SUMMARY
Guanosine tetraphosphate (ppGpp) is an alarmone that enables bacterial adaptation to their environment. It has been known for years that ppGpp acts directly on RNA polymerase (RNAP) to alter the rate of transcription, but its exact target site is still under debate. Here we report a crystal structure of Escherichia coli RNAP holoenzyme in complex with ppGpp at 4.5 Å resolution. The structure reveals that ppGpp binds at an interface between the shelf and core modules on the outer surface of RNA polymerase, away from the catalytic center and the nucleic acid-binding path. Bound ppGpp connects these two pivotal modules which may restrain the opening of the RNAP cleft. A detailed mechanism of action of ppGpp is proposed, in which ppGpp prevents the closure of the active center that is induced by the binding of NTP, which could slow down nucleotide addition cycles and destabilize the initial transcription complexes.
INTRODUCTION
The appropriate turning on and off of genes at the right time under various conditions plays a crucial role for the survival and proliferation of cellular organisms. Guanosine tetraphosphate (ppGpp) is a bacterial alarmone that enables bacterial cells to swiftly reprogram their gene expression in order to adapt to various stress conditions. ppGpp and its precursor guanosine pentaphosphate (pppGpp), collectively known as ppGpp or magic spot, were found some 40 years ago in Escherichia coli during amino acid starvation in a phenomenon that is called stringent response (Cashel et al., 1996). Subsequent studies demonstrated that ppGpp is induced in other bacteria and plants (Takahashi et al., 2004) and by a variety of other stress conditions, such as limiting phosphate, iron, and fatty acids (Cashel et al., 1996). Although ppGpp is broadly involved in bacterial physiology and has been shown to regulate replication and translation (Srivatsan and Wang, 2008), it is generally believed that regulation by ppGpp mostly involves transcription. ppGpp induces profound alterations in transcription, including the repression of stable RNA synthesis and the induction of stress response factors and genes that are required for amino acid biosynthesis and transport (Cashel et al., 1996). Genomic profiling showed that the stringent response alters the expression of hundreds of genes (Durfee et al., 2008).
Previous studies suggested that ppGpp destabilizes the open complexes (RPo) formed during transcription initiation by the RNA polymerase (RNAP) and DNA promoters (Barker et al., 2001a). It was also shown that all suppressors of the ppGpp deficient phenotype mapped with the rpoB, rpoC and rpoD subunit genes of the RNA polymerase (Cashel et al., 1996). The direct involvement of ppGpp in interactions with the RNA polymerase was supported by covalent cross-linking analyses (Chatterji et al., 1998; Toulokhonov et al., 2001), however, the exact binding site of ppGpp on the RNA polymerase could not be established due to inconsistent conclusions that are inferred from these studies. A later crystallographic study of ppGpp bound to Thermus thermophilus RNAP (ttRNAP) showed ppGpp to be positioned adjacent to the RNAP catalytic center, partially overlapping with the incoming NTP binding site at the bottom of the secondary channel (Artsimovitch et al., 2004). The ppGpp site identified in these co-crystals appeared inconsistent with it being the site responsible for rRNA transcription regulation, since mutation of the homologous E. coli RNAP (ecRNAP) amino acid residues that form this ppGpp binding site did not significantly weaken the ppGpp response (Vrentas et al., 2008). Therefore, the target location for ppGpp action has still been under debate.
Here, we present the crystal structure of the ppGpp complex with the E. coli RNA polymerase holoenzyme. The structure reveals a different location for the ppGpp binding site and one that is consistent with many published biochemical and genetic experiments. This site of ppGpp binding is away from the nucleic acid-binding path, in contrast with that of the traditional transcription regulators, which typically bind the DNA or target on the nucleic acid-binding path of the RNA polymerase, suggesting a different mechanism for regulating the activity of the RNA polymerase.
RESULTS
Structure determination
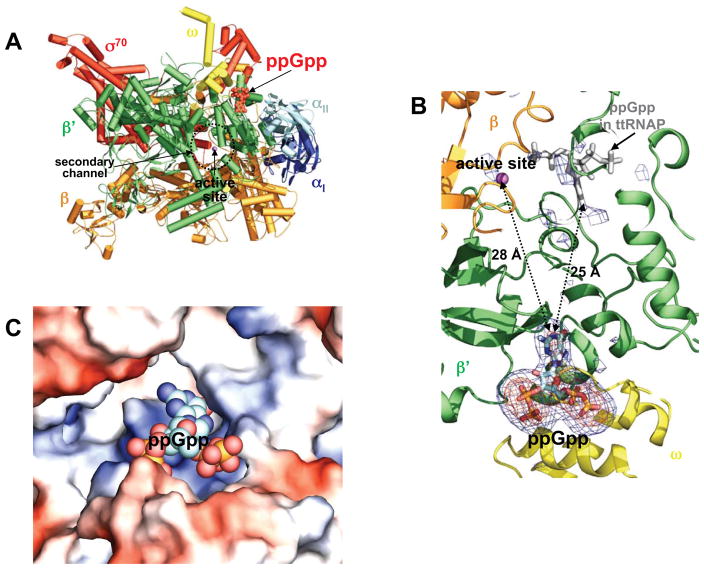
Recently we crystallized and solved the structure of the σ70 holoenzyme of the E. coli RNA polymerase at 3.8 Å resolution (Zuo and Steitz, to be published elsewhere). In the current study, we diffused ppGpp into the ecRNAP holoenzyme crystals and the initial difference Fourier map calculated using the Fobs-Fcalc amplitudes and phases obtained for the holoenzyme showed unambiguous difference electron density for the ppGpp. The final model was refined to a resolution of 4.2 Å (I/σI = 1.2) with an Rwork/Rfree of 0.243/0.314 (Table 1). The structure reveals that ppGpp binds to a single site on the outer surface of the RNA polymerase, more than 25 Å away from the catalytic center deep inside the claw cleft (Figure 1A and 1B). The difference electron density map exhibits no recognizable evidence for ppGpp binding near the catalytic center at the analogous site that was identified previously in the ttRNAP complex with ppGpp (Figure 1B), and no significant conformational changes from the apo form are observed in the RNA polymerase upon ppGpp binding.
TABLE 1.
Data Collection and Refinement Statistics
| Data Collection* | |
| Wavelength (Å) | 1.0078 |
| Space group | P212121 |
| Cell Dimensions (Å) | 187.6, 206.2, 311.1 |
| Resolution (Å) | 50–4.20 (4.27–4.20) |
| Rmerge | 0.149 (>1.0) |
| I/σI | 10.4 (1.2)** |
| Resolution | |
| Completeness (%) | 100 (100) |
| Redundancy | 13.2 (13.3) |
| Number of reflections | |
| Total | 1,157,327 |
| Unique | 87,378 |
|
| |
| Refinement | |
|
| |
| Rwork/Rfree (%) | 24.6/31.4 |
| Number of non-hydrogen atoms | 58,326 |
| Protein | 58,242 |
| Ions | 12 |
| ppGpp | 72 |
| R.m.s. deviation | |
| Bond lengths (Å) | 0.0077 |
| Bond angles (°) | 0.86 |
|
| |
| Ramachandran Plot (%) of Nonglycine and Nonproline Residues | |
|
| |
| Preferred regions | 91.9 |
| Allowed regions | 6.8 |
| Outliers | 1.2 |
Rmerge = (Σ;|I-<I>|)/(Σ;<I>), where I refers to the observed intensity and <I> refers to the average intensity of multiple measurements of the same reflection.
Rwork and Rfree = (Σ;|Fobs-Fcal|)/(Σ;|Fobs|). Rfree, was calculated using 5% of the data not included in the refinement.
Parentheses indicate the statistics from the outer shell.
I/σI = 2.0 at 4.5 Å.
Figure 1. ppGpp bound to E. coli RNA polymerase holoenzyme.
(A) An overview of ppGpp bound to the ecRNAP holoenzyme. The ecRNAP holoenzyme is shown in a tube and arrow cartoon representation with each subunit colored differently: α I – blue; α II – light blue; β - orange; β′ – green; ω - yellow; σ70 – red. A metal ion at the active site is shown as a magenta sphere, and ppGpp is shown in red as space-filling model.
(B) A close-up view of the ppGpp binding site and the active site. σA-weighted Fobs-Fcalc electron density maps contoured at 2.5σ (blue) and 5.0σ (orange) show ppGpp bound at the interface between the ω and β′ subunits. No recognizable electron density corresponding to ppGpp binding is seen near the catalytic center at the analogous site (grey model) identified previously in the ttRNAP complex with ppGpp; the σA-weighted Fobs-Fcalc electron density map is contoured at 2.5σ. The ppGpp molecules are shown in stick models, and a metal ion at the active site is shown as a magenta ball. EcRNAP subunits are shown in cartoon representation and colored as in (A).
(C) Electrostatic surface potential diagram of the ppGpp-binding pocket in ecRNAP with the positive potential in blue and the negative in red. ppGpp is shown as space-filling model with distinct colors for each atom type. The electrostatic surface potential diagram of the ecRNAP holoenzyme was generated using Pymol (Delano, 2002).
ppGpp-binding Site
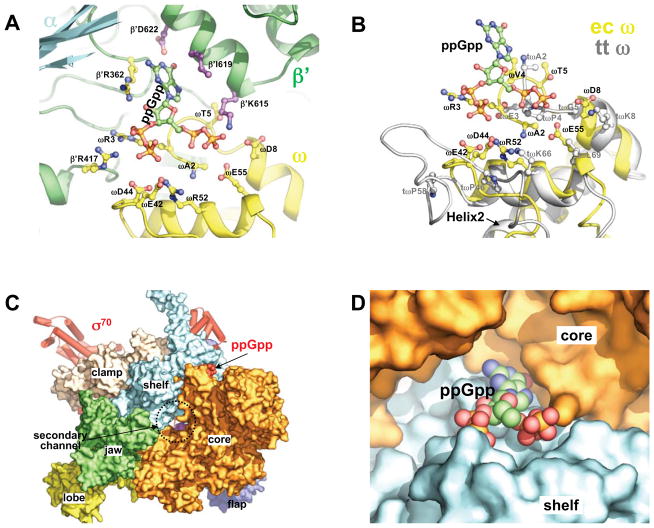
Although ppGpp was shown to regulate transcription at the initiation step (Barker et al., 2001a), its binding to the E. coli RNA polymerase holoenzyme does not involve the σ70 factor. ppGpp binds in a small, positively-charged pocket (Figure 1C) that lies on the edge of the interface between the β′ and ω subunits. This ppGpp binding site is part of a deep depression that is located between the β′ subunit and the N-terminal domain of one of the two α subunits. Both pyrophosphates of ppGpp interact mostly with the ω subunit, whereas the guanosine base interacts with the β′ subunit (Figure 2A). No close contacts were observed between ppGpp and the α subunit. The binding interactions between the ppGpp and both the β′ subunit and the ω subunit of the ecRNAP as inferred from the complex structure are consistent with previous experiments that demonstrated the requirement of the ω subunit for the RNA polymerase response to ppGpp (Vrentas et al., 2005).
Figure 2. ppGpp binding site.
(A) Details of the residues of ecRNAP that are contacting ppGpp. EcRNAP subunits are shown as cartoon representation with each subunit colored differently as in Figure 1A. ppGpp and the side chains of the residues that contact ppGpp are shown as ball-and-stick models.
(B) Structural comparison of the ω subunits of the E. coli (yellow) and Thermus thermophilus (grey) RNAPs at the ppGpp binding site. The side-chains of ecRNAP that contact ppGpp and their spatial counterparts in ttRNAP are shown as ball-and-stick models. The ω subunit structures are aligned on ω residues 17–32 in both sequences, the highly conserved second α-helix (Helix2) that is involved in interactions with the β′ subunit. The structural alignments were performed with the lsqkab program in the CCP4 suite (Winn et al., 2011).
(C) ppGpp bound on a modular interface of the ecRNAP holoenzyme. EcRNAP is shown in the same orientation as in Figure 1A, with the core enzyme shown in surface representation and colored differently for individual modules.
(D) A close-up view of ppGpp bound at the interface between the core and shelf modules. ppGpp is shown as a space-filling model, and the core (orange) and shelf (cyan) modules are shown as surface diagrams.
The difference electron densities for the two pyrophosphates appear to be asymmetric (Figure 1B), but the exact orientation of the bound ppGpp could not be firmly established at the present resolution. In the refined model presented here, the 5′ pyrophosphate interacts with the N-terminus (residue A2) and three hydrophilic residues (T5, D8 and E55) of the ω subunit, whereas the 3′ pyrophosphate is in close contact with four charged residues (R3, E42, D44 and R52) of the ω subunit (Figure 2A). Since several acidic residues are close to the highly negative-charged pyrophosphates, it is likely that the interactions between the pyrophosphates and the acidic residues are mediated by currently unresolved metal ions. Each pyrophosphate also interacts with one basic residue from the β′ subunit (K615 and R417 respectively). The alternative orientation of the ppGpp would switch the positions of the 5′ and 3′ pyrophosphates and their interactions with the RNAP, however, the ppGpp base would remain stacked between side chains of the I619 and R362 residues of the β′ subunit. The edge of the guanosine base lies near the carboxyl group of the D622 residue of the β′ subunit. Point mutations of many of these ppGpp-contacting residues abolish the ppGpp response of the RNA polymerase for rRNA transcription regulation (Ross et al., accompanying manuscript).
Lack of a similar ppGpp-binding site in the ttRNAP?
Although we could not explain the absence of ppGpp binding to ecRNAP at the active site location that was identified in the structure of the ttRNAP complexed with ppGpp, the structural and sequence differences between the E. coli and Thermus thermophilus ω subunits appear to nicely account for the absence of this ppGpp binding site in the ttRNAP. The ω subunit, which is the smallest RNAP core subunit, is conserved from bacteria to humans in both sequence and structure, and the conserved elements are mostly involved in the interactions between the ω and β′ subunits (Minakhin et al., 2001). Nevertheless, the ppGpp binding site in the ecRNAP contains regions that are less-conserved, including the very N-terminal region (residues 2–8) of the ω subunit, which displays a different fold in the ttRNAP. As shown in Figure 2B, this N-terminal part of the ω subunit of ttRNAP appears to be incompatible with the binding of ppGpp to ttRNAP in the way it interacts with the ecRNAP due to potential steric clashes between the 5′-pyrophosphate and a proline side chain.
DISCUSSION
Indirect regulation of transcription by ppGpp
Transcription, the process of RNA synthesis using a DNA template, is frequently the major target for regulating gene expression. Over the years, many features of transcription regulation have been revealed, and it has been apparent that many signals for transcription regulation are embedded in the DNA sequence, especially on the so-called promoter region preceding the transcribing unit. Many transcription factors bind to the promoter or other specific DNA sequences to regulate the rates of transcription. In addition to these traditional DNA-binding transcriptional factors, other trans-acting factors, including some antibiotics, regulate transcription by binding to the RNA polymerase (RNAP). These non-nucleic acid-binding factors typically target on the nucleic acid-binding path, including the entrances and the internal space of the primary, secondary, and RNA-exit channels of the RNA polymerase.
Regulatory signals also appear to be embedded in the promoter sequences of the genes whose transcription is regulated by ppGpp. These promoters, such as those of the rRNA operons, frequently contain a GC-rich discriminator −10 to +1 region and a shortened spacer (−35 to −10 region) and tend to form open complexes that are unstable and short-lived (Barker et al., 2001a; Park et al., 2002; Travers, 1980). Destabilization of these intrinsically short-lived open complexes is believed to be the mechanism by which ppGpp inhibits the RNA synthesis (Barker et al., 2001a). Interestingly, the structure presented here shows that ppGpp does not act directly on the promoter DNA, nor does it act near the nucleic acid-binding path on the RNA polymerase.
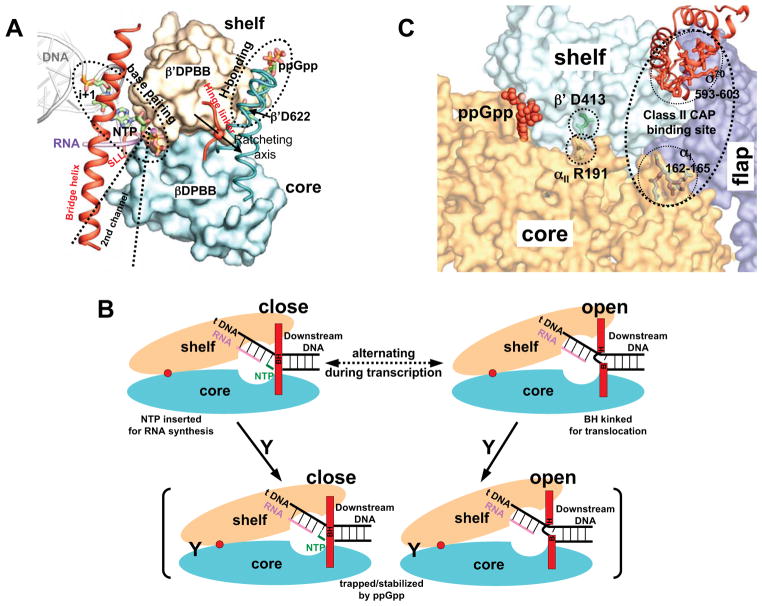
To understand the indirect effect of ppGpp on the open complex stability, we evaluated the ppGpp binding site in the context of the modular architecture of the RNA polymerase. The crab claw assembly of the cellular RNA polymerases consists of rigid modules (Cramer et al., 2001; Murakami et al., 2002; Tagami et al., 2010; Zuo and Steitz, unpublished results) that are mobile relative to each other. The catalytic center sits at the interface of the core and shelf modules deep inside the clamp cleft. Other modules, such as the β′ clamp, β1 protrusion and β2 lobe, are attached to the core and shelf modules to form the claw pincers. The core and shelf modules are connected via three peptidyl linkages and are the central part of the claw that shapes the primary, secondary and RNA-exit channels. The core and shelf modules were shown to make a ratcheting movement relative to each other (Tagami et al., 2010). This ratcheting movement affects the openness of the active center chamber of the primary channel, in which the template strand or template/RNA hybrid is held in a transcription bubble, as well as the gap between the clamp pincers.
ppGpp binding at a modular interface of RNA polymerase
The binding site of ppGpp lies at the interface between the core and shelf modules (Figure 2C and 2D). In contrast to the interface between the core and shelf that forms the catalytic center inside the cleft, the ppGpp-binding site is on the other side of the protein structure on the outer surface of the RNA polymerase (Figure 1). ppGpp interacts extensively with the shelf module as most of the residues that contact the pyrophosphates of ppGpp are from the ω subunit, which is an integral part of the shelf module (Figure 2). On the other hand, the guanosine base, which is sandwiched between the side chains of a shelf module residue (β′R362) and a core module residue (β′I619), appears likely to form hydrogen–bonding interactions with the carboxyl side chain of the core module residue β′D622. Hydrogen bonds between the ppGpp base and the carboxyl group of an aspartate or glutamate residue have been observed previously (Buglino et al., 2002; Rymer et al., 2012), and it appears that such interactions are a common theme in protein complexes with ppGpp.
In the context of a leverage system with hinged ratcheting between the core and shelf modules of the RNA polymerase (Figure 3A), the hydrogen bonding between ppGpp and D622 of the β′ subunit would exert an opposing force to the base pairing between the NTP and template DNA. While binding of the correct NTP induces cleft closure, ppGpp might act by stabilizing an open cleft conformation through the hydrogen bonding with β′D622. Consistent with this possibility, elimination of the hydrogen bond between ppGpp and the β′D622 by replacing β′D622 with an alanine residue abolishes the ppGpp effect on rRNA transcription in vitro (Ross et al., accompanying manuscript).
Figure 3. Mechanism of ppGpp action.
(A) ppGpp binding in the contexts of RNAP modular movement. The core and shelf modules pack against each other on two beta barrel domains (βDPBB and β′DPBB, shown as surface diagrams), and make a ratcheting movement relative to each other on a ratcheting axis (Tagami et al., 2010). Base pairing between the incoming NTP and the template DNA at the i+1 site and the putative ppGpp-β′D622 hydrogen bonding form a leverage system hinged on the core-shelf ratcheting axis. The three linkers (bridge helix, hinge linker and a switch-like linker, SLL), which restrain the core-shelf ratcheting, are shown in red as helix and loops. The secondary channel that provides access for NTP binding and the binding site of some regulatory proteins are marked for reference.
(B) Schematic representation of the proposed mechanism of ppGpp action. Dynamic cycles of ratcheting of the core and shelf modules between the open and closed states of the active site chamber that alternates between the nucleotide addition reaction and RNAP translocation. ppGpp binding on the outer surface of the cleft traps or stabilizes RNAP in certain state and slows down the ratcheting dynamics as well as RNA synthesis. ppGpp is represented by a letter Y, and the ratcheting axis is represented by a red dot. The bridge helix is BH and is separated into B and H for the kinked conformation.
(C) A proposed general mechanism of transcription regulation that involves the interface between the core and shelf modules. (i) ppGpp (red, space-filling model) binds at the core-shelf interface to exert stringent response (this study); (ii) substitution of αII R191 by an alanine, which breaks a salt bridge between the side chains of a core module residue (αII R191) and a shelf module residue (β′ D413), thereby stabilizing transcription open complexes (Ross et al., accompanying manuscript); (iii) Class II CAP actions involve the interface between the core and shelf modules. In addition to interacting with the CTD of the α subunit for RNAP recruiting, CAP interacts with RNAP at two other places. It interacts with residues 162–165 of the NTD of the αI subunit and residues 593–603 of the CTD of σ70, when it binds to a class II CAP-dependent promoters (Niu et al.,1996; Lonetto et al., 1998). These two sequence segments span a region that lies at the interface between the core, shelf and flap modules. The core, shelf and flap modules of the region of interest are shown in distinct colors as surface diagrams. The σ70 factor is shown in red as ribbon diagrams. Side chains of αII R191, β′ D413, and the two CAP-interacting sequence segments are shown as sticks.
A model for the mechanism of ppGpp action
On the basis of the structural observations presented here and the known biochemical and genetic features of ppGpp action, we propose a model for how the binding of ppGpp affects RNA polymerase activity (Figure 3B). The ratcheting movement between the core and shelf modules is an intrinsic feature of the RNA polymerase dynamics, which drives the cleft to alternate between the closed and open forms during catalysis and translocation. ppGpp binding tightens the clamp from the outside through its interactions with both the shelf and core modules, and would restrain the dynamic core-shelf ratcheting movement by stabilizing the RNA polymerase in a particular state. Consequently, this disruption of the ratcheting dynamics would slow down the rate of the nucleotide addition reaction. In addition, since the mobile modules are connected to the core and shelf modules, the binding of ppGpp could affect the cleft opening between the claw pincers, which controls loading of the template strand and the downstream DNA and plays a significant role in the stability of the transcription bubble.
For the open complexes which are intrinsically short-lived, the initial transcription bubbles are unstable, and the flexible template strand might easily cross some gate between the claw pincers to reverse the process of RPo formation. These complexes depend on rapid RNA synthesis to stabilize the bubbles for effective transcription initiation, since the formation of the DNA:RNA hybrid would significantly reduce the template strand flexibility and thereby prevent the escape of the template strand and bubble collapse. Therefore, if the effect of ppGpp binding is slowing down RNA synthesis as proposed above, it could potentially facilitate DNA escape in these complexes if the time required for nucleotide addition becomes longer than the life of the open complex, which would lead to an inhibitory effect on the initial RNA synthesis. If the open complexes are stable enough, ppGpp binding might have a negligible effect on the transcription initiation. It is conceivable that for some slow forming open complexes, ppGpp binding might even have an inductive effect on the gene expression as it could potentially widen the gap between the claw pincers to promote DNA loading and open complex formation.
Understanding stringent response mutants
ppGpp destabilizes the open complexes at all promoters (Barker et al., 2001a). This universal effect is the result of its indirect action and predetermines a weak effect so that RNA synthesis is specifically inhibited from very short-lived open complexes, which is consistent with the counter force caused by a single hydrogen bond as shown in Figure 3A. This weak effect of ppGpp could be partially compensated by many point mutations on the RNA polymerase. Over the years, many stringent mutations that produce the stringent response in the ppGpp-deficient cells were mapped to the RNA polymerase subunits (Cashel et al., 1996). These mutations mostly lie along the nucleic acid binding path on the inner surface of the primary channel (Trinh et al., 2006). Some of these mutations, such as the R1148C mutation in the β′ subunit (Barker et al., 2001b), weaken the interactions of the RNAP with the nucleic acid, while others, such as the P153L mutation in the β subunit (Trautinger and Lloyd, 2002), are expected to increase the flexibility of the pincers.
By contrast, a mutant RNAP with substitutions in the α subunit (αE188A, R191A) displays small defects in its ppGpp response (Ross et al., accompanying manuscript). Consistent with the module-bridging effect of ppGpp, these mutations, which are adjacent to the ppGpp binding site and disrupt a potential salt bridge between side chains of a core module residue (αII R191) and a shelf module residue (β′ D413) on the outer surface of ecRNAP (Figure 3C), increase the stability of the mutant open complex, which appears to be destabilized by ppGpp to the same extent as that of the wild-type RNAP (Ross et al., accompanying manuscript).
Synergistic effects of ppGpp and DksA
While ppGpp induces dramatic stringent regulation of many genes in vivo, only small effects on transcription were observed in highly purified in vitro systems. This apparent discrepancy is due to the protein factor DksA, which is required for the control of rRNA transcription by ppGpp in vivo and greatly amplifies inhibition of rRNA promoters by ppGpp in vitro (Paul et al., 2004). Similar to the GreA and GreB proteins that bind in the secondary channel, DksA contains a long helical hairpin and is expected to bind in the RNAP secondary channel in a similar manner (Perederina et al., 2004). The structure of ttRNAP bound to Gfh1, a GreA homolog in Thermus, shows that the binding of these proteins to the secondary channel could ratchet open the core-shelf cleft and the pincer gap (Tagami et al., 2010). Although we did not observe significant conformational changes upon ppGpp binding, possibly due to the constraints imposed by the crystal packing of the RNAP molecules, the synergistic action of ppGpp with a secondary channel binding protein strongly suggests that ppGpp binding promotes RNAP cleft opening.
A general allosteric regulation of transcription?
The structure presented here demonstrates that transcription regulation does not have to directly interfere with the interactions of RNAP with nucleic acids. Due to the modular structure of RNA polymerases, transcription regulation could be accomplished by ligands binding at a location on the surface of the RNA polymerases that is remote to the active site. This kind of allosteric regulation from remote binding sites that appears to be exhibited by ppGpp is presumably a general mechanism for the action of many transcription factors. For example, at class II catabolite activator protein (CAP) dependent promoters, in addition to its association with the C-terminal domains of the α subunits of RNAP for RNAP recruitment, CAP promotes transcription through interactions with two other regions on the RNAP, residues 162–165 of the N-terminal domain of one α subunit (Niu et al., 1996) and residues 593–603 of the σ70 factor (Lonetto et al., 1998). Although the detailed mechanism of this allosteric activation of polymerase activity by CAP is not understood, it may be that the binding of CAP at the interface between the core and shelf modules (Figure 3C) facilitates the opening of the cleft and thereby promotes the formation of the open complex in a way that is similar to the action of ppGpp.
EXPERIMENTAL PROCEDURES
Protein preparation and crystallization
The E. coli RNA polymerase (RNAP) core enzyme and N-terminal his-tagged s70 factor were over-expressed and purified separately, and the RNAP holoenzyme was prepared by mixing the purified core enzyme with s70 factor in excess followed by size exclusion chromatography, as to be published elsewhere (Zuo and Steitz, unpublished results).
Crystals of E. coli RNAP holyenzyme were grown by vapor diffusion (Zuo and Steitz, unpublished results). After crystals grew to full size at room temperature in about a week, they were transferred to a soaking buffer containing the crystallization mother liquor as well as 100uM ppGpp (TriLink BioTech Inc.). After incubation at room temperature for 30 minutes, the crystals were flash-frozen by plunging them into liquid nitrogen using pump oil Fomblin Y HVAC 25/9 (Sigma-Aldrich) as the cryoprotectant.
Data collection and structural refinement
X-ray diffraction data were collected at the beamline X25 at Brookhaven National Laboratory (Upton, NY). All data collection was carried out at 100 K. Diffraction data were integrated and scaled with HKL2000 (Otwinowski and Minor, 1997). The crystal belongs to the orthorhombic space group P212121 and has two RNAP holoenzymes per asymmetric unit.
The structure was solved by molecular replacement with PHASER (McCoy et al., 2007) using the structure of the E. coli RNAP holoenzyme (Zuo and Steitz, unpublished results) as a starting model. Initially, rigid body refinement was carried out using Refmac5 (Murshudov et al., 2011) in the CCP4 suite (Winn et al., 2011) with the RNAP holoenzyme split into multiple domains. The difference electron density corresponding to one ppGpp per holoenzyme became clearly visible in the (Fobs-Fcalc) difference electron density map after rigid body refinement. After examining several ppGpp models from the protein data bank, the ppGpp model from 1LNZ (Buglino et al., 2002) was used as the starting model as it fits the difference density perfectly with minimal adjustments of the phosphate linkages. After manually fitting ppGpp into the difference density in Coot (Emsley and Cowtan, 2004), ten cycles of TLS and restrained refinement were performed to generate the final structural model. Data collection and structural refinement statistics are summarized in Table 1.
HIGHLIGHTS.
A co-crystal structure of the E. coli RNA polymerase in complex with ppGpp
No ppGpp binding seen in the vicinity of the RNAP active site
ppGpp binds at an interface between modules on the outer surface of the RNAP
An allosteric mechanism is proposed for ppGpp regulation of RNAP activity
Acknowledgments
We thank Wilma Ross and Richard Gourse for helpful discussions. We thank the staffs of the National Synchrotron Light Source beamline X25 for help during data collection and the Center for Structural Biology facility at Yale University for computational support. This work was supported by NIH grant GM057510 to T.A.S.
Footnotes
ACCESSION NUMBER
The coordinates and structural factor files have been deposited in the Protein Data Bank with accession code 4JKR.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Hosaka T, Ochi K, Yokoyama S, Vassylyev DG. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117:299–310. doi: 10.1016/s0092-8674(04)00401-5. [DOI] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001a;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. II Models for positive control based on properties of RNAP mutants and competition for RNAP. J Mol Biol. 2001b;305:689–702. doi: 10.1006/jmbi.2000.4328. [DOI] [PubMed] [Google Scholar]
- Buglino J, Shen V, Hakimian P, Lima CD. Structural and biochemical analysis of the Obg GTP binding protein. Structure. 2002;10:1581–1592. doi: 10.1016/s0969-2126(02)00882-1. [DOI] [PubMed] [Google Scholar]
- Cashel M, Gentry DR, Hernandez VH, Vinella D. In: The stringent response Escherichia coli and Salmonella. Neidhardt FC, editor. ASM Press; Washington, DC: 1996. pp. 1458–1496. [Google Scholar]
- Chatterji D, Fujita N, Ishihama A. The mediator for stringent control, ppGpp, binds to the β-subunit of Escherichia coli RNA polymerase. Genes Cells. 1998;3:279–287. doi: 10.1046/j.1365-2443.1998.00190.x. [DOI] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 Ångstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS. ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev. 2010;74:171–199. doi: 10.1128/MMBR.00046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delano WL. The PYMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. http://www.pymol.org. [Google Scholar]
- Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190:1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Jain V, Kumar M, Chatterji D. ppGpp: stringent response and survival. J Microbiol. 2006;44:1–10. [PubMed] [Google Scholar]
- Lonetto MA, Rhodius V, Lamberg K, Kiley P, Busby S, Gross C. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase σ70 subunit. J Mol Biol. 1998;284:1353–1365. doi: 10.1006/jmbi.1998.2268. [DOI] [PubMed] [Google Scholar]
- Magnusson LU, Gummesson B, Joksimović P, Farewell A, Nyström T. Identical, independent, and opposing roles of ppGpp and DksA in Escherichia coli. J Bacteriol. 2007;189:5193–5202. doi: 10.1128/JB.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minakhin L, Bhagat S, Brunning A, Campbell EA, Darst SA, Ebright RH, Severinov K. Bacterial RNA polymerase subunit ω and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc Natl Acad Sci USA. 2001;98:892–897. doi: 10.1073/pnas.98.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. Refmac5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Kim Y, Tau G, Heyduk T, Ebright RH. Transcription activation at class II CAP-dependent promoters: two interactions between CAP and RNA polymerase. Cell. 1996;87:1123–1134. doi: 10.1016/s0092-8674(00)81806-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Park JW, Jung Y, Lee SJ, Jin DJ, Lee Y. Alteration of stringent response of the Escherichia coli rnpB promoter by mutations in the −35 region. Biochem Biophys Res Commun. 2002;290:1183–1187. doi: 10.1006/bbrc.2001.6331. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell. 2004;118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Perederina A, Svetlov V, Vassylyeva MN, Tahirov TH, Yokoyama S, Artsimovitch I, Vassylyev DG. Regulation through the secondary channel—structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- Potrykus K, Murphy H, Philippe N, Cashel M. ppGpp is the major source of growth rate control in E. coli. Environ Microbiol. 2011;13:563–575. doi: 10.1111/j.1462-2920.2010.02357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. (Accompanying manuscript) [Google Scholar]
- Rymer RU, Solorio FA, Tehranchi AK, Chu C, Corn JE, Keck JL, Wang JD, Berger JM. Binding mechanism of metal-NTP substrates and stringent-response alarmones to bacterial DnaG-type primases. Structure. 2012;20:1478–1489. doi: 10.1016/j.str.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivatsan A, Wang JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol. 2008;11:100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Tagami S, Sekine S, Kumarevel T, Hino N, Murayama Y, Kamegamori S, Yamamoto M, Sakamoto K, Yokoyama S. Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein. Nature. 2010;468:978–982. doi: 10.1038/nature09573. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Kasai K, Ochi K. Identification of the bacterial alarmone gunanosine 5′-diphosphate 3′-diphosphate (ppgpp) in plants. Proc Natl Acad Sci USA. 2004;101:4320–4324. doi: 10.1073/pnas.0308555101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulokhonov II, Shulgina I, Hernandez VJ. Binding of the transcription effector ppGpp to Escherichia coli RNA polymerase is allosteric, modular and occurs near the N terminus of the β′-subunit. J Biol Chem. 2001;276:1220–1225. doi: 10.1074/jbc.M007184200. [DOI] [PubMed] [Google Scholar]
- Trautinger BW, Lloyd RG. Modulation of DNA repair by mutations flanking the DNA channel through RNA polymerase. EMBO J. 2002;21:6944–6953. doi: 10.1093/emboj/cdf654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers AA. Promoter sequence for stringent control of bacterial ribonucleic acid synthesis. J Bacteriol. 1980;141:973–976. doi: 10.1128/jb.141.2.973-976.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinh V, Langelier MF, Archambault J, Coulombe B. Structural perspective on mutations affecting the function of multisubunit RNA polymerases. Microbiol Mol Biol Rev. 2006;70:12–36. doi: 10.1128/MMBR.70.1.12-36.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrentas CE, Gaal T, Berkmen MB, Rutherford ST, Haugen SP, Vassylyev DG, Ross W, Gourse RL. Still looking for the magic spot: the crystallographically defined binding site for ppGpp on RNA polymerase is unlikely to be responsible for rRNA transcription regulation. J Mol Biol. 2008;377:551–564. doi: 10.1016/j.jmb.2008.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrentas CE, Gaal T, Ross W, Ebright RH, Gourse RL. Response of RNA polymerase to ppGpp: requirement for the ω subunit and relief of this requirement by DksA. Genes Dev. 2005;19:2378–2387. doi: 10.1101/gad.1340305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3′,5′-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]