Abstract
SNARE proteins drive membrane fusion by assembling into a four-helix bundle in a zippering process. Here we used optical tweezers to observe in real time a long-sought SNARE assembly intermediate in which only the membrane-distal N-terminal half of the bundle is assembled. Our finding supports the zippering hypothesis, but suggests that zippering proceeds through three sequential binary switches, not continuously, in the N- and C-terminal halves of the bundle and the linker domain. The half-zippered intermediate was stabilized by externally applied force which mimicked the repulsion between apposed membranes being forced to fuse. This intermediate then rapidly and forcefully zippered, delivering free energy of 36 kBT to mediate fusion.
SNARE proteins mediate membrane fusion in the cell, and in particular the fusion of vesicles stored at nerve endings to release neurotransmitters for synaptic transmission (1, 2). The neuronal SNAREs consist of VAMP2 (synaptobrevin) on the vesicle membrane (v-SNARE) and syntaxin 1 and SNAP-25 on the plasma membrane forming a binary complex (t-SNARE) (3, 4). Together these SNAREs drive membrane fusion by joining into a parallel four-helix bundle (4), which is envisioned to zipper progressively towards the membranes (5), providing force that overcomes an estimated energy barrier of over 40 kBT (6). Considerable indirect evidence has accumulated in favor of the zippering hypothesis (7–12). Direct observation of the assembly intermediates and accurate characterization of the zippering energy and kinetics have been lacking.
We developed a single-molecule manipulation assay to investigate SNARE assembly based on high-resolution dual-trap optical tweezers (Fig. 1A). We cross-linked the N-termini of syntaxin and VAMP2 by a disulfide bridge, and attached syntaxin by its C-terminus to one bead and VAMP2 to another through a DNA handle (13). The experiment was started with a single pre-assembled SNARE complex containing truncated syntaxin (187–265) and VAMP2 (25–92) and full-length SNAP-25 (4, 14) to avoid mis-assembled SNARE byproducts (10, 15).
Fig. 1.
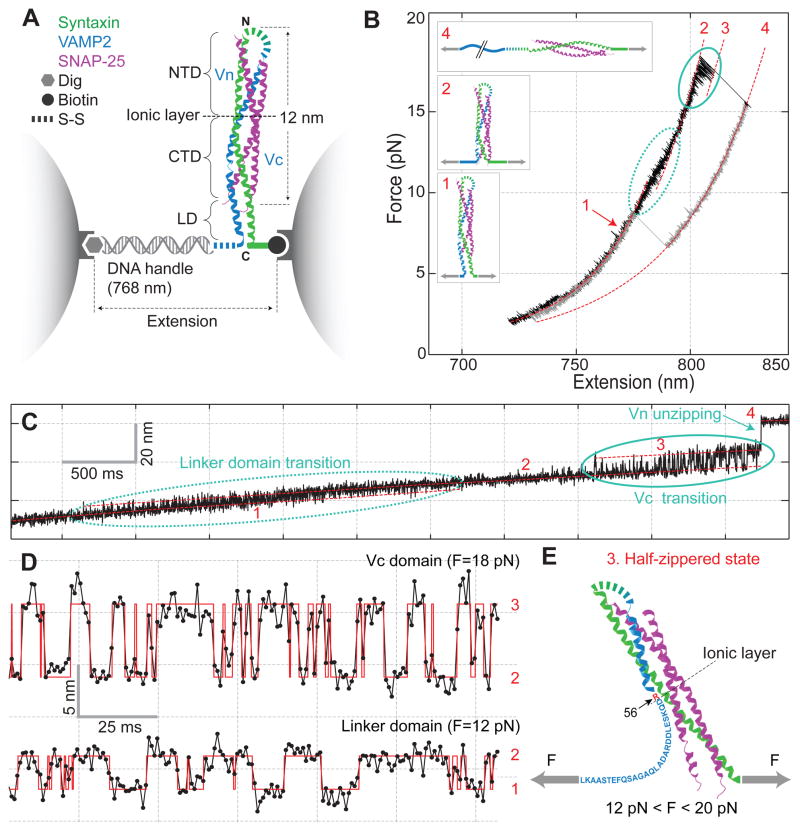
Dynamic disassembly and reassembly of the single SNARE complex. (A) Experimental setup. The SNARE complex contains the N-terminal (NTD) and C-terminal (CTD) SNARE domains, with the corresponding VAMP2 regions designed as Vn and Vc, respectively, separated by the ionic layer and the linker domain (LD). (B) Force-extension curve (FEC) of the SNARE-DNA conjugate. The FEC corresponds to the first of 5 cycles of pull (black) and relaxation (gray) shown in fig. S2. Different segments of the FEC can be fit by the worm-like chain model (red dashed lines), revealing the structures of SNARE assembly states (inset, same red numbering throughout the figures). The LD and CTD transitions are marked by dashed and solid ovals, respectively. (C) Time-dependent extension corresponding to the pulling phase from 8.6 pN to 17.5 pN (fig. S3). (D) Extension transitions of LD (bottom panel) and CTD (top panel) with their idealized transitions determined by the HMM analysis (red traces). The histogram distributions of extension are shown in fig. S5. (E) Structural model of the force-dependent half-zippered state with Vc unzipped to the ionic layer (red).
The protein-DNA conjugate extended with the increasing pulling force in a nonlinear manner predicated by the worm-like chain model (Fig. 1B and fig. S2). However, the monotonic force and extension curves were interrupted by abrupt changes caused by SNARE disassembly or reassembly (Fig. 1C). Fast reversible transitions appeared in two force regions, the first in 8–13 pN with ~3 nm average extension change (Fig. 1D, fig. S3) and the second in 14–19 pN with ~7 nm extension change (Fig. 1D, fig. S4). Both transitions occurred between two states (fig. S5 and Table S1), manifesting two distinct binary switches in SNAREs. When the linker domain (LD) of VAMP2 was deleted, the first transition disappeared whereas the second transition remained (fig. S6). Thus, the first transition is caused by reversible folding and unfolding of the LD alone. The average size of the extension change suggests that a total of 22 (±3, s.d.) amino acids (a.a.) or 10 a.a. in VAMP2 (83–92) participated in the transition (fig. S7A). This observation is consistent with a fully zippered LD in a coiled-coil conformation in solution (Fig. 1B, state 1) as seen in the crystal structure of the SNARE complex (4, 16). Further deletion into the C-terminal SNARE domain of VAMP2 (Vc) abolished the second transition (fig. S8), which suggests that this VAMP2 region is involved in the transition.
The additional ~7 nm extension increase from the LD unfolded state (Fig. 1B, state 2) leads to a partially zippered SNARE state (state 3, Fig. 1E). To derive the structure, energy, and kinetics associated with this state, we measured the real-time transition involving Vc at different mean forces (Fig. 1D). The fast two-state transition was confirmed by hidden-Markov modeling (17) and the histogram distribution of extension (figs. S5B & S9). Based on the measured extension change and an asymmetrical transition model (fig. S10) (18), we found that 26 (±3) a.a. in VAMP2 (57–82) were unzipped in the partial SNARE complex (Fig. 1E). This places the interface of the unzipped Vc and the zippered N-terminal VAMP2 (Vn) at residue 56 (±5, s.d.; ±1, s.e.m.), or at the central ionic layer of the bundle. This ionic layer is one of the most evolutionally conserved features of all SNAREs (19), yet with unclear functions. Thus, our result suggests a possible role of the ionic layer in stabilizing the half-zippered neuronal SNARE structure crucial for regulation of membrane fusion (7, 8, 12, 18, 20).
The half unfolding probability of Vc (Fig. 2A) determines an average equilibrium force (feq) of 17 (±2 s.d., N=76) pN, which can be defined as the maximum force output of Vc zippering averaged over the accompanying extension change. This force is the highest equilibrium force reported for any coiled-coil proteins (17), including the designed strongest known coiled coil designated as pIL with an equilibrium force of 12.4 pN (21). The unfolding probability could be extrapolated to zero force (14, 17, 22) to reveal the Vc zippering free energy of 28 (±3) kBT, higher than the folding energy of pIL 24 (±1) kBT despite pIL’s greater length (33 a.a.) (21).
Fig. 2.
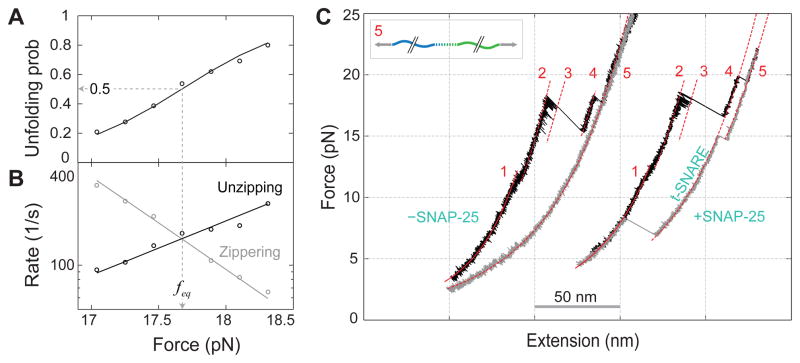
SNAP-25-dependent SNARE assembly. (A) Force-dependent unzipping probability of CTD measured on a single SNARE complex. (B) Corresponding CTD transition rates. Theoretical predications are shown in lines. (C) FECs measured in the absence (− SNAP-25) and presence (+ SNAP-25) of SNAP-25 in solution.
The average Vc zippering rate at the maximum force output (~160 s−1) (Fig. 2B) is also greater than the equilibrium rate of pIL (~10 s−1) (21). Yet the rate is much less than the estimated SNARE zippering rate of 4×104 s−1 commensurate with the time required for synaptic vesicle fusion (~60 μs) (23). However, the Vc zippering rate increases exponentially as the opposing force drops (Fig. 2B) (17), reaching the required rate for synaptic fusion at a predicted force of 14 pN. This means that a single SNARE complex in principle can generate a high average force of 14 pN during membrane fusion. When further extrapolated to zero force (14), the Vc zippering becomes down-hill and barrier-less, with a rate estimated to be ~1×106 s−1 (21) corresponding to the rate of the coil-to-helix transition and diffusion of the Vc polypeptide (24). This fast zippering rate is partially attributed to the largely ordered t-SNARE complex that serves as a template for Vc zippering (figs. S10 & S11) (18). In conclusion, the extraordinarily high force output, energy, and rate qualify Vc zippering as the major power stroke for the fast synaptic fusion (11).
Similarly, we measured a folding energy of 8 (±2) kBT for LD (fig. S7B–C) with a maximum force output close to that of pIL (~12 pN). An energy barrier of 5 (±2) kBT was derived, corresponding to a zipping rate of 7×103 (9×102–5×104) s−1 at zero force. LD also strongly and rapidly zippers, which may transduce and augment the Vc zippering energy to drive membrane fusion. LD’s zippering rate and energy may be increased by trans-membrane domains (25) and proteins such as synaptotagmin and Munc18-1 (3, 26).
The Vc transition generally ended with an irreversible extension jump of 15 (±1) nm (Fig. 1B–C, from states 3 to 4), corresponding to complete Vn unzipping (fig. S12). When immediately relaxed, the SNARE complex first followed a different force-extension curve (FEC) to reach lower force (Fig. 1B). As the force was lowered to 3–10 pN, the complex fully and cooperatively reassembled.
The dynamic disassembly and reassembly of a single SNARE complex could be repeated for many cycles of pull and relaxation (fig. S2). However, when the t-SNARE complex was pulled to a higher force in the range of 15–28 pN, one additional irreversible extension jump was observed (Fig. 2C, from states 4 to 5, fig. S11). No more transitions were seen when further pulling to even higher forces (>30 pN), indicating complete unfolding of the SNARE complex (state 5). Thus, this jump may correspond to unfolding of the remaining t-SNARE complex. When relaxed to low forces (<3 pN), a majority (~85%) of syntaxin and VAMP2 conjugates could not reassemble (Fig. 2C and fig. S2), suggesting dissociation of the SNAP-25 molecule.
To further test the dependence of SNARE assembly on SNAP-25, we repeated the above experiment by adding SNAP-25 in the solution. Single SNARE complexes were fully disassembled by being pulled to high forces and then relaxed to detect SNARE reassembly. At 0.2 μM SNAP-25, the ternary SNARE complex could reassemble at a low force (3–10 pN) with a probability of ~0.8 for each round of relaxation (Fig. 2C). This reassembly was always followed by formation of the t-SNARE complex in a higher force range (>12 pN). The SNAP-25 molecule in solution could bind to the syntaxin-VAMP2 conjugate and initiate de novo SNARE assembly. Comparing this to the experiment in the absence of SNAP-25 suggests that the binary t-SNARE is required for SNARE zippering (27). In contrast, the affinity between syntaxin and VAMP2 in the absence of SNAP-25 is minimal and could not be detected in our experiments (Fig. 2C) (10, 28).
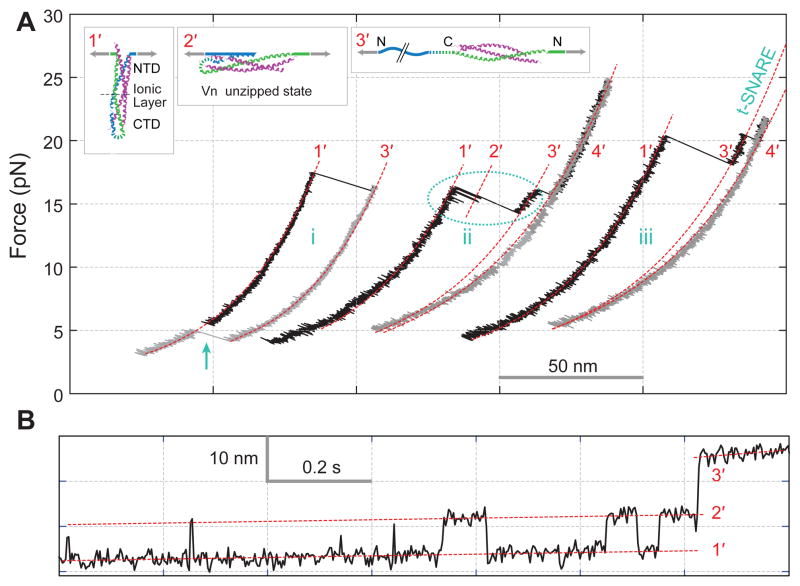
The assembly and disassembly of NTD occurred with a large force hysteresis, indicating a large energy barrier for NTD transitions and preventing us from measuring its association energy. To gain insight into NTD’s assembly mechanism, we pulled a single SNARE complex from the N-termini of syntaxin and VAMP2 (Fig. 3A) (14). The SNARE complex unzipped into t- and v-SNAREs (from states 1′ to 3′) in two parallel pathways. About two thirds of the 53 complexes tested showed one-step unzipping (FECs i & iii), whereas other complexes exhibited two-step unzipping via a transient intermediate state (state 2′). Reversible transitions were often observed between this state and the fully zippered state 1′ (Fig. 3B), and lasted long enough for the equilibrium transition force and the average extension change to be determined. This partially zippered SNARE complex had Vn unzipped to residue 57 (±3, s.e.m.), or approximately the ionic layer. The Vn domain unzipped at a higher average equilibrium force of 18.5 (±0.4, s.e.m.) pN than Vc from C-terminal pull at 16.7 (±0.2, s.e.m.) pN. In addition, the average extension change of Vn (8.3±0.2, s.e.m., nm) was greater than that of Vc (7.1 ±0.1, s.e.m., nm). These measurements allow an estimation of the folding free energy of 35 (±4) kBT for NTD assembly in the presence of the pre-assembled CTD (14, 17). However, Vn assembled much more slowly than Vc (Fig. 3B).
Fig. 3.
Disassembly and reassembly of the SNARE complex under N-terminal pulling force. (A) FECs and their segmental fit (red dashed line) showing different assembly states (inset) including the completely unfolded state 4′. The full SNARE reassembly (cyan arrow) is t-SNARE dependent. (B) Extension-time trace corresponding to the region marked in the dashed oval in A.
The t-SNARE complex again remained intact after VAMP2 was pulled out (Fig. 3, state 3′). This complex has similar structure to those obtained from the C-terminal pull (Table S1). On average, the t-SNARE complex contained 61 (±4) helical amino acids in the syntaxin molecule, suggesting an approximately fully assembled three-helix bundle for the t-SNARE (figs. S10 & S11). Upon relaxation the ternary complex could reassemble at an average force of 3.2 (±0.2, s.e.m.) pN (Fig. 3, FEC i), compared to 5.0 (±0.2, s.e.m.) pN from the C-terminal pull. The more force-sensitive assembly of the SNARE complex from the N-terminal pull indicates that initiation of the assembly occurs in NTD (10, 17, 29).
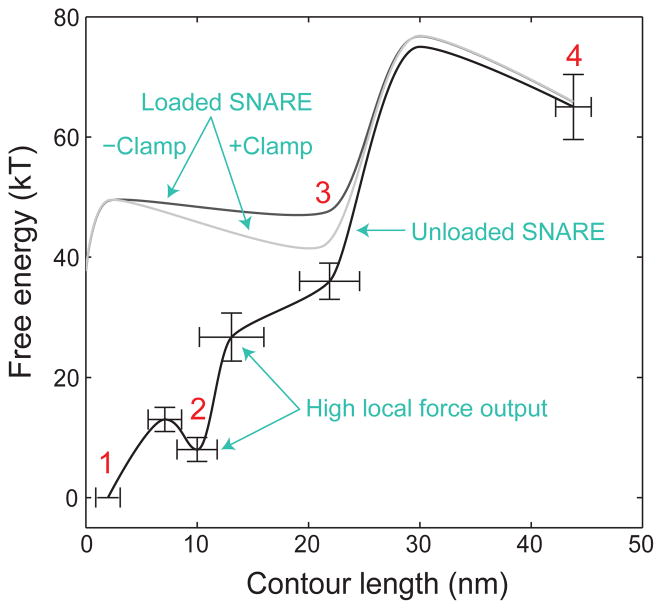
We suggest a model for three-stage neuronal SNARE assembly through sequential zippering of NTD, CTD, and LD (fig. S13). Overall, SNARE assembly outputs free energy of 65 (±6) kBT (14). This energy is much higher than previous reports (28, 30, 31), but justified by our equilibrium measurements required for any thermodynamic quantifications (14). These measurements lead to the energy landscape of SNARE zippering (Fig. 4). The CTD zippering energy is enriched at its C-terminal end or in VAMP2 (74–82), leading to a higher local force generation (~34 pN) (Fig. S8) (30).
Fig. 4.
Sketches of the energy landscapes for SNARE zippering in the absence (black) and presence of the opposing force load from membranes with (light gray) and without (gray) complexin clamp. The contour length of the SNARE complex between the C-termini of syntaxin (residue 265) and VAMP2 (residue 92) is chosen as a reaction coordinate. Error bars show the standard deviations of the measurements.
The half-zippered state appeared to only exist in the force range of 12–20 pN. Below this force range, the state collapsed into the 4-helix bundle, consistent with its down-hill folding at zero force. Thus, SNARE domain zippering became a two-state process limited by the slow NTD assembly (10, 28). Above this range, the half-zippered state completely unzipped. The half-zippered state could be further stabilized by synaptotagmin and complexin (12, 18, 20) (Fig. 4).
The elegant molecular logic of the SNARE complex thus revealed perfectly fits to precise regulation of neurotransmitter release (3), with slow intrinsic assembly of NTD to allow control of vesicle priming by regulatory factors that accelerate NTD assembly, thereby creating the readily-releasable pool of neurotransmitters (11); an intrinsic pause near the ionic layer stabilized by the repulsion between membranes being forced to fuse to enable clamping by complexin at this stage of zippering (17); and unobstructed, fast zippering of CTD and LD to open the fusion pore once the clamp is removed, enabling neurotransmitters to be rapidly released. The pore then expands as the trans-membrane domains of VAMP2 and syntaxin dimerize (32).
Supplementary Material
Acknowledgments
We thank H. Ji and W. Xu for technical assistance and F. Pincet, T. Melia, and E. Karatekin for valuable discussion. This work is supported by the NIH grants GM093341 to Y.Z. and DK027044 to J.E.R. Y.Z. designed the experiments; Y.G., S.Z., G.G., Z.X., L.M., and G.S. performed the experiments; Y.Z. and Y.G. analyzed the data; and Y.Z. and J.E.R. wrote the paper. Matlab programs and original data are available upon request.
References and Notes
- 1.Sollner T, et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 2.Weber T, et al. SNAREpins: Minimal machinery for membrane fusion. Cell. 1998;92:759. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 3.Sudhof TC, Rothman JE. Membrane fusion: Grappling with SNARE and SM proteins. Science. 2009;323:474. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 angstrom resolution. Nature. 1998;395:347. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 5.Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell. 1997;90:523. doi: 10.1016/s0092-8674(00)80512-7. [DOI] [PubMed] [Google Scholar]
- 6.Cohen FS, Melikyan GB. The energetics of membrane fusion from binding, through hemifusion, pore formation, and pore enlargement. J Membr Biol. 2004;199:1. doi: 10.1007/s00232-004-0669-8. [DOI] [PubMed] [Google Scholar]
- 7.Xu T, et al. Inhibition of SNARE complex assembly differentially affects kinetic components of exocytosis. Cell. 1999;99:713. doi: 10.1016/s0092-8674(00)81669-4. [DOI] [PubMed] [Google Scholar]
- 8.Hua SY, Charlton MP. Activity-dependent changes in partial VAMP complexes during neurotransmitter release. Nat Neurosci. 1999;2:1078. doi: 10.1038/16005. [DOI] [PubMed] [Google Scholar]
- 9.Melia TJ, et al. Regulation of membrane fusion by the membrane-proximal coil of the t-SNARE during zippering of SNAREpins. J Cell Biol. 2002;158:929. doi: 10.1083/jcb.200112081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pobbati AV, Stein A, Fasshauer D. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science. 2006;313:673. doi: 10.1126/science.1129486. [DOI] [PubMed] [Google Scholar]
- 11.Walter AM, Wiederhold K, Bruns D, Fasshauer D, Sorensen JB. Synaptobrevin N-terminally bound to syntaxin-SNAP-25 defines the primed vesicle state in regulated exocytosis. J Cell Biol. 2010;188:401. doi: 10.1083/jcb.200907018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kyoung M, et al. In vitro system capable of differentiating fast Ca2+-triggered content mixing from lipid exchange for mechanistic studies of neurotransmitter release. Proc Natl Acad Sci USA. 2011;108:E304. doi: 10.1073/pnas.1107900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 14.Materials and Methods in the Supplementary Materials.
- 15.Weninger K, Bowen ME, Chu S, Brunger AT. Single-molecule studies of SNARE complex assembly reveal parallel and antiparallel configurations. Proc Natl Acad Sci USA. 2003;100:14800. doi: 10.1073/pnas.2036428100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stein A, Weber G, Wahl MC, Jahn R. Helical extension of the neuronal SNARE complex into the membrane. Nature. 2009;460:525. doi: 10.1038/nature08156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao Y, Sirinakis G, Zhang YL. Highly anisotropic stability and folding kinetics of a single coiled coil protein under mechanical tension. J Am Chem Soc. 2011;133:12749. doi: 10.1021/ja204005r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kummel D, et al. Complexin cross-links prefusion SNAREs into a zigzag array. Nat Struct Mol Biol. 2011;18:927. doi: 10.1038/nsmb.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA. 1998;95:15781. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li F, et al. Complexin activates and clamps SNAREpins by a common mechanism involving an intermediate energetic state. Nat Struct Mol Biol. 2011;18:941. doi: 10.1038/nsmb.2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xi ZQ, Gao Y, Sirinakis G, Guo HL, Zhang YL. Direct observation of helix staggering, sliding, and coiled coil misfolding. Proc Natl Acad Sci USA. 2012;109:5711. doi: 10.1073/pnas.1116784109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodside MT, et al. Direct measurement of the full, sequence-dependent folding landscape of a nucleic acid. Science. 2006;314:1001. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabatini BL, Regehr WG. Timing of neurotransmission at fast synapses in the mammalian brain. Nature. 1996;384:170. doi: 10.1038/384170a0. [DOI] [PubMed] [Google Scholar]
- 24.Kubelka J, Hofrichter J, Eaton WA. The protein folding ‘speed limit’. Curr Opin Struct Biol. 2004;14:76. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 25.Ellena JF, et al. Dynamic structure of lipid-bound synaptobrevin suggests a nucleation-propagation mechanism for trans-SNARE complex formation. Proc Natl Acad Sci USA. 2009;106:20306. doi: 10.1073/pnas.0908317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu Y, Su LJ, Rizo J. Binding of Munc18-1 to synaptobrevin and to the SNARE four-helix bundle. Biochemistry-Us. 2010;49:1568. doi: 10.1021/bi9021878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karatekin E, et al. A fast, single-vesicle fusion assay mimics physiological SNARE requirements. Proc Natl Acad Sci USA. 2010;107:3517. doi: 10.1073/pnas.0914723107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiederhold K, Fasshauer D. Is assembly of the snare complex enough to fuel membrane fusion? J Biol Chem. 2009;284:13143. doi: 10.1074/jbc.M900703200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weninger K, Bowen ME, Choi UB, Chu S, Brunger AT. Accessory proteins stabilize the acceptor complex for synaptobrevin, the 1 : 1 syntaxin/SNAP-25 complex. Structure. 2008;16:308. doi: 10.1016/j.str.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu W, Montana V, Parpura V, Mohideen U. Single molecule measurements of interaction free energies between the proteins within binary and ternary SNARE complexes. J Nanoneurosci. 2009;1:120. doi: 10.1166/jns.2009.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li F, et al. Energetics and dynamics of SNAREpin folding across lipid bilayers. Nat Struct Mol Biol. 2007;14:890. doi: 10.1038/nsmb1310. [DOI] [PubMed] [Google Scholar]
- 32.Shi L, et al. SNARE proteins: one to fuse and three to keep the nascent fusion pore open. Science. 2012;335:1355. doi: 10.1126/science.1214984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trimble WS, Cowan DM, Scheller RH. VAMP-1 - a synaptic vesicle-associated integral membrane-protein. Proc Natl Acad Sci USA. 1988;85:4538. doi: 10.1073/pnas.85.12.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett MK, Calakos N, Scheller RH. Syntaxin - a synaptic protein implicated in docking of synaptic vesicles at presynaptic active zones. Science. 1992;257:255. doi: 10.1126/science.1321498. [DOI] [PubMed] [Google Scholar]
- 35.Oyler GA, et al. The identification of a novel synaptosomal-associated protein, SNAP-25, differentially expressed by neuronal subpopulations. J Cell Biol. 1989;109:3039. doi: 10.1083/jcb.109.6.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carrico IS, Carlson BL, Bertozzi CR. Introducing genetically encoded aldehydes into proteins. Nat Chem Biol. 2007;3:321. doi: 10.1038/nchembio878. [DOI] [PubMed] [Google Scholar]
- 37.Cecconi C, Shank EA, Dahlquist FW, Marqusee S, Bustamante C. Protein-DNA chimeras for single molecule mechanical folding studies with the optical tweezers. Eur Biophys J. 2008;37:729. doi: 10.1007/s00249-007-0247-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moffitt JR, Chemla YR, Izhaky D, Bustamante C. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc Natl Acad Sci USA. 2006;103:9006. doi: 10.1073/pnas.0603342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sirinakis G, et al. The RSC chromatin remodeling ATPase translocates DNA with high force and small step size. EMBO J. 2011;30:2364. doi: 10.1038/emboj.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759. [Google Scholar]
- 42.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic Elasticity of Lambda-Phage DNA. Science. 1994;265:1599. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 43.Stigler J, Ziegler F, Gieseke A, Gebhardt JCM, Rief M. The complex folding network of single calmodulin molecules. Science. 2011;334:512. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 44.Yang WY, Gruebele M. Folding at the speed limit. Nature. 2003;423:193. doi: 10.1038/nature01609. [DOI] [PubMed] [Google Scholar]
- 45.Zimm BH, Bragg JK. Theory of the phase transition between helix and random coil in polypeptide chains. J Chem Phys. 1959;31:526. [Google Scholar]
- 46.Brooks CL. Helix-coil kinetics: Folding time scales for helical peptides from a sequential kinetic model. J Phys Chem-Us. 1996;100:2546. [Google Scholar]
- 47.Jackson MB. SNARE complex zipping as a driving force in the dilation of proteinaceous fusion pores. J Membr Biol. 2010;235:89. doi: 10.1007/s00232-010-9258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rabiner LR. A tutorial on hidden Markov-models and selected applications in speech recognition. Proc IEEE. 1989;77:257. [Google Scholar]
- 49.McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91:1941. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Syed S, Mullner FE, Selvin PR, Sigworth FJ. Improved hidden Markov models for molecular motors. Part 1: Basic theory. Biophys J. 2010;99:3684. doi: 10.1016/j.bpj.2010.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qin F, Auerbach A, Sachs F. A direct optimization approach to hidden Markov modeling for single channel kinetics. Biophys J. 2000;79:1915. doi: 10.1016/S0006-3495(00)76441-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fasshauer D, Antonin W, Subramaniam V, Jahn R. SNARE assembly and disassembly exhibit a pronounced hysteresis. Nat Struct Biol. 2002;9:144. doi: 10.1038/nsb750. [DOI] [PubMed] [Google Scholar]
- 53.Yersin A, et al. Interactions between synaptic vesicle fusion proteins explored by atomic force microscopy. Proc Natl Acad Sci USA. 2003;100:8736. doi: 10.1073/pnas.1533137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu W, Montana V, Bai JH, Chapman ER, Parpura V. Single molecule mechanical probing of the SNARE protein interactions. Biophys J. 2006;91:744. doi: 10.1529/biophysj.105.073312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parpura V, Mohideen U. Molecular form follows function: (un)snaring the SNAREs. Trends Neurosci. 2008;31:435. doi: 10.1016/j.tins.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 56.Abdulreda MH, et al. Pulling force generated by interacting SNAREs facilitates membrane hemifusion. Integr Biol. 2009;1:301. doi: 10.1039/b900685k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marqusee S, Baldwin RL. Helix stabilization by Glu− … Lys+ salt bridges in short peptides of de novo design. Proc Natl Acad Sci USA. 1987;84:8898. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gebhardt JCM, Bornschlogla T, Rief M. Full distance-resolved folding energy landscape of one single protein molecule. Proc Natl Acad Sci USA. 2010;107:2013. doi: 10.1073/pnas.0909854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Methods. 2008;5:491. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fowler SB, et al. Mechanical unfolding of a titin Ig domain: Structure of unfolding intermediate revealed by combining AFM, molecular dynamics simulations, NMR and protein engineering. J Mol Biol. 2002;322:841. doi: 10.1016/s0022-2836(02)00805-7. [DOI] [PubMed] [Google Scholar]
- 61.Williams PM, et al. Hidden complexity in the mechanical properties of titin. Nature. 2003;422:446. doi: 10.1038/nature01517. [DOI] [PubMed] [Google Scholar]
- 62.Woodside MT, et al. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc Natl Acad Sci USA. 2006;103:6190. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Junker JP, Ziegler F, Rief M. Ligand-dependent equilibrium fluctuations of single Calmodulin molecules. Science. 2009;323:633. doi: 10.1126/science.1166191. [DOI] [PubMed] [Google Scholar]
- 64.Bornschlog T, Rief M. Single-molecule dynamics of mechanical coiled-coil unzipping. Langmuir. 2008;24:1338. doi: 10.1021/la7023567. [DOI] [PubMed] [Google Scholar]
- 65.Bornschlogl T, Rief M. Single molecule unzipping of coiled coils: Sequence resolved stability profiles. Phys Rev Lett. 2006;96 doi: 10.1103/PhysRevLett.96.118102. [DOI] [PubMed] [Google Scholar]
- 66.Celik E, Moy VT. Nonspecific interactions in AFM force spectroscopy measurements. J Mol Recognit. 2012;25:53. doi: 10.1002/jmr.2152. [DOI] [PubMed] [Google Scholar]
- 67.Parlati F, et al. Rapid and efficient fusion of phospholipid vesicles by the alpha-helical core of a SNARE complex in the absence of an N-terminal regulatory domain. Proc Natl Acad Sci USA. 1999;96:12565. doi: 10.1073/pnas.96.22.12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Greenleaf WJ, Woodside MT, Abbondanzieri EA, Block SM. Passive all-optical force clamp for high-resolution laser trapping. Phys Rev Lett. 2005;95:2081021. doi: 10.1103/PhysRevLett.95.208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.