Abstract
Rational
Although the fibroblast growth factor (FGF) signaling axis plays important roles in heart development, the molecular mechanism by which the FGF regulates cardiogenesis is not fully understood.
Objective
To investigate the mechanism by which FGF signaling regulates cardiac progenitor cell differentiation.
Methods and results
Using mice with tissue-specific ablation of FGF receptors and FGF receptor substrate 2α (Frs2α) in heart progenitor cells, we demonstrate that that disruption of FGF signaling leads to premature differentiation of cardiac progenitor cells in mice. Using embryoid body (EB) cultures of mouse embryonic stem cells (ESCs), we reveal that FGF signaling promotes mesoderm differentiation in ESCs, but inhibits cardiomyocyte differentiation of the mesoderm cells at later stages. Furthermore, we also report that inhibiting FRS2α-mediated signals increases autophagy and that activating autophagy promotes myocardial differentiation and vice versa.
Conclusions
The results indicate that the FGF/FRS2α-mediated signals prevent premature differentiation of heart progenitor cells through suppressing autophagy. The findings provide the first evidence that autophagy plays a role in heart progenitor differentiation.
Keywords: FGF, autophagy, heart development, second heart field, premature differentiation, heart defect
Introduction
Cardiovascular disease is the most prevalent disease, affecting over 80 million people in America alone.1 Mammalian hearts only have a very limited ability to regenerate, which prevents functional repair and restoration of diseased hearts. Stem cell therapies, including using embryonic stem cells (ESCs), induced pluripotent stem cells (iPS), and cardiac progenitor cells, have been implicated in heart repairs. Yet, how to induce differentiation of these cells into needed, fully functional cell types remains a major challenge. Understanding the underlying molecular mechanisms by which the cardiac progenitor cell formation, self-renewal, and differentiation are controlled is essential for developing stem cell therapy for cardiovascular diseases. In mammals, the majority of heart cells are derived from two heart fields, the first and second heart fields (FHF and SHF). Initially, the cardiomyocytes originated from the FHF form a single heart tube, which then undergoes rightward looping, expansion, and formation of recognizable cardiac chambers. At the start of looping and throughout the process, cells from the pharyngeal and splanchnic mesoderm (SM) are recruited to the heart, which are collectively designated as the SHF.2-4
The fibroblast growth factor (FGF) family consists of 18 receptor-binding members that regulate a broad spectrum of cellular activities.5 The FGF signaling axis has been implicated in heart progenitor development, recruitment, and differentiation; disruption of FGF signaling leads to severe defects in heart development.6-12 The FGF elicits its regulatory signals via activating the FGF receptor (FGFR) tyrosine kinases encoded by four highly homologous genes. FGF receptor substrate 2α (FRS2α) is a broadly expressed membrane-anchored adaptor protein that is required for the FGFR to activate the MAP and PI3 kinase pathways, the two major pathways in the FGF signaling cascade.13-15 Frs2α null embryos die between embryonic (E) 7.0-7.5 days.16 Ablation of Frs2α, or double ablation of Fgfr1/Fgfr2, in heart progenitor cells disrupts the endothelial-to-mesenchymal transition (EMT) of the endocardium and the deployment of neural crest cells (NCC) to the outflow tract (OFT), resulting in OFT alignment and septation defects.9 Similarly, ablation of Fgf8 also leads to OFT alignment and septation defects.8 Recently, we also showed that FGF signaling in the OFT myocardium controls NCC differentiation in OFT cushions and regulates the formation of OFT valves.10 Disrupting FGF signaling in the anterior heart field (AHF) in chicken causes premature differentiation of the AHF.17, 18
In addition, a number of signaling molecules or transcriptional factors, including BMP, Wnt, Tbx1, Notch, Hh, Nkx2.5, FAK, Vangl2, and Isl1 have been implicated in SHF development.2, 3, 19 Among them, Tbx1, Notch1, Isl1, BMP, and Wnt have been shown to regulate SHF progenitor cell differentiation.20-24 The FGF signaling axis has been shown to serve as a downstream pathway of Tbx1, Notch1, and Wnt signaling during SHF development,25-27 or upstream regulators for Isl1 and BMP4 in the SHF.9 These studies demonstrate a central role of FGF signaling in regulating SHF progenitor cell differentiation, although how FGF signals regulate cardiac differentiation is still not well understood.
Autophagy is a lysosomal-mediated “self-digestion” process for degrading and recycling various cellular constituents, such as long-lived proteins and entire organelles. Autophagy initiates with the formation of autophagosomes by fusing double-membrane vesicles with sequestrated cellular constituents.28, 29 Autophagosomes then fuse with lysosomes to form autolysosomes where the contents are degraded via acidic lysosomal hydrolases. As a self-digestion system, autophagy may influence cell differentiation by providing new building blocks for formation of new protein or organelle, making ways for new cellular apparatus, or accelerating turnover of old protein or organelles.30 However, no direct evidence of autophagy in regulating cell differentiation is available.
Here, we showed that ablation of the FGFR1/2-FRS2α signaling axis in mouse heart progenitor cells led to premature differentiation of SHF cells. By using embryoid bodies (EBs) derived from mouse ESCs and in vitro cultures of embryonic heart explants, we demonstrated that FGF signaling promoted ESCs undergoing mesoderm differentiation in early stages, but inhibited cardiomyocyte differentiation at late stages. We further demonstrated that FGF signaling suppressed cardiac progenitor cell differentiation by inhibiting autophagy. The results demonstrate a novel mechanism by which the FGF regulates cardiogenesis and shed new light on understanding how stem/progenitor cells undergoing cardiomyocytes differentiation is regulated.
Materials and Methods
Animals
All animals were housed in the Program for Animal Resources of the Institute of Biosciences and Technology, and were handled in accordance with the principles and procedures of the Guide for the Care and Use of Laboratory Animals. All experimental procedures were approved by the Institutional Animal Care and Use Committee. The mice carrying Frs2αflox, Fgfr1flox, Fgfr2flox, Mef2cCre, Nkx2.5Cre, and GFP-LC3 transgenic alleles, as well as the R26R knock-in alleles, were bred and genotyped as described.31-37 The embryos or hearts were excised at the indicated stages, fixed with 4% paraformaldehyde-PBS for 0.5-2 hours, and paraffin embedded. The sections were rehydrated and H&E stained for histological analyses. For chloroquine suppression assay, pregnant females were i.p. injected with 4.5 mg/100g body weight of chloroquine (Sigma, St. Louis, MO) at 9.5 days post copulation as described.38 The mice were sacrificed two hours after the injection; the embryos were collected and fixed with 4% paraformaldehyde-PBS for 0.5-2 hours followed by paraffin embedding.
Short-term whole-embryo or heart cultures
E8.5 mouse embryos were dissected in PBS, leaving the yolk sac intact. Embryos were cultured (37 °C, 5% CO2) in 12-well plates containing 1 ml medium (DMEM: FBS = 1:1) for 16-24 hours, and then dissected in PBS, fixed in methanol/DMSO (4:1) for whole-mount immunostaining. For embryonic heart cultures, the hearts were dissected at E11.5 in PBS and cultured at 37 °C in 12-well plates containing 1 ml medium (DMEM: FBS = 9:1) as described.39 Two days later, the hearts were collected for RNA isolation. Inhibitors for FGFR (PD166866) and ERK1/2 kinase (SL327) were from EMD Chemicals Inc. (Gibbstown, NJ). PI3K inhibitor (LY294002) was from Cell Signaling (Beverly, MA). Bafilomycin A1 was from LC Laboratories (Woburn MA). Although the embryo morphology often had slight changes after inhibitor treatments, the embryos were mounted and sectioned at the same orientation for analyzing the structures of the OFT, ventricle, splanchnic mesoderm, and pharyngeal endoderm.
Immunostaining and lacZ staining
Immunostaining was performed on 5 μm sections mounted on Superfrost/Plus slides (Fisher Scientific, Pittsburgh, PA). The antigens were retrieved by incubating in the citrate buffer (10 mM) for 20 minutes at 100 °C or as suggested by manufacturers of the antibodies. The sources and concentrations of primary antibodies are: anti-smooth muscle actin (1:1) from Sigma (St. Louis, MO); anti-SRF (1:1000); anti-p-Histone H3 (1:500) and anti-cyclin D2 (1:500) from Santa Cruz (Santa Cruz, CA); anti-sarcomere myosin MF20 (1:50) and anti-Isl1 (1:500) from Developmental Studies Hybridoma Bank (Iowa City, IA); anti-Complex V beta subunit (1:100) from Invitrogen (Carlsbad, CA). The specifically bound antibodies were detected with HRP-conjugated secondary antibody (Bio-Rad Co., Hercules, CA) and visualized using TSA™ Plus Fluorescence Systems from PerkinElmer (Boston, MA) on a Zeiss LSM 510 Confocal Microscope. For lacZ staining, the cells were first lightly fixed with 0.2% glutaraldehyde for 30 minutes and then incubated overnight with 1mg/ml X-Gal at room temperature. For whole-mount immunostaining, embryos were fixed and permealized in methanol/DMSO (4:1) overnight at 4 °C. Embryos were then rehydrated and treated with PBT solution (PBS+ 0.3% Triton X-100). MF20 antibody was applied and incubated overnight at 4 °C with rocking. The embryos were then washed with the PBT.3 solution and incubated with peroxidase conjugated second antibody. After being washed with PBT solution, specifically bound antibodies were visualized by peroxidase staining. At least three samples were used for each group.
Embryoid body differentiation
The EB culturing wae carried out as described elsewhere.20 Briefly, mouse embryonic stem cells (1,000) were cultured in hanging drops (20 μl) as aggregates in DMEM containing 10% FBS placed in drops on the lid of petri dishes. The lids were inverted and placed on the dishes filled with PBS. After 2-day culturing, the formed EBs were resuspended in Ultra-low attachment dishes, followed by suspension culture for 4 days. The EBs were then transferred onto a gelatin-coated plate for further differentiation cultures. For siRNA knockdown assays, the EBs were treated with 25nM BECN-1 (CAG TTT GGC ACA ATC AAT A) or ATG7 (GTT TGT AGC CTC AAG TGT T) siRNA at day 6, followed by cultured at 37 °C for 2 more days. The cell were lysed for real-time RT-PCR analyses. All RNAi oligoribonucleotides and DharmaFECT transfection reagents were purchased from Dharmacon (Boulder, CO).
Quantitative RT-PCR analyses
Total RNA was extracted from fresh or in vitro cultured tissues with the Bioopure Kit (Bioo Scientific Co., Austin, TX). Reverse transcriptions were carried out with SuperScript II (GIBCO/BRL, Life Technologies, Grand Island, NY) enzymes and random primers according to manufacturer’s protocols. Real-time RT-PCR analyses were carried out with the SYBR Green JumpStart Taq ReadyMix (Sigma, St. Louis, MO) as suggested by the manufacturer. The primer sequences are:α-MHC-1 (TGT TAA GGC CAA GGT CGT G) and α-MHC-2 (GCA TGT ACT GAT AGG CGT TGT); SMA-1 (TGA CGC TGA AGT ATC CGA TAG) and SMA-2 (GCC AAG TCC AGA CGC ATG A); SKA-1 (ACT TCC TAC CCT CGG CAC CC) and SKA-2 (AGT CAT CTT CTC CCG GTT AGC); CAA-1 (AGA GCT GTC TTC CCG TCC AT) and CAA-2(ATG AGT TAC ACC ATC GCC AGA); SRF-1 (GCA CAG ACC TCA CGC AGA CCT) and SRF-2 (AGC TTG CTG CCC TAT CAC A); Nkx2.5-1 (CAA AGA CCC TCG GGC GGA TAA) and Nkx2.5-2 (GCA TTG AGA CCC ACG CCG TAG); GATA4-1 (GGG ACG GGA CAC TAC CTG T) and GATA4-2 (AGT GGC ATT GCT GGA GTT ACC); BECN1-1 (GTA CCG ACT TGT TCC CTA TG) and BECN1-2 (TGC CTC CAG TGT CTT CAA TC); ATG7-1 (GCC TGT TCA CCC AAA GTT CT) and ATG7-2 (TTC AGA CAG TCT CCT CGT CA). Relative abundances of the mRNA were calculated using the comparative threshold (CT) cycle method and normalized with GADPH. Data derived from at least three independent experiments are expressed as fold of differences between experimental and control samples.
Transwell migration assay
The pharyngeal arches dissected from E8.75 embryos were minced in the MSS buffer, followed by extensively rinsing with the same buffer. After being digested with 0.25% trypsin in PBS for 10 minutes at 37 °C, the samples were triturated to obtain single-cell suspensions, followed by filtering with cell strainer. The cells were pelleted by centrifugation at 300 g for 2 minutes and resuspended in DMEM with 15% FBS. The cells were then placed on transwell filters (Corning Costar, Corning, NY) and cultured at 37 °C for 24 hours. The medium was replaced with serum-free DMEM in transwell inserts and serum-containing medium in the chamber to provide a chemoattractant gradient. One or two days later, LacZ staining was performed. LacZ+ cells were counted as the SHF derived cells. The non-migrated cells on the upper surface of the filter were then removed with a cotton swab; the migrated MF20 positive cells on the lower surface were counted.
Statistical analysis
All experiments were repeated at least three times. Data are expressed as means and standard deviations. Student’s t-test (Tails=2, Type=1) was performed based on at least 3 individual experiments and p 0.05 was considered significant.
Results
Disruption of FGF signaling leads to premature differentiation of SHF progenitor cells
Tissue specific ablation of the FGF signaling axis in heart progenitor cells with the Nkx2.5Cre or Mef2cCre driver compromises participation of the SHF in the OFT, reduces the size of the OFT, and affects OFT valve formation.9, 10 To investigate the molecular mechanism by which the FGF signaling axis regulated heart progenitor cell patterning in the OFT, the embryos with Fgfr1/Fgfr2 double conditional inactivation by the Nkx2.5Cre driver (Fgfr1/r2Nkx) were subjected to detailed immunohistological analyses. In control embryos, the cardiomyocyte differentiation markers, sarcomeric α-actin (Sar) and smooth muscle α-actin (SMA), were not expressed in the SHF cells until they migrated into the OFT myocardium. In mutant embryos, however, both Sar and SMA were expressed in SHF progenitor cells that still resided in the SM (Fig. 1A). The results are in line with recent reports that FGF signaling is required to prevent chick SHF premature differentiation.17, 18 Individual ablation of Fgfr1 or Fgfr2 did not change SMA and Sar expression patterns (Fig. 1B), indicating that FGFR1 and FGFR2 redundantly mediated FGF signaling to maintain SHF progenitor cells in the undifferentiated stage.
Fig. 1. Double ablation of Fgfr1/r2 causes premature differentiation of SHF progenitor cells.
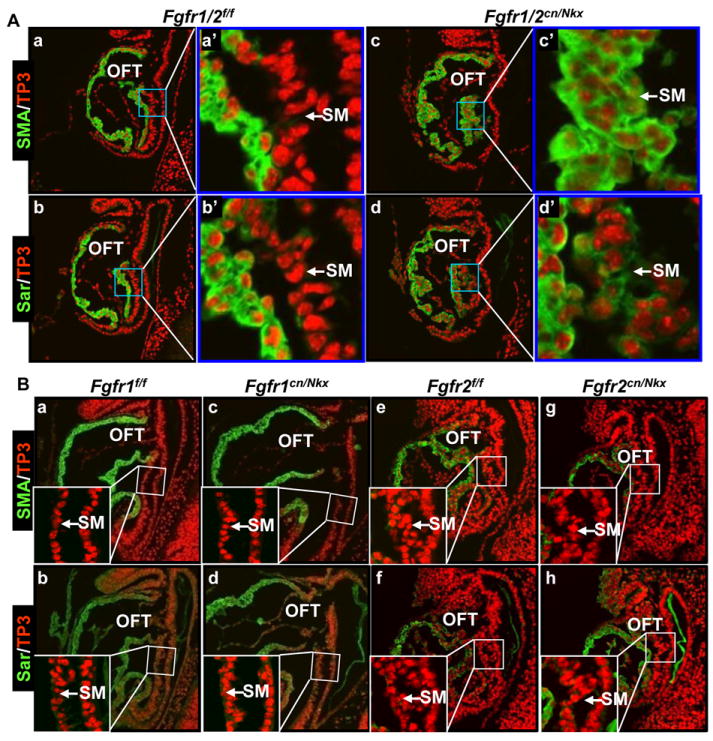
A&B, Sagittal sections of E8.5 embryos were immunostained with anti-smooth muscle α-actin (SMA) and sarcomeric α-actin (Sar) antibodies. Nuclei were counter-stained with To-Pro3 iodine (TP3). Inserts in (B) were high magnification views of the boxed areas. SM, splanchnic mesoderm; OFT, outflow tract; f, floxed alleles; cn, conditional null alleles; Nkx, Nkx2.5Cre.
FRS2α is a common adaptor protein for FGFRs, which links multiple downstream pathways to FGFR kinases. To investigate whether FRS2α was required to repress differentiation of SHF cells, Nkx2.5Cre-mediated Frs2α conditional null (Frs2αcn/Nkx) embryos were subjected to the same analyses to determine whether Sar and SMA were expressed in the SHF at the same stage. Similar to Fgfr1/Fgfr2 double ablation, ablation of Frs2α in heart progenitors caused ectopic expression of Sar and SMA in the SHF, suggesting that FRS2α mediated FGF signals to regulate SHF differentiation (Fig. 2A). The serum response factor (SRF) plays key roles in cardiomyocyte differentiation, and its expression is increased after onset of the cardiomyocyte differentiation.40 Consistent with ectopic expression of Sar and SMA, SRF expression was also increased in the SM of Frs2αcn/Nkx embryos (Fig. 2A). Furthermore, whole-mount immunostaining with the MF20 monoclonal antibody that recognized both light and heavy chains of myosin revealed that myosin expression in the SHF was dramatically increased in Frs2αcn/Nkx embryos (Fig. 2B). Quantitative real-time RT-PCR analyses with RNA isolated from E9.5 pharyngeal arches revealed that SMA, cardiac α-actin (CAA), skeletal α-actin (SKA), and GATA4 were significantly increased in Frs2αcn/Nkx embryos (Fig. 2C). Since these molecules are associated with cardiomyocyte differentiation, the results suggest that FRS2α is required for preventing cardiac differentiation of the SHF.
Fig. 2. Ablation of Frs2α leads to premature differentiation of SHF progenitor cells.
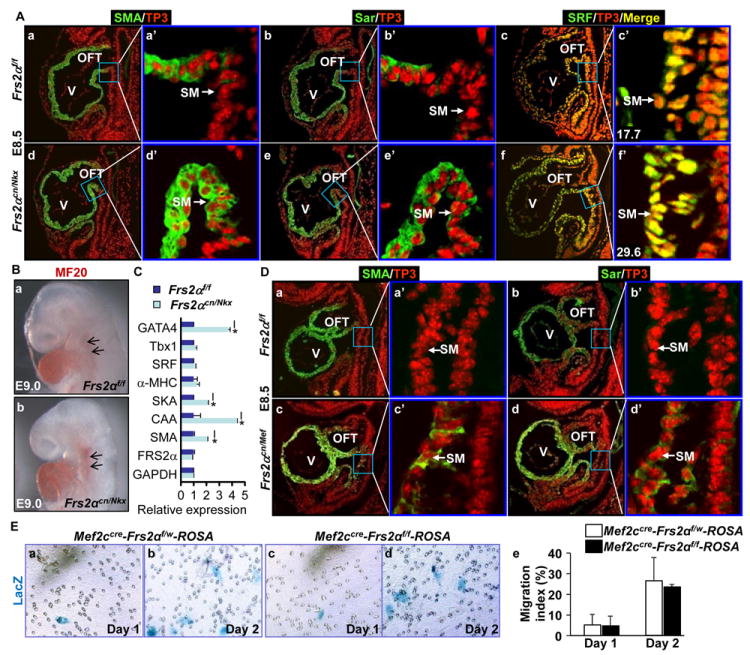
A&D. Sagittal sections of E8.5 embryos were immunostained with the antibody against SMA, Sar, and serum response factor (SRF). Numbers in panels c’ and f’ are fluorescent intensities quantitated by the Image J software, which represent SRF expression levels in the SM. B. Whole-mount immunostaining of E9.0 embryos with the MF20 antibody. C. Real-time RT-PCR analyses of cells isolated from E9.5 pharyngeal arches. Data are normalized to GAPDH and expressed as folds of changes over wildtype samples. D. Sagittal sections of E8.5 embryos were immunostained with anti-SMA or Sar antibodies as indicated. E. The migration activity of cells isolated from E8.75 pharyngeal arches were assessed with the Transwell assays. SHF cells were identified with X-Gal staining. The ratios of migrated SHF cells over total SHF cells were calculated from 3 individual samples and expressed as mean±sd. Mef, Mef2CCre; SKA, skeletal α-actin; CAA, cardiac α-actin; w, wildtype Frs2α allele; *, p≤0.05.
Ablation of Frs2α in the SM with Mef2cCre also led to expression of Sar and SMA in the SHF (Fig. 2D), indicating that FRS2α-mediated signals prevented SHF differentiation in a cell autonomous mode. Ablation of Frs2α compromises the cell migration activity at E12.5 during OFT cushion remodeling.10 To determine whether FRS2α-mediated signals were required for cell migration activity in the SHF at early stages, pharyngeal arch cells were isolated from E8.75 Mef2cCre-ROSA26 embryos for cell migration assays. Since the ROSA26 reporter allele remained silent until it was activated by Mef2cCre in the SHF, only SHF-derived cells were stained blue in the assay. Transwell analyses showed the deletion of Frs2α did not significantly reduce the number of cells migrating through the membrane at this stage (Fig. 2E), indicating the migration activity was not affected. Thus, the results demonstrate that ectopic appearance of cardiac marker-expressing cells in the SHF is not caused by migration failure of differentiated SHF cells, but is caused by premature differentiation of SHF progenitors prior to migrating to the OFT.
FRS2α-mediated signals regulate SHF progenitor cell differentiation independent of proliferation activity
The Frs2αcn/Nkx embryos exhibit low proliferation activities in the SHF.9 To determine whether the SHF premature differentiation was caused by the reduced proliferation activity, ex vivo cultured embryos were treated with excessive thymidine to induce cell cycle arrest at the beginning of the S phase. Immunostaining revealed that the thymidine treatment significantly reduced expression of cyclin D2 required for the G1/S transition in the cell cycle (Fig. 3A). Yet, the treatment did not induce premature myosin expression in the SHF, as revealed by immunostaining with the MF20 antibody (Fig. 3B), indicating that suppressing cell proliferation did not induce SHF differentiation. Thus, it appears that FGF signals regulate SHF progenitor cell differentiation independent of cell cycle progression.
Fig. 3. SHF differentiation is independent of its proliferation activity and is negatively regulated by both MAP kinase and PI3K/AKT pathways.
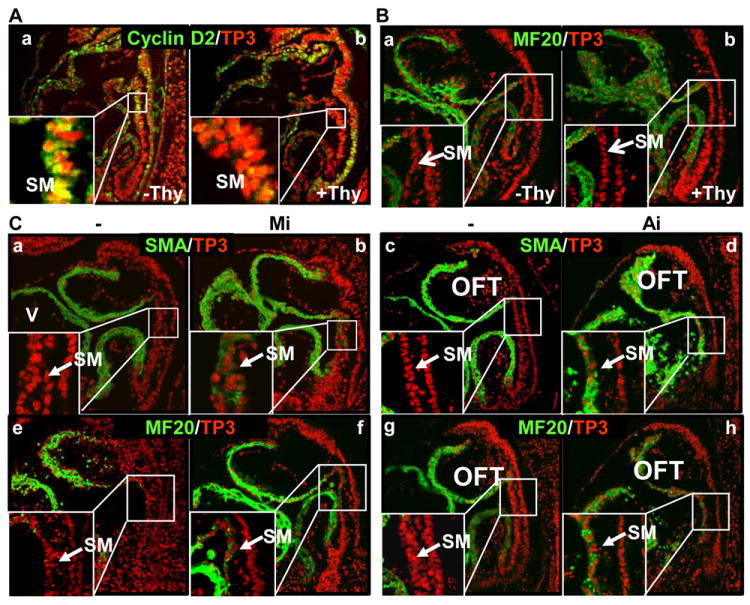
A&B. The embryo cultures were treated with 2 mM excessive thymidine. Sagittal sections were immunostained with anti-cyclin D2 (A) or MF20 antibodies (B) to demonstrate that cell cycle arrest did not affect SHF differentiation. C. The embryos were cultured in the presence or absence of 5 μM MEK or 3 μM PI3K inhibitors. Sagittal sections were immunostained with the indicated antibodies demonstrating that inhibition of MEK or PI3K induced SHF differentiation. -, DMSO as a solvent control; Mi, MEK inhibitor; Ai, PI3K/AKT inhibitor.
FRS2α has multiple tyrosine phosphorylation sites and links FGF signaling to the MAPK and AKT pathways. To test whether the two pathways were involved in regulating SHF differentiation, ex vivo cultures of E8.5 embryos were treated with MEK1/2 or PI3K inhibitors. Immunostaining of embryo sections demonstrated that both MEK and PI3K inhibitions induced ectopic expression of SMA and myosin in the SHF (Fig. 3C), suggesting that both MAPK and PI3K/AKT pathways were required to prevent SHF premature differentiation.
To further investigate the mechanisms by which FGF signaling regulated SHF differentiation, SHF cells were isolated from E9.5 pharyngeal arches for in vitro differentiation assays in the presence or absence of FGF2 treatment. Isl1 is a transcription factor transiently expressed in SHF progenitors. Since Isl1 expression is quickly down regulated in differentiating SHF cells, it is often used to identify undifferentiated SHF cells.41, 42 Immunostaining showed that no Isl1+ progenitor cells were detected after three-day culture in the absence of FGF2 (Fig. 4Aa), indicating that no undifferentiated SHF cells remained in the culture. However, in the presence of FGF2, there were still many Isl1+ undifferentiated SHF cells in the culture (Fig. 4Ab). On the other hand, treating the cells with FGFR, MAPK or PI3/AKT inhibitors significantly increased myosin expression in the SHF cultures as revealed by immunostaining with MF20 antibody, indicating that inhibition of FGFR, MAPK or PI3K/AKT promoted SHF undergoing myocardial differentiation (Fig. 4B&C). Collectively, the data demonstrate that FGF signaling inhibits cardiac differentiation of heart progenitor cells.
Fig. 4. The FGF suppresses cultured SHF progenitor cells differentiation.
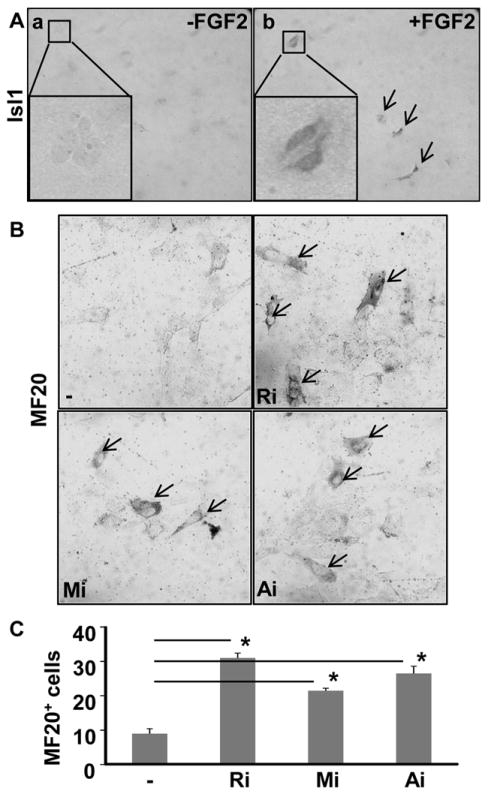
Cells isolated from E9.5 pharyngeal arches were cultured in the presence or absence of FGF2 (25 ng/ml), MEK inhibitor (5 μM), PI3K inhibitor (3 μM), or FGFR inhibitors (250 nM) as indicated, and then were immunostained with anti-Isl1 (A) or MF20 (B) antibodies as indicated. The positively stained cells are indicated with arrows. Statistical analyses of MF20 positive cells in three independent experiments were presented in (C) as mean±sd. Ri, FGFR inhibitor; *, P≤0.05
FGF signaling promotes ESCs to undergo mesoderm differentiation, but inhibits the mesoderm cells to undergo cardiac differentiation
Since it was difficult to expand the SHF progenitor cells in vitro for further experiments, the well-established hanging-drop culture of ESC-derived EBs was used to investigate the mechanism by which the FGF regulated cardiac differentiation. In this culture system, the ESCs undergo mesenchymal differentiation and commit to the cardiac lineage within the first four days; by day 5, the cells start to undergo cardiac differentiation and form beating foci at later stages.20 To investigate the role of FGF signaling in these two stages, the EB cultures were treated with the FGFR inhibitor at the early mesenchymal commitment stage or at the late cardiac differentiation stage. About 25% of EBs exhibited spontaneous contraction at day 8. The ratio of beating foci was gradually increased over the time. When treated with the FGFR inhibitor at day 0 to 4, the number of beating foci was significantly reduced, although formation of EBs was not affected (Fig. 5A). As expected, the expression of mesoderm marker Brachyury (T), as well as early cardiac markers Nkx2.5, Tbx5, and Tbx1 were reduced by treating with the FGFR inhibitor (Fig. 5B), which was consistent with the previous report.43 Together, the data indicate that FGF signaling promotes the mesoderm formation and cardiac lineage commitment.
Fig. 5. The FGF signaling axis regulates cardiogenesis in a developmental stage-specific manner.
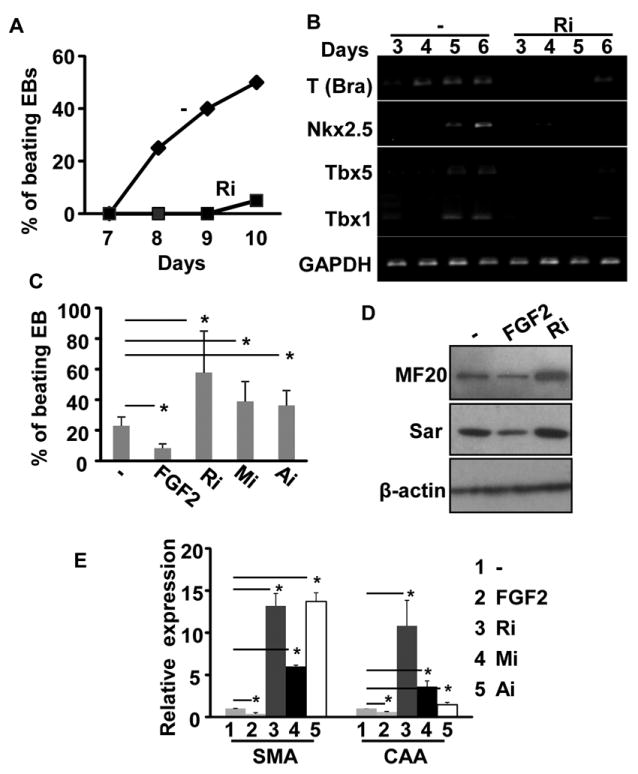
A&B. Embryo bodies (EBs) were treated with FGFR inhibitor (250 nM) from day 0 to day 4, and the percentage of beating embryoid bodies (EBs) was counted and presented as means±sd of three independent experiments (A); expression of mesoderm markers and early cardiac markers was analyzed with RT-PCR at the indicated time points (B). C-E. EB cultures were treated with 25 ng/ml FGF2, the MEK inhibitor (5 μM), PI3K inhibitor (3 μM), or FGFR inhibitors (250 nM) at day 6 to day 10. The percentage of beating EBs were scored and presented as mean±sd from three independent experiments (C); expression of cardiac markers in day 10 EBs was assessed by Western blot (D) or real-time RT-PCR analyses (E). β-actin was used as a loading control; *, p<0.05.
However, treating the EB cultures with FGF2 at day 6 and forward significantly inhibited beating focus formation, and treating the EBs with the FGFR, MEK, or PI3K inhibitor at this stage increased the number of beating foci (Fig. 5C). Moreover, Western analyses revealed that expression of myosin and Sar in EBs was reduced by FGF2 and increased by FGFR inhibitor treatment at this stage (Fig. 5D). Real-time RT-PCR analysis further confirmed that SMA and CAA expression was increased by FGFR, MAPK, or PI3K inhibitors, but decreased by FGF2, treatments (Fig. 5E). Thus, similar to in vivo cardiogenesis results, the data again indicate that the FGF signaling axis suppresses cardiac differentiation of cardiac progenitor cells.
FGF signaling suppresses differentiation of cardiac progenitor cells during cardiogenesis through inhibition of autophagy
We next tried to identify downstream effectors of the FGF signaling axis involved in suppressing cardiac progenitor premature differentiation. Tbx1 is negative regulators of SHF differentiation,20 but its expression in the pharyngeal arches was not affected by Frs2α deletion (Fig. 2C). Isl1 is transiently expressed in undifferentiating SHF cells41 and promotes SHF cell differentiation21, however, Isl1 expression in decreased in the SHF of Frs2αcn embryos.9 Although BMP4 promotes SHF differentiation,17, 18, 24 expression of BMP4 is reduced in the Frs2α mutant SHF.9 This suggests that FGF suppresses SHF differentiation independent of these molecules/pathways. Autophagy, which can be down-regulated by the FGF in mouse embryo fibroblasts via the mTOR pathway,44 has been proposed to regulate cell differentiation.30 The conversion of microtubule associated light chain 3 (LC3) from the unlipidated LC3-I to lipidated LC3-II is a broadly used autophagy indicator.45, 46 To test whether FGF signaling regulated autophagy in EBs, the abundance of LC3-II in EBs was evaluated in the presence or absence of FGFR inhibitors. Apparently, inhibiting FGFR activity increased the abundance of LC3-II (Fig. 6Aa). Increased LC3-II abundance can be resulted either from enhanced LC3-I to LC3-II conversion or from reduced degradation of LC3-II, which reflects enhanced autophagic initiation or reduced autophagosome turnover, respectively.45, 46 To determine the nature of changes in LC3-II abundance, the EB cultures were treated with NH4Cl, a lysozyme inhibitor, to block LC3-II degradation. The results showed that inhibiting FGFR kinase increased LC3-II abundance when LC3-II turnover was blocked (Fig. 6Aa). The results ruled out the possibility that increased LC3-II abundance was due to decreased degradation, and indicated that LC3-II conversion, and therefore autophagy initiation, was increased by inhibiting FGF signaling. In addition, the abundance of two key autophagy regulators, Beclin1 and p27, were increased in the presence of the FGFR inhibitor (Fig. 6Ab). The results indicate that FGF signaling suppresses autophagy in EBs.
Fig. 6. The FGF signaling axis inhibits ESCs undergoing myocardiac differentiation via suppressing autophagy.
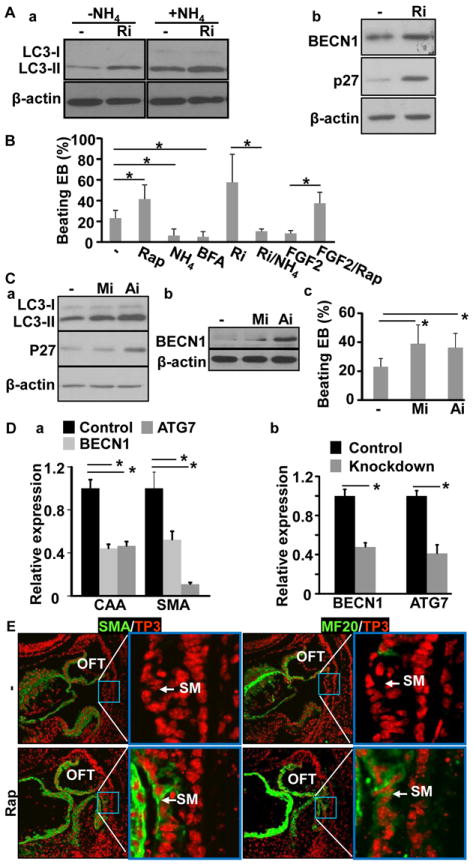
A. EB cultures of mouse ESCs were treated with 250 nM FGFR inhibitor from day 6. The abundance of LC3, Beclin 1, and P27 was analyzed by Western blot at day 7. B, EB cultures were treated with 100 nM rapamycin, 5 mM NH4Cl, 5 nM bafilomycin A1, 250 nM FGFR inhibitor, 25 ng/ml FGF2 as indicated, or the indicated combinations, and the beating foci were scored at day 8. Data were mean±sd of three independent experiments. C. EB cultures were treated with 5 μM MEK inhibitor or 3 μM PI3K inhibitor from day 6. The abundance of LC3, Beclin 1, and P27 was assessed with Western blot (panels a&b), and the beating foci were counted from three independent experiments and expressed as mean±sd (panel c). D. EB cultures were treated with siRNA at day 6 and the cells were lysed at day 8. Expression of the indicated molecules were assessed by real-time RT-PCR analyses. E. E8.5 embryos were cultured in the presence or absence of 100 nM rapamycin, and the expression of SMA and myosin was analyzed with immunostaining. β-actin was used as a loading control; Rap, rapamycin; NH4, NH4Cl; BFA, bafilomycin A; *, p≤0.05; BECN1, Beclin 1.
To determine whether autophagy played a role in myocardial differentiation of EB cells, the EB cultures were treated with autophagy inhibitors, NH4Cl or bafilomycin A1, or the autophagy promoter, rapamycin, and the myocardial differentiation was assessed (Fig. 6B). The data showed that the treatment of autophagy inhibitor significantly inhibited formation of beating foci, while the treatment of autophagy promoter increased formation of beating foci. Furthermore, treating with NH4Cl blocked the enhancement of beating focus formation by FGFR inhibitors, and treating with rapamycin abolished the suppression activity of FGF2 on beating focus formation.
To determine which pathway mediated FGFR signals to regulate autophagy in the EBs, the EBs were treated with either ERK or PI3K inhibitors at day 6. Western blot revealed that treating with either ERK or PI3K inhibitor increased autophagy activities, evidenced by increased LC3-II abundance. Interestingly, treating the EBs with the PI3K, but not ERK, inhibitor up-regulated expression of Beclin1 and p27, although inhibition of either the MAPK or AKT pathway promoted beating focus formation (Fig. 6C). Knockdown of Beclin1 or ATG7 in EBs suppressed expression of cardiac α-actin and smooth muscle α-actin (Fig. 6D). Treating ex vivo cultured E8.5 embryos with rapamycin induced premature expression of SMA and myosin in the SHF (Fig. 6E). Consistently, ablation of Frs2α significantly increased numbers of green fluorescent protein (GFP)-LC3 positive punctate foci in the OFT myocardium of Frs2αcn/Nkx embryos carrying a GFP-LC3 transgenic allele encoding a GFP-LC3 fusion protein (Fig. 7A), especially in the presence of chloroquine, a lysosome inhibitor (Fig. 7B). The results demonstrate that the formation of autophagosomes is increased in differentiating cardiomyocytes of mutant embryos. Double immunostaining with GFP-LC3 and Complex V beta subunit antibodies revealed that only a fraction of autophagosomes contained mitochondria (Fig. 7C). Further efforts are needed to characterize the contents of other autophagosomes. Together, the results suggest that FGF signaling suppresses cardiomyocyte differentiation by inhibiting autophagy.
Fig. 7. Ablation of Frs2α promotes autophagy in the OFT myocardium.
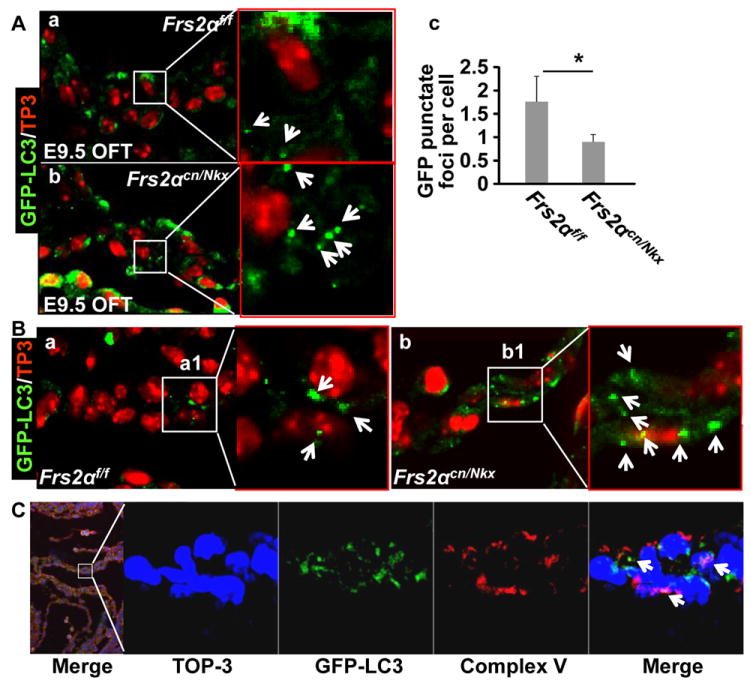
A. Sagital sections of E9.5 Frs2αcn/Nkx or Frs2αf/f embryos expressing the GFP-LC3 transgene were immunostained with anti-GFP antibody. Data are confocal images demonstrating increased LC3 positive punctate foci that represented autophagosomes (arrows) in the Frs2αcn/Nkx OFT myocardium. Average numbers of punctate foci per cell were presented as mean±sd in panel c. B. Frs2αcn/Nkx/ or Frs2αf/f/GFP-LC3 embryos from chloroquine injected dam were analyzed as in A, further demonstrating increase autophagy influx in Frs2αcn/Nkx OFT myocardium. C. Double immunostaining of E9.5 wildtype/GFP-LC3 embryos demonstrating that only a fraction of autophagosomes contained mitochondria (arrows).
FRS2α-mediated signals also inhibit maturation of ventricular cardiomyocytes and differentiation of cardiac mesenchymal cells
It has been shown that ablation of multiple FGFs in the heart suppresses proliferation and promotes myocardial differentiation.39 Consistently, immunostaining analyses also demonstrated that ablation of Frs2α significantly reduced proliferation in the ventricular myocardium at E12.5 (Fig. 8A), resulting in a thinner ventricular wall in Frs2αcn/Nkx embryos than in the controls (Fig. 8B). Furthermore, both immunostaining and Western blot analyses revealed that ablation of Frs2α resulted in increased Sar and myosin expression, indicating that FGF signaling suppressed cardiomyocyte maturation (Fig. 8C&D). To investigate whether autophagy regulated myocardium maturation, the heart explants were treated with autophagy inhibitor or activator. Activation of autophagy by rapamycin enhanced expression of MHC, CAA, SMA, SRF, and GATA4 (Fig. 8E). On the other hand, inhibition of autophagy by NH4Cl decreased expression of these molecules. Consistently, treating Frs2αcn/Nkx hearts with NH4Cl reduced expression of MHC and CAA, indicating that the myocardial maturation was suppressed (Fig. 8F). In addition, FGF2 also exhibited a similar inhibitory function on cardiac mesenchymal cell differentiation, which was isolated from newborn hearts (Fig. 8G). Together, the results indicate that FRS2α-mediated signaling also inhibits ventricular myocardium maturation through regulating autophagy.
Fig. 8. The FRS2α-mediated signals in the myocardium promote proliferation and suppress maturation.
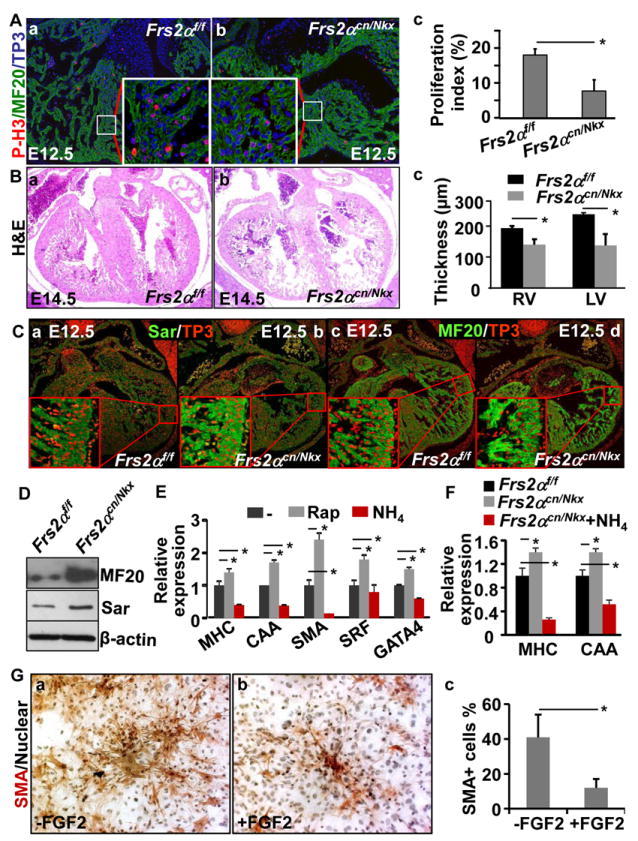
A. E12.5 embryos were subjected to immunostaining analyses with anti-phosphorylated histone H3 and MF20 antibodies. Statistical data from three independent experiments were presented in panel c. Nuclei were counterstained with To-Pro3. Inserts were high magnification views of the boxed areas. B. Heart sections of E14.5 embryonic were subjected to H&E staining. The average thickness of compact zones of the ventricle from 5 individual hearts was shown in panel c. C. Sections from E12.5 embryos were immunostained with anti-Sar and MF20 antibodies. Nuclei were counterstained with To-Pro3. D. Western blot analyses of E12.5 wildtype or Frs2αcn/Nkx hearts showed increased expression of Sarcomeric actin and myosin in mutant hearts. β-actin was used as a loading control. E&F. E11.5 heart cultures from Frs2α floxed (E), or indicated genotypes embryos were treated with 100 nM rapamycin or 5 mM NH4Cl as indicated, and total RNAs were extracted for real-time RT-PCR analyses. Data were normalized with GAPDH and expressed as folds of changes from control samples. G. Cardiac mesenchymal cells isolated from neonatal hearts were cultured in the presence or absence of FGF2. The cells were then immunostained with anti-SMA antibodies. Nuclei were counterstained with hematoxylin. Average ratios of SMA positive cells from three individual experiments were shown in panel c as means±sd. MHC, myosin heavy chain; *, p≤0.05.
Discussion
Although the FGF is a key regulator in cardiogenesis, how FGF signaling regulates cardiac stem/progenitor cell differentiation remains to be elucidated, and the role of FGF signaling in cardiogenesis is also controversial. FGF2 has been shown to promote cardiac precursors to differentiate into functional cardiomyocytes in neonatal mouse;47 EBs from murine Fgfr1-/- ES cells fail to form contractile cardiomyocyte foci and do not express early and late cardiac markers.43 On the other hand, FGF signals are required for preventing premature differentiation of cells either in the AHA or in the ventricle myocardium.17, 18, 39 Here we report that FGF signaling promotes mesoderm differentiation in EBs, but prevents premature differentiation in heart progenitors. We further demonstrate that FRS2α-mediated pathways are required for the FGF to suppress premature differentiation of heart progenitor cells. Moreover, the activity of the FGF signaling axis to suppress cardiac differentiation is independent of cell proliferation activity and requires autophagy. Suppression of autophagy inhibits cardiomyocyte differentiation of heart progenitors in the SHF and EBs.
The FGF-ERK signaling axis has been shown to inhibit differentiation of chicken AHF progenitors.17, 18 However, mouse and chicken heart progenitor cells have different FGF expression patterns.17 Also, it was not clear whether the function of FGF signaling in heart progenitor cells is species-specific. Here we used a mouse model to demonstrate that FGF signaling has similar function in mouse heart progenitor cell differentiation as in chicken. Interestingly, although PI3K/AKT signaling is not compromised by ablation of Frs2α in the SHF,9 the data herein demonstrate that PI3K/AKT signaling also inhibits mouse SHF differentiation, suggesting that other signaling may also redundantly activate the PI3K/AKT pathway. The results are different from those derived from chicken SHF progenitor cells.18 It appeared that inhibiting the ERK or PI3K/AKT individually was less potent than inhibiting the FGFR (Fig. 5C). Also, the abundance of Beclin 1 and p27 in EBs was increased by PI3K/AKT, but not by ERK, inhibition (Fig. 6C), indicating that the two pathways likely regulated autophagy/differentiation in heart progenitor cells via non-redundant mechanisms. Indeed, it has been shown that PI3K/AKT regulates autophagy through the mTOR pathway and MAPK through transcription factor EB (TFEB). 44, 48
The data herein suggest that FGF signaling plays biphasic roles in cardiogenesis depending on the stage of differentiation. In the early stage, FGF signaling promotes mesoderm formation and cardiac lineage commitment, evidenced by that inhibition of FGFR at early stage suppresses cardiac differentiation marker expression. The notion is also supported by previous studies that Fgf8-/-, Fgfr1-/- or Frs2α-/- embryos exhibit abnormal mesoderm formation and that FGF10 increases mouse iPS cell differentiation into the cardiac lineage.49-52 In addition, inactivation of Fgfr1 dramatically affects the expression of early cardiac transcription factors Nkx2.5, d-Hand, and other mesoderm-related early genes in ES cells.43 Deletion of the Fgfr homologue, heartless (htl), in Drosophila disrupts the cardiac mesoderm formation.53, 54 After cardiac lineage commitment, FGF signaling inhibits cardiac progenitor cell differentiation, which is supported by the most recent study that FGF signaling inhibits chick AHF differentiation.17, 18 Interestingly, early work also demonstrates that cardiomyocyte development during tubular stages of cardiogenesis is FGF signaling dependent, but becomes FGF-independent after the second week of embryogenesis.55 In consistence, it has been reported that Wnt signaling, which acts as the upstream regulator of FGF signaling during SHF development, also has similar biphasic effects in regulating ESC differentiation.22, 56
Autophagy is one of the major cellular pathways to degrade bulky subcellular organelles and macromolecules to make way for new organelles or to provide nutrition and needed building blocks for formation of new cellular apparatus, and plays important roles in development, metabolism, tumorigenesis and cardiovascular diseases.57 However, how autophagy contributes to development is not understood, although it has been proposed that autophagy may influence cell differentiation either by impairing new protein synthesis or new organelle formation or by accelerating turnover of old proteins or organelles. Although detailed molecular mechanism still needs to be investigator, here we provided the first evidence that autophagy positively regulated cardiac progenitor cell differentiation and cardiomyocyte maturation.
In summary, the FGF signaling axis plays a biphasic function in cardiogenesis: it promotes cardiac lineage determination at early stages and suppresses premature differentiation at late stages. This is the first report showing that the autophagy plays a crucial role in mediating growth factor signaling pathways in regulating heart progenitor differentiation. Thus, this study will facilitate the developments of novel strategies to manipulate cardiomyocyte development from ES cells for therapeutic applications.
Novelty and Significance.
What is known?
Stem cell therapy is a promising method being actively pursued for cardiac repair.
A major challenge of stem cell therapy is how to efficiently induce the stem cells differentiate into fully functional cardiomyocytes.
Fibroblast grow factor (FGF) signaling has been shown to regulate cardiac progenitor cell differentiation, although the underlying mechanism is unclear.
What is new?
FGF signaling promotes the differentiation of embryonic stem cells (ESCs) into cardiac progenitor cells but inhibits progenitor cell differentiation into cardiomyocytes.
FGF signaling suppresses cardiac progenitor cells differentiation though regulating autophagy activity.
Manipulating FGF signaling and autophagy enhances the cardiogenesis of ESCs.
These findings provide new insights into the molecular mechanism by which the differentiation of cardiac progenitor cells is regulated. Our study suggests that regulating FGF signaling and autophagy can be potential venues for improving cardiac stem cell therapy.
Acknowledgments
We thank Drs. Noboru Mizushima, Robert J Schwartz, Juha Patanen, and David Ornitz for their generosity to share the GFP-LC3 transgenic mice, the Nkx2.5Cre knock-in mice, and the Fgfr1-floxed and Fgfr2-floxed mice, respectively; Kerstin McKeehan for her excellent technical support, and Mary Cole for critical reading of the manuscript.
Source of Funding
NIH-CA96824 and CA140388 from the NCI to FW; R01 HL093484-02 and R01 DE12324-13 from NIH to JFM, and AHA0655077Y to FW and 09PRE2010130 predoctoral fellowship to JZ from The American Heart Association.
Nonstandard Abbreviations
- FGF
fibroblast growth factor
- FGFR
FGF receptor
- FRS2α
FGFR substrate 2α
- FHF
first heart field
- NCCs
neural crest cells
- OFT
outflow tract
- SHF
second heart field
- SM
splanchnic mesoderm
Footnotes
Disclosures
None
References
- 1.Roger VL, Go AS, Lloyd-Jones DM, Adams RJ, Berry JD, Brown TM, Carnethon MR, Dai S, de Simone G, Ford ES, et al. Heart disease and stroke statistics--2011 update: A report from the american heart association. Circulation. 2011;123:e18–e209. doi: 10.1161/CIR.0b013e3182009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buckingham M, Meilhac S, Zaffran S. Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet. 2005;6:826–835. doi: 10.1038/nrg1710. [DOI] [PubMed] [Google Scholar]
- 3.Srivastava D. Making or breaking the heart: From lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Kelly RG, Brown NA, Buckingham ME. The arterial pole of the mouse heart forms from fgf10-expressing cells in pharyngeal mesoderm. Developmental cell. 2001;1:435–440. doi: 10.1016/s1534-5807(01)00040-5. [DOI] [PubMed] [Google Scholar]
- 5.McKeehan WL, Wang F, Luo Y. The fibroblast growth factor (fgf) signaling complex. Handbook of cell signaling. New York: Academic/Elsevier Press; 2009. [Google Scholar]
- 6.Chien KR, Domian IJ, Parker KK. Cardiogenesis and the complex biology of regenerative cardiovascular medicine. Science (New York, N Y. 2008;322:1494–1497. doi: 10.1126/science.1163267. [DOI] [PubMed] [Google Scholar]
- 7.Park EJ, Ogden LA, Talbot A, Evans S, Cai CL, Black BL, Frank DU, Moon AM. Required, tissue-specific roles for fgf8 in outflow tract formation and remodeling. Development. 2006;133:2419–2433. doi: 10.1242/dev.02367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park EJ, Watanabe Y, Smyth G, Miyagawa-Tomita S, Meyers E, Klingensmith J, Camenisch T, Buckingham M, Moon AM. An fgf autocrine loop initiated in second heart field mesoderm regulates morphogenesis at the arterial pole of the heart. Development. 2008;135:3599–3610. doi: 10.1242/dev.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Lin Y, Zhang Y, Lan Y, Lin C, Moon AM, Schwartz RJ, Martin JF, Wang F. Frs2alpha-deficiency in cardiac progenitors disrupts a subset of fgf signals required for outflow tract morphogenesis. Development. 2008;135:3611–3622. doi: 10.1242/dev.025361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Chang JY, Huang Y, Lin X, Luo Y, Schwartz RJ, Martin JF, Wang F. The fgf-bmp signaling axis regulates outflow tract valve primordium formation by promoting cushion neural crest cell differentiation. Circulation research. 2010;107:1209–1219. doi: 10.1161/CIRCRESAHA.110.225318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank DU, Fotheringham LK, Brewer JA, Muglia LJ, Tristani-Firouzi M, Capecchi MR, Moon AM. An fgf8 mouse mutant phenocopies human 22q11 deletion syndrome. Development. 2002;129:4591–4603. doi: 10.1242/dev.129.19.4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abu-Issa R, Kirby ML. Heart field: From mesoderm to heart tube. Annual review of cell and developmental biology. 2007;23:45–68. doi: 10.1146/annurev.cellbio.23.090506.123331. [DOI] [PubMed] [Google Scholar]
- 13.Kouhara H, Hadari YR, Spivak-Kroizman T, Schilling J, Bar-Sagi D, Lax I, Schlessinger J. A lipid-anchored grb2-binding protein that links fgf-receptor activation to the ras/mapk signaling pathway. Cell. 1997;89:693–702. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 14.Xu H, Lee KW, Goldfarb M. Novel recognition motif on fibroblast growth factor receptor mediates direct association and activation of snt adapter proteins. J Biol Chem. 1998;273:17987–17990. doi: 10.1074/jbc.273.29.17987. [DOI] [PubMed] [Google Scholar]
- 15.Ong SH, Lim YP, Low BC, Guy GR. Shp2 associates directly with tyrosine phosphorylated p90 (snt) protein in fgf-stimulated cells. Biochemical and biophysical research communications. 1997;238:261–266. doi: 10.1006/bbrc.1997.7272. [DOI] [PubMed] [Google Scholar]
- 16.Hadari YR, Gotoh N, Kouhara H, Lax I, Schlessinger J. Critical role for the docking-protein frs2 alpha in fgf receptor-mediated signal transduction pathways. Proc Natl Acad Sci U S A. 2001;98:8578–8583. doi: 10.1073/pnas.161259898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tirosh-Finkel L, Zeisel A, Brodt-Ivenshitz M, Shamai A, Yao Z, Seger R, Domany E, Tzahor E. Bmp-mediated inhibition of fgf signaling promotes cardiomyocyte differentiation of anterior heart field progenitors. Development. 2010;137:2989–3000. doi: 10.1242/dev.051649. [DOI] [PubMed] [Google Scholar]
- 18.Hutson MR, Zeng XL, Kim AJ, Antoon E, Harward S, Kirby ML. Arterial pole progenitors interpret opposing fgf/bmp signals to proliferate or differentiate. Development. 2010;137:3001–3011. doi: 10.1242/dev.051565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garry DJ, Olson EN. A common progenitor at the heart of development. Cell. 2006;127:1101–1104. doi: 10.1016/j.cell.2006.11.031. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Fulcoli FG, Tang S, Baldini A. Tbx1 regulates proliferation and differentiation of multipotent heart progenitors. Circulation research. 2009;105:842–851. doi: 10.1161/CIRCRESAHA.109.200295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon C, Qian L, Cheng P, Nigam V, Arnold J, Srivastava D. A regulatory pathway involving notch1/beta-catenin/isl1 determines cardiac progenitor cell fate. Nature cell biology. 2009;11:951–957. doi: 10.1038/ncb1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qyang Y, Martin-Puig S, Chiravuri M, Chen S, Xu H, Bu L, Jiang X, Lin L, Granger A, Moretti A, et al. The renewal and differentiation of isl1+ cardiovascular progenitors are controlled by a wnt/beta-catenin pathway. Cell Stem Cell. 2007;1:165–179. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 23.Jia Q, McDill BW, Li SZ, Deng C, Chang CP, Chen F. Smad signaling in the neural crest regulates cardiac outflow tract remodeling through cell autonomous and non-cell autonomous effects. Developmental biology. 2007;311:172–184. doi: 10.1016/j.ydbio.2007.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Greene SB, Bonilla-Claudio M, Tao Y, Zhang J, Bai Y, Huang Z, Black BL, Wang F, Martin JF. Bmp signaling regulates myocardial differentiation from cardiac progenitors through a microrna-mediated mechanism. Developmental cell. 2010;19:903–912. doi: 10.1016/j.devcel.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vitelli F, Taddei I, Morishima M, Meyers EN, Lindsay EA, Baldini A. A genetic link between tbx1 and fibroblast growth factor signaling. Development. 2002;129:4605–4611. doi: 10.1242/dev.129.19.4605. [DOI] [PubMed] [Google Scholar]
- 26.Cohen ED, Wang Z, Lepore JJ, Lu MM, Taketo MM, Epstein DJ, Morrisey EE. Wnt/beta-catenin signaling promotes expansion of isl-1-positive cardiac progenitor cells through regulation of fgf signaling. The Journal of clinical investigation. 2007;117:1794–1804. doi: 10.1172/JCI31731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.High FA, Jain R, Stoller JZ, Antonucci NB, Lu MM, Loomes KM, Kaestner KH, Pear WS, Epstein JA. Murine jagged1/notch signaling in the second heart field orchestrates fgf8 expression and tissue-tissue interactions during outflow tract development. The Journal of clinical investigation. 2009;119:1986–1996. doi: 10.1172/JCI38922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cecconi F, Levine B. The role of autophagy in mammalian development: Cell makeover rather than cell death. Developmental cell. 2008;15:344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soriano P. Generalized lacz expression with the rosa26 cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 32.Lin Y, Zhang J, Zhang Y, Wang F. Generation of an frs2alpha conditional null allele. Genesis. 2007;45:554–559. doi: 10.1002/dvg.20327. [DOI] [PubMed] [Google Scholar]
- 33.Trokovic R, Trokovic N, Hernesniemi S, Pirvola U, Vogt Weisenhorn DM, Rossant J, McMahon AP, Wurst W, Partanen J. Fgfr1 is independently required in both developing mid- and hindbrain for sustained response to isthmic signals. Embo J. 2003;22:1811–1823. doi: 10.1093/emboj/cdg169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, Towler DA, Ornitz DM. Conditional inactivation of fgf receptor 2 reveals an essential role for fgf signaling in the regulation of osteoblast function and bone growth. Development. 2003;130:3063–3074. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- 35.Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an nkx2-5/cre gene using rosa26 reporter mice. Genesis. 2001;31:176–180. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- 36.Dodou E, Verzi MP, Anderson JP, Xu SM, Black BL. Mef2c is a direct transcriptional target of isl1 and gata factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- 37.Xie R, Nguyen S, McKeehan K, Wang F, McKeehan WL, Liu L. Microtubule-associated protein 1s (map1s) bridges autophagic components with microtubules and mitochondria to affect autophagosomal biogenesis and degradation. The Journal of biological chemistry. 2011;286:10367–10377. doi: 10.1074/jbc.M110.206532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwai-Kanai E, Yuan H, Huang C, Sayen MR, Perry-Garza CN, Kim L, Gottlieb RA. A method to measure cardiac autophagic flux in vivo. Autophagy. 2008;4:322–329. doi: 10.4161/auto.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived fgf signals regulate myocardial proliferation and differentiation in vivo. Developmental cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Niu Z, Li A, Zhang SX, Schwartz RJ. Serum response factor micromanaging cardiogenesis. Current opinion in cell biology. 2007;19:618–627. doi: 10.1016/j.ceb.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Developmental cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moretti A, Caron L, Nakano A, Lam JT, Bernshausen A, Chen Y, Qyang Y, Bu L, Sasaki M, Martin-Puig S, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 43.Dell’Era P, Ronca R, Coco L, Nicoli S, Metra M, Presta M. Fibroblast growth factor receptor-1 is essential for in vitro cardiomyocyte development. Circulation research. 2003;93:414–420. doi: 10.1161/01.RES.0000089460.12061.E1. [DOI] [PubMed] [Google Scholar]
- 44.Lin X, Zhang Y, Liu L, McKeehan WL, Shen Y, Song S, Wang F. Frs2alpha is essential for the fibroblast growth factor to regulate the mtor pathway and autophagy in mouse embryonic fibroblasts. International Journal of Biological Sciences. 2011;7:1114–1121. doi: 10.7150/ijbs.7.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanida I, Ueno T, Kominami E. Lc3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barth S, Glick D, Macleod KF. Autophagy: Assays and artifacts. The Journal of pathology. 2010;221:117–124. doi: 10.1002/path.2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenblatt-Velin N, Lepore MG, Cartoni C, Beermann F, Pedrazzini T. Fgf-2 controls the differentiation of resident cardiac precursors into functional cardiomyocytes. The Journal of clinical investigation. 2005;115:1724–1733. doi: 10.1172/JCI23418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. Tfeb links autophagy to lysosomal biogenesis. Science (New York, N Y. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gotoh N, Manova K, Tanaka S, Murohashi M, Hadari Y, Lee A, Hamada Y, Hiroe T, Ito M, Kurihara T, et al. The docking protein frs2alpha is an essential component of multiple fibroblast growth factor responses during early mouse development. Mol Cell Biol. 2005;25:4105–4116. doi: 10.1128/MCB.25.10.4105-4116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan SS, Li HJ, Hsueh YC, Lee DS, Chen JH, Hwang SM, Chen CY, Shih E, Hsieh PC. Fibroblast growth factor-10 promotes cardiomyocyte differentiation from embryonic and induced pluripotent stem cells. PLoS One. 2010;5:e14414. doi: 10.1371/journal.pone.0014414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamaguchi TP, Harpal K, Henkemeyer M, Rossant J. Fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes & development. 1994;8:3032–3044. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- 52.Sun X, Meyers EN, Lewandoski M, Martin GR. Targeted disruption of fgf8 causes failure of cell migration in the gastrulating mouse embryo. Genes & development. 1999;13:1834–1846. doi: 10.1101/gad.13.14.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beiman M, Shilo BZ, Volk T. Heartless, a drosophila fgf receptor homolog, is essential for cell migration and establishment of several mesodermal lineages. Genes & development. 1996;10:2993–3002. doi: 10.1101/gad.10.23.2993. [DOI] [PubMed] [Google Scholar]
- 54.Gisselbrecht S, Skeath JB, Doe CQ, Michelson AM. Heartless encodes a fibroblast growth factor receptor (dfr1/dfgf-r2) involved in the directional migration of early mesodermal cells in the drosophila embryo. Genes & development. 1996;10:3003–3017. doi: 10.1101/gad.10.23.3003. [DOI] [PubMed] [Google Scholar]
- 55.Mima T, Ueno H, Fischman DA, Williams LT, Mikawa T. Fibroblast growth factor receptor is required for in vivo cardiac myocyte proliferation at early embryonic stages of heart development. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:467–471. doi: 10.1073/pnas.92.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Naito AT, Shiojima I, Akazawa H, Hidaka K, Morisaki T, Kikuchi A, Komuro I. Developmental stage-specific biphasic roles of wnt/beta-catenin signaling in cardiomyogenesis and hematopoiesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19812–19817. doi: 10.1073/pnas.0605768103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]


