factors that portend a poor outcome of patients with PC (e.g., lymph node status, tumor size, stage). The ability to clinically predict survival based on genotype after surgical resection would be helpful as we may be able to determine who would benefit from either conservative therapy or more aggressive therapeutic strategies such as surgery with chemoradiation therapy.
An important limitation of this study is that the results have yet to be validated in an independent dataset. Due to the relatively low frequency of patients with surgically treated PC (15%–20% of all patients diagnosed with the disease), additional time is required to collect an adequate validation sample. This work is presently underway. Another important limitation is that our study was not powered to address the impact of radiation and chemotherapy in the cohorts however the median survival of carriers of the minor alleles in the resected group did not seem to be driven solely by radiation and chemotherapy as patients had higher median survival whether or not they were treated with radiation and chemotherapy.
Conclusion
Variants in genes in the inflammation related pathway may inform clinical prognosis and aid in treatment planning for patients who are candidates for resection with PC. Prognostic information for patients with locally advanced and metastatic disease may potentially also be informed by genotype.
Supplementary Material
Acknowledgments
Source Funding
This publication was made possible by the Mayo Clinic SPORE in Pancreatic Cancer Grant (P50 CA 102701), Grant Number 1 UL1 RR024150 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), CA15083 (Mayo Clinic Comprehensive Cancer Center Grant), and the NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR or NIH. Information on NCRR is available at http://www.ncrr.nih.gov/. Information on Reengineering the Clinical Research Enterprise can be obtained from http://nihroadmap.nih.gov.
Footnotes
Conflicts of Interest:
There are no conflicts of interest to disclose by any author.
Oral Presentation at the 97th Annual Meeting of the American College of Surgery in San Francisco, CA October 26, 2011
References
- 1.Howlader N, Noone AM, Krapcho M, Neyman N, Aminou R, Altekruse SF, Kosary CL, Ruhl J, Tatalovich Z, Cho H, Mariotto A, Eisner MP, Lewis DR, Chen HS, Feuer EJ, Cronin KA (eds). SEER Cancer Statistics Review, 1975-2009 (Vintage 2009 Populations), National Cancer Institute. Bethesda, MD, http://seer.cancer.gov/csr/1975_2009_pops09/, based on November 2011 SEER data submission, posted to the SEER web site, 2012
- 2.Niederhuber JE, Brennan MF, Menck HR. The National Cancer Data Base report on pancreatic cancer. Cancer. 1995;76(9):1671–7. doi: 10.1002/1097-0142(19951101)76:9<1671::aid-cncr2820760926>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 3.Winter JM, Cameron JL, Campbell KA, et al. 1423 pancreaticoduodenectomies for pancreatic cancer: A single-institution experience. J Gastrointest Surg. 2006;10(9):1199–210. doi: 10.1016/j.gassur.2006.08.018. discussion 1210–1. [DOI] [PubMed] [Google Scholar]
- 4.Bilimoria KY, Bentrem DJ, Ko CY, et al. Validation of the 6th edition AJCC Pancreatic Cancer Staging System: report from the National Cancer Database. Cancer. 2007;110(4):738–44. doi: 10.1002/cncr.22852. [DOI] [PubMed] [Google Scholar]
- 5.Artinyan A, Hellan M, Mojica-Manosa P, et al. Improved survival with adjuvant external-beam radiation therapy in lymph node-negative pancreatic cancer: a United States population-based assessment. Cancer. 2008;112(1):34–42. doi: 10.1002/cncr.23134. [DOI] [PubMed] [Google Scholar]
- 6.Fatima J, Schnelldorfer T, Barton J, et al. Pancreatoduodenectomy for ductal adenocarcinoma: implications of positive margin on survival. Arch Surg. 2010;145(2):167–72. doi: 10.1001/archsurg.2009.282. [DOI] [PubMed] [Google Scholar]
- 7.Kazanjian KK, Hines OJ, Duffy JP, et al. Improved survival following pancreaticoduodenectomy to treat adenocarcinoma of the pancreas: the influence of operative blood loss. Arch Surg. 2008;143(12):1166–71. doi: 10.1001/archsurg.143.12.1166. [DOI] [PubMed] [Google Scholar]
- 8.Miller RC, Iott MJ, Corsini MM. Review of adjuvant radiochemotherapy for resected pancreatic cancer and results from Mayo Clinic for the 5th JUCTS symposium. Int J Radiat Oncol Biol Phys. 2009;75(2):364–8. doi: 10.1016/j.ijrobp.2008.11.069. [DOI] [PubMed] [Google Scholar]
- 9.Tsai S, Choti MA, Assumpcao L, et al. Impact of obesity on perioperative outcomes and survival following pancreaticoduodenectomy for pancreatic cancer: a large single-institution study. J Gastrointest Surg. 2010;14(7):1143–50. doi: 10.1007/s11605-010-1201-3. [DOI] [PubMed] [Google Scholar]
- 10.Yeo CJ, Cameron JL. Prognostic factors in ductal pancreatic cancer. Langenbecks Arch Surg. 1998;383(2):129–33. doi: 10.1007/s004230050104. [DOI] [PubMed] [Google Scholar]
- 11.Talamini G, Falconi M, Bassi C, et al. Incidence of cancer in the course of chronic pancreatitis. American Journal of Gastroenterology. 1999;94(5):1253–60. doi: 10.1111/j.1572-0241.1999.01075.x. [DOI] [PubMed] [Google Scholar]
- 12.Boyle P, Maisonneuve P, Bueno de Mesquita B, et al. Cigarette smoking and pancreas cancer: a case control study of the search programme of the IARC. International Journal of Cancer. 1996;67(1):63–71. doi: 10.1002/(SICI)1097-0215(19960703)67:1<63::AID-IJC12>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka M, Okazaki T, Suzuki H, et al. Association of multi-drug resistance gene polymorphisms with pancreatic cancer outcome. Cancer. 2011;117(4):744–51. doi: 10.1002/cncr.25510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reid-Lombardo KM, Fridley BL, Cunningham JM, et al. Inflammation-Related Gene Variants as Risk Factors for Pancreatic Cancer. Cancer Epidemiol Biomarkers Prev. 2011;20(6):1251–4. doi: 10.1158/1055-9965.EPI-11-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McWilliams RR, Rabe KG, Olswold C, et al. Risk of malignancy in first-degree relatives of patients with pancreatic carcinoma. Cancer. 2005;104(2):388–94. doi: 10.1002/cncr.21166. [DOI] [PubMed] [Google Scholar]
- 16.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 17.SeattleSNPs [database online] Insert City of Publication Here see notes. National Heart, Lung and Blood Institute (NHLBI); 2000. [Google Scholar]
- 18.NIEHS SNPs. Insert City of Publication Here see notes. 2006. NIEHS Environmental Genome Project, University of Washington, Seattle, WA [database online] [Google Scholar]
- 19.Carlson CS, Eberle MA, Rieder MJ, et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74(1):106–20. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graziadei IW, Schwaighofer H, Koch R, et al. Long-term outcome of endoscopic treatment of biliary strictures after liver transplantation. Liver Transpl. 2006;12(5):718–25. doi: 10.1002/lt.20644. [DOI] [PubMed] [Google Scholar]
- 21.Lesina M, Kurkowski MU, Ludes K, et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19(4):456–69. doi: 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 22.Ebrahimi B, Tucker SL, Li D, et al. Cytokines in pancreatic carcinoma: correlation with phenotypic characteristics and prognosis. Cancer. 2004;101(12):2727–36. doi: 10.1002/cncr.20672. [DOI] [PubMed] [Google Scholar]
- 23.Falconer JS, Fearon KC, Plester CE, et al. Cytokines, the acute-phase response, and resting energy expenditure in cachectic patients with pancreatic cancer. Annals of Surgery. 1994;219(4):325–31. doi: 10.1097/00000658-199404000-00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waeber G, Delplanque J, Bonny C, et al. The gene MAPK8IP1, encoding islet-brain-1, is a candidate for type 2 diabetes. Nature Genetics. 2000;24(3):291–5. doi: 10.1038/73523. [DOI] [PubMed] [Google Scholar]
- 25.Bonny C, Nicod P, Waeber G. IB1, a JIP-1-related nuclear protein present in insulin-secreting cells. Journal of Biological Chemistry. 1998;273(4):1843–6. doi: 10.1074/jbc.273.4.1843. [DOI] [PubMed] [Google Scholar]
- 26.Yuan HY, Chiou JJ, Tseng WH, et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic acids research. 2006;34(Web Server issue):W635–41. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grabe N. AliBaba2: context specific identification of transcription factor binding sites. In silico biology. 2002;2(1):S1–15. [PubMed] [Google Scholar]
- 28.Ursing BM, van Enckevort FH, Leunissen JA, et al. EXProt--a database for EXPerimentally verified Protein functions. In silico biology. 2002;2(1):1–4. [PubMed] [Google Scholar]
- 29.Dong X, Javle M, Hess KR, et al. Insulin-like growth factor axis gene polymorphisms and clinical outcomes in pancreatic cancer. Gastroenterology. 2011;139(2):464–73. 473, e1–3. doi: 10.1053/j.gastro.2010.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nair PN, De Armond DT, Adamo ML, et al. Aberrant expression and activation of insulin-like growth factor-1 receptor (IGF-1R) are mediated by an induction of IGF-1R promoter activity and stabilization of IGF-1R mRNA and contributes to growth factor independence and increased survival of the pancreatic cancer cell line MIA PaCa-2. Oncogene. 2001;20(57):8203–14. doi: 10.1038/sj.onc.1205044. [DOI] [PubMed] [Google Scholar]
- 31.Tomizawa M, Shinozaki F, Sugiyama T, et al. Insulin-like growth factor-I receptor in proliferation and motility of pancreatic cancer. World Journal of Gastroenterology. 2010;16(15):1854–8. doi: 10.3748/wjg.v16.i15.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chae YS, Kim JG, Sohn SK, et al. RIPK1 and CASP7 polymorphism as prognostic markers for survival in patients with colorectal cancer after complete resection. Journal of cancer research and clinical oncology. 2011;137(4):705–13. doi: 10.1007/s00432-010-0929-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.