Abstract
Introduction
This study focuses on the implementation of modulated modularity clustering (MMC) a new cluster algorithm for the identification of molecular signatures of preeclampsia and intrauterine growth restriction (IUGR), and the identification of affected microRNAs
Methods
Eighty-six human placentas from normal (40), growth-restricted (27), and preeclamptic (19) term pregnancies were profiled using Illumina Human-6 Beadarrays. MMC was utilized to generate modules based on similarities in placental transcriptome. Gene Set Enrichment Analysis (GSEA) was used to predict affected microRNAs. Expression levels of these candidate microRNAs were investigated in seventy-one human term placentas as follows: control (29); IUGR (26); and preeclampsia (16).
Results
MMC identified two modules, one representing IUGR placentas and one representing preeclamptic placentas. 326 differentially expressed genes in the module representing IUGR and 889 differentially expressed genes in a module representing preeclampsia were identified. Functional analysis of molecular signatures associated with IUGR identified P13K/AKT, mTOR, p70S6K, apoptosis and IGF-1 signaling as being affected. Analysis of variance of GSEA-predicted microRNAs indicated that miR-194 was significantly down-regulated both in preeclampsia (p=0.0001) and IUGR (p=0.0304), and miR-149 was significantly down-regulated in preeclampsia (p=0.0168).
Discussion
Implementation of MMC, allowed identification of genes disregulated in IUGR and preeclampsia. The reliability of MMC was validated by comparing to previous linear modeling analysis of preeclamptic placentas.
Conclusion
MMC allowed the elucidation of a molecular signature associated with preeclampsia and a subset of IUGR samples. This allowed the identification of genes, pathways, and microRNAs affected in these diseases.
Keywords: Placenta, preeclampsia, IUGR, miR-194, miR-149
1. Introduction
Preeclampsia (PE) is a pregnancy associated syndrome characterized by hypertension and proteinuria during pregnancy, which is a consequence of diverse pathophysiological processes involving impaired implantation, endothelial dysfunction, and systemic inflammation [1–4]. Intrauterine growth restriction (IUGR), of diverse causes, refers to the poor growth of a fetus that has not reached its growth potential while in the mother’s uterus during pregnancy [5]. This study describes the genome-wide gene expression analysis of a large (n=86) set of human placentas in order to uncover expression patterns (or molecular signatures) associated with preeclampsia and IUGR. Previously, we successfully identified, using a linear model analysis, genes disregulated in preeclampsia [6]. However, a similar analysis of IUGR samples was less effective, likely due to the high heterogeneity of the IUGR samples. As a result, we used an alternative method of analysis referred to as modulated modularity clustering (MMC) [7] that identifies unique expression signatures in a heterogeneous sample population. MMC is analogous to K-means clustering [8] with the exception that the numbers of clusters or modules are independently identified by MMC, not arbitrarily selected by the investigator. Using MMC we were able to identify unique placental gene expression signatures for both preeclampsia and a subset of IUGR subjects and utilized those expression profiles to identify, using gene set enrichment analysis (GSEA) [9], microRNA candidates disregulated in IUGR and/or preeclampsia. In the case of PE we compared the MMC-generated results with our previously published linear model analysis of PE placentas [6].
2. Materials and methods
2.1 Study design
An initial study population, consisting of 86 Caucasian and African-American subjects collected during 2004–2008, was utilized for gene expression analysis. Initially a small subset of 14 subjects (batch #1) was analyzed using Illumina arrays to ensure the quality of the RNA before proceeding further. Once quality was confirmed the remaining 72 samples were analyzed (batch #2). Principle component analysis identified one sample as an outlier and that sample was discarded from the analysis. For the linear modeling batch #1 and batch #2 samples were included and the data was corrected for batch effect using a set of 8 technical replicates. For the MMC analysis only the second batch was utilized as preliminary analysis indicated that batch effect negatively affected the performance of the MMC analysis. Thus, a subset population of 71 subjects in the following groups: (1) preeclampsia (n=16); (2) IUGR (n=26); and (3) control group (n=29), was used for MMC and microRNA qRT-PCR validation.
2.2 Subjects and sample collection
Preeclampsia was diagnosed when both pregnancy-induced hypertension and proteinuria were present according to American College of Obstetricians and Gynecologists 2000 guidelines [10]. Pregnancy-induced hypertension was defined as a sustained (≥ 2 measures 6 hours apart) blood pressure elevation (> 140/90 mm Hg) > 20 weeks of gestation. Proteinuria was defined as a sustained (≥2 measures 4 hours apart) presence of elevated protein in the urine (> 30 mg/dL or > 1+ on a urine dipstick). IUGR was defined when the estimated weight of a fetus was below the 10th percentile for its gestational age and whose abdominal circumference was below the 2.5th percentile. Subjects were enrolled at the Duke University Medical Center Obstetric Clinic starting August 1, 2003. The criteria for subject enrollment and procedure for sample collection and storage were described in our previous paper [6]. Summary characteristics of the studied population are presented in (Table 1). The study was approved by the Duke University Medical Center Institutional Review Board (IRB 00016065).
Table 1.
Demographic information of study population
| Variablesa | Group
|
p-valueb
|
||||
|---|---|---|---|---|---|---|
| Control (n=40) | PE (n=19) | IUGR (n=27) | Control vs. PE | Control vs. IUGR | PE vs. IUGR | |
| Fetal estimated Gestational Age (weeks) | 37.7 ± 1.9 | 33.6 ± 3.7 | 35.0 ± 3.9 | <0.0001 | 0.0006 | 0.1365 |
| Female % | 42.9 | 57.9 | 59.3 | 0.2761 | 0.1835 | 0.9263 |
| Placental weight (g) | 491.1 ± 141.3 | 448.2 ± 237.8 | 329.8 ± 114.7 | 0.3424 | <0.0001 | 0.0166 |
| Birth weight (g) | 3263.7 ± 625.8 | 2278.4 ± 998.9 | 1936.8 ± 676.6 | <0.0001 | <0.0001 | 0.1244 |
| Corrected percentile birth weight | 49.6 ± 3.8 | 40.4 ± 5.7 | 6.7 ± 4.8 | 0.1831 | <0.0001 | <0.0001 |
| Maternal parity | 2.4 ± 1.7 | 0.4 ± 0.7 | 1.1 ± 1.6 | <0.0001 | 0.0008 | 0.1464 |
| Maternal weight (lb) | 215.9 ± 57.6 | 209.2 ± 45.5 | 153.1 ± 35.0 | 0.6207 | <0.0001 | 0.0003 |
| Induced % | 16.7 | 73.7 | 48.2 | <0.0001 | 0.0049 | 0.0833 |
Continuous data are presented as mean ± SD; categorical data as percentage
Obtained by student t test and Chi-square test for continuous and categorical variables, respectively (JMP 9, SAS Institute, Cary, NC)
2.3 RNA isolation from human placenta
Total RNA was isolated from term human placentas using the Totally RNA kit (Ambion). Small RNA used for microRNA qPCR validation, was isolated using the mirVana miRNA isolation kit (Ambion). Only samples with an OD260:OD280 ≥ 2.0 were used.
2.4 Real-time Quantitative RT-PCR
Small RNA-containing total RNA was converted into cDNA using the miScript Reverse Transcription Kit (Qiagen). EvaGreen [11] based qRT-PCR was performed to profile miRNA levels in 71 placentas from healthy or case (PE or IUGR) complicated pregnancies. The fold change between the experimental sample and the calibration sample was calculated using the Pfaffl method [12] (See supplemental method for detailed information).
2.5 Statistical analysis
Transcript data was log2 transformed, and quantile normalized as described previously [6]. Principle components analysis [13] was performed to calculate the contribution of each of the factors to the measured transcriptional variation: classification (or module), gender, induction of labor, their pair-wise two-way interactions, and estimated gestational age, by using JMP Genomics 5.0 (SAS Institute, Cary, NC). Since no significant effect of induction of labor was detected in our previous study [6], we chose to use a model without induction of labor to perform gene-specific analysis of variance (ANOVA) using PROC MIXED in SAS (SAS Institute, Cary, NC): expression = μ + classification + gender + gender × classification + batch + ε, treating classification, gender, and batch as fixed effects. Custom hypothesis tests were constructed to test for differential expression between case (PE or IUGR) and control or between different modules predicted by MMC (with module in place of classification). Raw p-values were corrected for multiple comparisons via Benjamini-Hochberg FDR at α < 0.05 (for pathway analysis) and Bonferonni at α < 0.05 methods [14] as implemented in PROC MULTTEST in SAS (SAS Institute, Cary NC).
miRNA-specific analysis of variance (ANOVA) was performed using PROC MIXED in SAS 9.2 (SAS Institute, Cary, NC), treating classification as fixed effect. The difference of miRNA levels in three groups was also profiled by fitting relative expression unit to classification in JMP 9 (SAS Institute, Cary, NC). Student t test was performed to evaluate the module effect by using SAS 9.2 (SAS Institute, Cary, NC). Differences in demographics between the control and case groups were tested using one-way ANOVA and Chi-square test for continuous and discrete variables respectively, by using JMP 9 (SAS Institute, Cary, NC). Validation of microarray results using RT-PCR was previously reported [6]
2.6 Modulated Modularity Clustering and Gene Set Enrichment Analysis
Modulated Modularity Clustering (MMC)[7] was used to separate placentas on the basis of the gene expression profiles. This method compares the gene expression profiles of input samples, correlates them to each other, and creates modules based on an overall correlation index, with no information provided as to which samples were classified as control, IUGR or preeclampsia. The program does not know how many modules exist a priori, but determines how many different molecular signatures are there in the sample population. A normalized and quality filtered transcript data for 34471 probes in 71 human placenta samples was submitted to the online software for clustering (http://mmc.gnets.ncsu.edu/). Validation of the MMC procedure was provided by comparison of previous linear model analysis of preeclamptic samples [6] with results obtained from MMC clustering.
Gene Set Enrichment Analysis (GSEA) [9] was performed on the ranked list (based on p-value) of differentially expressed genes to identify functionally enriched gene sets. Curated microRNA targets from the Molecular Signature Database (MSigDB, Borad Institute) were analyzed for enrichment.
2.7 Pathway analysis
Genes from the data set that met the FDR<0.05 and were annotated as high-quality (“perfect” or “good”) in an updated Illumina probe set annotation [15] were considered for ingenuity pathways analysis (IPA). The significance of the association between the data set and the canonical pathway was measured by using Benjamini-Horhberg multiple testing corrected p-value.
2.8 Decision tree construction
The relative expression level of all the candidate miRNAs was used to construct the decision tree. We used a well-known decision tree algorithm, C4.5, implemented in an open-source software library [16] (WEKA, J48, University of Waikato, New Zealand). This analysis uses recursive partitioning methods to separate the patients into distinct sub-sets by identifying the value of miRNA relative expression level and automatically constructing the decision branches. The corresponding breakpoints were selected with the criterion of maximization of the purity of the group of patients after splitting. The algorithm also includes a “pruning” procedure to reflexively eliminate unnecessary branches, reduce the estimated errors, and generalize the model. The outcome is a set of probabilities associated with the likelihood that the different microRNAs can be used to predict whether an individual is a case (IUGR or PE) or a control based on the expression pattern of the selected microRNAs. The potential predictive performance of the tree was evaluated using 5-fold cross-validation, where the model was built on 4/5 of the data, and tested for its prediction error on the withheld 1/5 of the data. This was repeated using every possible 4/5 and 1/5 split of the data, and an average prediction error was calculated.
3. Results
3.1 Demographics of study population
Study population characteristics are described in Table 1. Preeclampsia and IUGR cases were more likely to deliver early due to the induction of labor and had a lower infant birth weight compared to controls (p<0.01). Lower maternal parity was found to be associated with preeclampsia and IUGR samples (p<0.01). Placental weight, corrected birth weight percentile and maternal weight of IUGR were significantly lower than that of control and preeclampsia (p<0.05). There were no significant differences in infant sex among the three groups.
3.2 Variance component analysis
Variance component analysis indicated that classification (Control, IUGR, or PE) is the main contributing effect on expression variation, with minor contributing effects of labor and estimated gestational age. Gender has no significant contribution to the principle components of expression variation (Supplemental figure 1A). Additionally, when we performed this variance component analysis with module (from MMC analysis) in place of classification, the contribution of module to the observed transcriptional variation is greater than classification (Supplemental figure 1B). The dataset is freely available under GEO Series accession number GSE35574.
3.3 Differentially expressed genes between control and IUGR placentas
Linear model analysis estimating placental gene expression differences between normal and IUGR placentas found only 1 differentially expressed gene at a conservative Bonferroni significance level. When we reexamined the data using a less conservative false discovery rate (FDR) set at <0.1, we were able to identify 26 differentially expressed genes (Supplemental figure 2A).
3.4 Modulated Modularity Clustering of patients based on placental gene expression profiles
To get a clear module structure, 71 human term placentas composed of 29 control, 26 IUGR and 16 PE were analyzed by MMC and classified into 6 modules based on the expression profile. Unlike other modules that were a mix of PE and/or IUGR and control samples, module 3 consisted of only IUGR samples, and module 5 consisted of only PE and IUGR samples (Table 2). The gene expression differences between modules 3 (or module 5) and the remaining modules were then compared by linear model analysis as described above. Comparison of module 3 vs all identified 326 differentially expressed genes at a conservative Bonferroni significance level and 2100 differentially expressed genes at FDR<0.05; in the test of module 5 vs all, we found 889 differentially expressed genes at a conservative Bonferroni significance level and 5184 differentially expressed genes at FDR<0.05 (Supplemental figure 2B, 2C and supplemental table 1, 2).
Table 2.
Patient composition of modules generated by MMC
| Module | No. | Classification
|
Mean ± S.D.
|
||||
|---|---|---|---|---|---|---|---|
| Control | IUGR | PE | Weight percentile | Placental Weight | Gestational Age | ||
| 1 | 2 | 1 | 1 | 0 | 19.00 ± 22.63 | 365.00 ± 63.64 | 33.50 ± 2.12 |
| 2 | 8 | 6 | 2 | 0 | 50.38 ± 31.85 | 461.00 ± 165.32 | 35.63 ± 4.27 |
| 3 | 7 | 0 | 7 | 0 | 2.43 ± 1.81 | 367.71 ± 101.48 | 37.43 ± 0.79 |
| 4 | 18 | 14 | 4 | 0 | 32.61 ± 29.72 | 465.18 ± 162.10 | 38.00 ± 1.19 |
| 5 | 18 | 0 | 7* | 11 | 19.44 ± 18.04 | 271.35 ± 118.65 | 31.50 ± 3.62 |
| 6 | 18 | 8 | 5 | 5 | 39.72 ± 38.25 | 469.94 ± 146.58 | 36.94 ± 2.51 |
4 out of 7 IUGR were also diagnosed as PE
All the 128 differentially expressed genes previously identified as disregulated in preeclamptic samples [6] were represented at the top position in the analysis of module 5, including the known preeclampsia associated genes ENG (endoglin), FLT1 (fms-related tyrosine kinase 1), INHA (inhibin alpha), PAPPA2 (pappalysin-2), and RDH13 (retinol dehydrogenase 13) (Supplemental figure 2C).
The differentially expressed genes (FDR<0.05) in “module 3 vs all” and “module 5 vs all” were used to search for affected canonical pathways. Significant pathways are listed in Table 3. Pathways that are enriched in “module 3 vs all” are associated with cellular growth, proliferation and development. Significant pathways affected included P13K/AKT/mTOR/eIF4/p70S6K signaling, apoptosis signaling and IGF-1 signaling. Pathways that are enriched in “module 5 vs all” are associated with cellular growth, proliferation and development (e.g. EIF2 signaling, RhoGDI signaling, and mTOR signaling) and immune responses (e.g. leukocyte extravasation signaling, Fcg receptor-mediated phagocytosis in macrophages and monocytes, and CXCR4 signaling). All of the eight pathways we previously identified as disregulated in preeclampsia using linear modeling analysis [6] match those we identified in “module 5 vs all” using MMC (Table 3), supporting the validity of the MMC clustering analysis method.
Table 3.
Ingenuity pathways significantly enriched for differentially expressed genes in module 3 and module 5
| Ingenuity Canonical Pathways | Label | FDR (B-H p-value) |
|---|---|---|
| P13K/AKT Signaling | Module 3 vs all | 0.0176 |
| Regulation of eIF4 and p70S6K Signaling | Module 3 vs all | 0.0203 |
| Molecular Mechanisms of Cancer | Module 3 vs all | 0.0324 |
| mTOR Signaling | Module 3 vs all | 0.0324 |
| Neuregulin Signaling | Module 3 vs all | 0.0412 |
| Apoptosis Signaling | Module 3 vs all | 0.0412 |
| IGF-1 Signaling | Module 3 vs all | 0.0420 |
| Amyloid Processing | Module 3 vs all | 0.0425 |
| Cleavage and Polyadenylation of Pre-mRNA | Module 3 vs all | 0.0425 |
| JAK/Stat Signaling | Module 3 vs all | 0.0499 |
| EIF2 Signaling | Module 5 vs all | 0.0002 |
| Leukocyte Extravasation Signaling* | Module 5 vs all | 0.0009 |
| RhoGDI Signaling | Module 5 vs all | 0.0015 |
| mTOR Signaling | Module 5 vs all | 0.0015 |
| Androgen Signaling | Module 5 vs all | 0.0015 |
| Huntington’s Disease Signaling | Module 5 vs all | 0.0015 |
| Sertoli Cell-Sertoli Cell Junction Signaling | Module 5 vs all | 0.0015 |
| Mitochondrial Dysfunction | Module 5 vs all | 0.0015 |
| Ephrin Receptor Signaling | Module 5 vs all | 0.0016 |
| CXCR4 Signaling* | Module 5 vs all | 0.0017 |
| RhoA Signaling | Module 5 vs all | 0.0017 |
| Regulation of Actin-based Motility by Rho* | Module 5 vs all | 0.0017 |
| Fcγ Receptor-mediated Phagocytosis in Macrophages and Monocytes* | Module 5 vs all | 0.0018 |
| Gap Junction Signaling | Module 5 vs all | 0.0018 |
| Breast Cancer Regulation by Stathmin1 | Module 5 vs all | 0.0018 |
| Axonal Guidance Signaling | Module 5 vs all | 0.0018 |
| Regulation of eIF4 and p70S6K Signaling | Module 5 vs all | 0.0018 |
| Molecular Mechanisms of Cancer | Module 5 vs all | 0.0026 |
| α-Adrenergic Signaling | Module 5 vs all | 0.0028 |
| Tight Junction Signaling | Module 5 vs all | 0.0032 |
| fMLP Signaling in Neutrophils | Module 5 vs all | 0.0052 |
| Cell Cycle: G1/S Checkpoint Regulation | Module 5 vs all | 0.0052 |
| Phospholipase C Signaling | Module 5 vs all | 0.0052 |
| Signaling by Rho Family GTPases | Module 5 vs all | 0.0058 |
| Oxidative Phosphorylation | Module 5 vs all | 0.0063 |
| Germ Cell-Sertoli Cell Junction Signaling* | Module 5 vs all | 0.0068 |
| Mismatch Repair in Eukaryotes | Module 5 vs all | 0.0078 |
| Mechanisms of Viral Exit from Host Cells | Module 5 vs all | 0.0093 |
| Antiproliferative Role of TOB in T Cell Signaling | Module 5 vs all | 0.0093 |
| NRF2-mediated Oxidative Stress Response* | Module 5 vs all | 0.0172 |
| Semaphorin Signaling in Neurons* | Module 5 vs all | 0.0221 |
| N-Glycan Biosynthesis* | Module 5 vs all | 0.0499 |
Pathways identical with the ones that enriched in differentially expressed genes between preeclamptic and normal placentas in our previous report [6]
3.5 Gene Set Enrichment Analysis
A ranked list of differentially expressed genes was generated according to the log transformed p-value in comparison of module 3 vs. all and module 5 vs. all respectively, and was subsequently used as the input file for GSEA to identify functionally enriched gene sets.
From the enrichment of curated microRNA targets in the Molecular Signature Database (MsigDB, Borad Institute), a set of microRNAs was predicted to target the differentially expressed genes. Considering the normalized enrichment score (NES), false discovery rate (FDR) and nominal p-value, six leading candidates (miR-520A-5p, miR-194, miR-412, miR-149, miR-483, and miR-503) were selected for microRNA profiling (Table 4).
Table 4.
Candidate microRNAs predicted by GSEA
| Gene Sets | Module Affected | ES1 | NES2 | NOM3 p-value | FDR q- value | microRNA specific primer (5′-3′) |
|---|---|---|---|---|---|---|
| MIR-520A-5p | Module 3 | 0.646 | 1.404 | 0.000 | 0.038 | CTCCAGAGGGAAGTACTTTCT |
| MIR-194 | Module 3 | 0.655 | 1.407 | 0.000 | 0.039 | TGTAACAGCAACTCCATGTGG |
| MIR-412 | Module 3 | 0.676 | 1.401 | 0.006 | 0.039 | ACTTCACCTGGTCCACTAGC |
| MIR-503 | Module 5 | 0.727 | 1.414 | 0.016 | 0.158 | TAGCAGCGGGAACAGTTCTG |
| MIR-483 | Module 5 | 0.669 | 1.378 | 0.000 | 0.150 | TCACTCCTCTCCTCCCGTCT |
| MIR-149 | Module 5 | 0.648 | 1.371 | 0.000 | 0.114 | TCTGGCTCCGTGTCTTCACT |
ES: Enrichment score
NES: Normalized enrichment score
NOM: Nominal p-value
3.6 Comparison of miRNA levels between MMC modules and between PE, IUGR, and Control groups
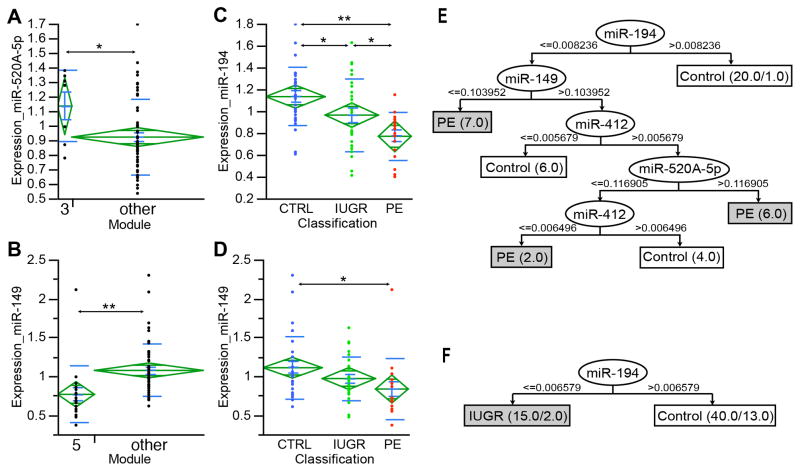
Results showed that the expression level of miR-520A-5p (p=0.0427) and miR-149 (p=0.0017) were significantly different in module 3 and module 5 respectively, when compared to all other modules (Table 5 and Figure 1A, B). Similarly, miR-194 and miR-149 were significantly different between PE, IUGR, and control groups (Table 5 and Figure 1C, D). The microRNA qPCR results were used to build a decision tree in order to determine their potential usefulness as diagnostic biomarkers. As shown in Figure 1E, for preeclampsia, a decision tree with an average predictive accuracy of 66.7% was identified. For IUGR, a single variable decision tree with an average predictive accuracy of 58.2% was identified (Fig. 1F)
Table 5.
Analysis of variance of expression levels of candidate microRNAs
| Human microRNAs | Module affected | Module t-test p-value | Classification F-test p-value | Control - PE p-value | Control - IUGR p-value | IUGR-PE p-value |
|---|---|---|---|---|---|---|
| miR-194 | Module 3 | 0.2119 | 0.0004** | 0.0001** | 0.0304* | 0.0344* |
| miR-412 | Module 3 | 0.9812 | 0.2085 | 0.1091 | 0.1794 | 0.6629 |
| miR-520A-5p | Module 3 | 0.0427* | 0.9175 | 0.6801 | 0.8455 | 0.8113 |
| miR-149 | Module 5 | 0.0017** | 0.0507 | 0.0168* | 0.1542 | 0.2432 |
| miR-483 | Module 5 | 0.7375 | 0.6126 | 0.7851 | 0.4506 | 0.3643 |
| miR-503 | Module 5 | 0.1722 | 0.4062 | 0.2272 | 0.3056 | 0.7523 |
Significant at p < 0.05
Significant at p < 0.01
Figure 1.
4. Discussion
Intrauterine growth restriction (IUGR), as reflected by the birth of small for gestational age (SGA) infants, has a complex etiology including maternal smoking, undernutrition, infection or congenital abnormalities. IUGR can occur alone or associated with preeclampsia [17]. In a previous study [6] we had reported 128 differentially expressed genes (conservative Bonferroni-corrected p-value<0.05) and 2109 differentially expressed genes (FDR<0.05) between preeclamptic and normal placentas. In this study, in contrast, only 1 differentially expressed gene was identified between IUGR and normal placentas at a conservative Bonferroni significance level, supporting the high degree of heterogeneity of IUGR. As a result, we utilized a new clustering method, Modulated Modularity Clustering (or MMC) in an attempt to identify subsets of IUGR and/or preeclamptic samples that had similar molecular signatures.
There is a vast array of clustering methods available, and most methods are in fact a family of approaches that require the user to supply additional arbitrary specifications. We compared MMC to other approaches, and considered a number of clustering methods including hierarchical clustering [18] and k-mean clustering [8]. Both of these approaches require the user to specify the number of clusters, either directly (for k-means) or indirectly (for hierarchical clustering) by choosing where to sever the tree. Two cases were considered: to parallel the a priori category grouping, three clusters were chosen, whereas to parallel the MMC results, six cluster were selected. A correlation-based distance was used for both methods, and additionally squared Euclidean distance for k-means was considered. In total, we compared MMC to six other clustering scenarios. None of the tested methods generated biologically meaningful clusters except k-mean clustering when specifying six clusters a priori, for which the results are essentially the same as those obtained with MMC clustering (supplemental figure 4). The difference being that for K-means we has to arbitrarily select the number of clusters thus biasing the results. MMC in contrasts used the data to determine the number of “natural” cluster in the dataset. This makes MMC a stronger approach than k-means or hierarchical clustering.
MMC successfully identified two groups of placentas that had unique molecular signatures. One of these modules (module 3) is associated with IUGR and the other (module 5) with both IUGR and preclampsia. Essentially, what MMC accomplished was to take a pool of highly heterogeneous IUGR placentas and identify within that group a subset that had a common gene expression signature. While this does not resolve the issue of high heterogeneity of the disease it does, at the very least, allow for a greater understanding of a subset of IUGR patients. As shown in Table 2, the percentile birth weight represented by placentas in Module 3 was 2.4% suggesting that these are placentas from severally growth-restricted babies. Examination of other clinical parameter did not show anything remarkable about these cases compared to other severely restricted IUGR cases in our sample population, so at this time we have no clear explanation as to why this group of placentas is unique compared to other IUGR cases.
The validity of the MMC result is supported by the following: a) 11/16 preeclamptic placentas and a smaller subset (7) of IUGR placentas clustered in module 5. Examination of the clinical data for these IUGR placentas indicated that 4/7 had been classified originally as both IUGR and preeclamptic. b) The differentially expressed genes we had previously identified in preeclamptic placentas [6] are essentially the same as those identified in Module 5 including known preeclampsia associated genes. c) All eight significant pathways we previously reported as disregulated in preeclampsia [6] are represented in the pathway analysis of module 5. d) When “classification” is replaced with “module” in the variance component analysis, the independent contribution of module to gene expression variation is larger than that of classification (Supplemental figure. 1). This reinforces the hypothesis that IUGR may be a highly heterogeneous disease, and that the currently used binary classification may not be capturing the underlying biological variability.
Moreover, linear model comparison of module 3 patients resulted in the identification of 326 differentially expressed genes that are associated with this subset of IUGR placentas. Included in the top ten differentially expressed genes are: ARPC3 (actin related protein 2/3 complex, subunit 3) essential for trophoblast outgrowth and implantation [19]; TGM2, transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase), involved in trophoblast intercellular fusion [20]; EVI5 (ecotropic viral integration site 5), involved in collective cell migration [21]; IL18 (interleukin 18 ; interferon-gamma-inducing factor), involved in the regulation of placental inflammation and a known contributor to the pathogenesis of preeclampsia [22,23]; and DKFZP564K142, an implantation-associated protein [24,25]. Also, many transcripts revealed in module 3 are involved in migration, invasion, allergy or hypertension disorders. Similarly, pathway analysis identified P13K/AKT signaling, mTOR and p70S6K signaling as being affected and these pathways are known to be involved in IUGR and placental growth [26–28]. Also, IGF-1 signaling, a well-known regulator of fetal growth, was also affected [29,30]. Our results provides for the first time an opportunity to closely examine the gene expression profile unique to a subset of IUGR patients.
We were also able to utilize the MMC results to identify a set of microRNAs, miR-149, miR-194, and miR-520a, associated with preeclampsia and/or IUGR. miR-149 has been detected previously in the plasma of pregnant women [31] but no association with either PE or IUGR has been reported. miR-194 has been implicated in cancer metastasis/cell migration [32,33] but there are no previous reports of association with PE or IUGR. miR-520a has been detected in the maternal plasma at 35 weeks of gestation and was found to be unaffected by placental insufficiency-related complications [34].
In addition, putative targets of differentially expressed miRNAs (miR-194 and miR-149) were analyzed based on the gene set information from GSEA. Target genes that are core enriched in GSEA process are shown in supplemental table 3 and 4, with the expression profiles in placenta. Most of these targets are represented in the differentially expressed genes by classification or by module (supplemental figure 3), and several were implicated in the pathogenesis of preeclampsia. SLC6A8, a putative target of miR-149 and up-regulated in our preeclampsia samples, was functionally related to preeclampsia by transporting creatine into and out of cells, since creatine is an important predictor for preeclampsia [35]. Another putative target of miR-149, IGFBP5, was functionally associated with preeclampsia by the interaction with PAPPA2, a known preeclampsia related gene [36]. Similarly, BTBD7 and ARHGAP21, putative targets of miR-194, may be functionally associated with preeclampsia. BTBD7 is reported to promote epithelial tissue remodeling and formation of branched organs [37]. ARHGAP21 (also known as ARHGAP10), promotes activation of RhoA [38], which is in the center of RhoA signaling enriched in differentially expressed genes of module 5 and has a major role in the mechanisms of enhanced vascular reactivity in preeclampsia [39]. Finally, NFAT5 (nuclear factor of activated T cells), putative target of miR-194, was reported to regulate placental osmolytes inositol and sorbitol in the ovine model of IUGR [40].
The decision tree modeling was used to explore the potential of these expression variables as predictors of disease outcomes. We used internal model validation to estimate the potential predictive accuracy of the resulting models, but future studies should evaluate the performance on independent data.
In short, the evidence shows that the MMC analysis was capable of identifying unique molecular signatures, and that in two cases (modules 3 and 5) those molecular signatures are associated with disease.
Conclusions
MMC allowed the elucidation of a molecular signature associated with a subset of IUGR samples. This allowed the identification of genes, pathways, and microRNAs affected in this disease.
Supplementary Material
Acknowledgments
The authors thank all the women who agreed to participate in this study, which was supported by NIH Grant HD048510 to JP and AJ, and is part of an initiative from the Center for Comparative Medicine and Translational Research at the North Carolina State University Collage of Veterinary Medicine.
Abbreviations
- IUGR
Intrauterine Growth Restriction
- PE
Preeclampsia
- GSEA
Gene Set Enrichment Analysis
- MMC
Modulated Modularity Clustering
Footnotes
Author’s contributions
LG, performed the microRNA experiment, helped analyze the data, and drafted the manuscript. ST, NH, ES and CD assisted with experimental design and data analysis. AM assisted with data analysis and manuscript writing. BT, ST coordinated study and collected patient samples. JP and AJ designed and coordinated experiments and led the manuscript preparation. All authors read and approved of the final manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Redman CW, Sargent IL. Latest advances in understanding preeclampsia. Science. 2005;308(5728):1592–4. doi: 10.1126/science.1111726. [DOI] [PubMed] [Google Scholar]
- 2.Sibai B, Dekker G, Kupferminc M. Pre-eclampsia. Lancet. 2005;365:785–99. doi: 10.1016/S0140-6736(05)17987-2. [DOI] [PubMed] [Google Scholar]
- 3.Grill S, Rusterholz C, Zanetti-Dälenbach R, Tercanli S, Holzgreve W, Hahn S, et al. Potential markers of preeclampsia--a review. Reprod Biol Endocrinol. 2009;7:70. doi: 10.1186/1477-7827-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duley L. The global impact of pre-eclampsia and eclampsia. Semin Perinatol. 2009;33(3):130–7. doi: 10.1053/j.semperi.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Haram K, Søfteland E, Bukowski R. Intrauterine growth restriction. Int J Gynaecol Obstet. 2006;93(1):5–12. doi: 10.1016/j.ijgo.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 6.Tsai S, Hardison NE, James AH, Motsinger-Reif AA, Bischoff SR, Piedrahita JA, et al. Transcriptional profiling of human placentas from pregnancies complicated by preeclampsia reveals disregulation of sialic acid acetylesterase and immune signalling pathways. Placenta. 2011;32(2):175–82. doi: 10.1016/j.placenta.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stone EA, Ayroles JF. Modulated modularity clustering as an exploratory tool for functional genomic inference. PLoS Genet. 2009;5(5):e1000479. doi: 10.1371/journal.pgen.1000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinley D. K-means clustering: A half-century synthesis. British Journal of Mathematical and Statistical Psychology. 2006;59(Pt 1):1–34. doi: 10.1348/000711005X48266. [DOI] [PubMed] [Google Scholar]
- 9.Bild A. Application of a priori established gene sets to discover biologically important differential expression in microarray data. Proc Natl Acad Sci U S A. 2005;102(43):15278–9. doi: 10.1073/pnas.0507477102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Report of the National High Blood Pressure Education Program Working Group on High Blood Pressure in Pregnancy. Am J Obstet Gynecol. 2000;183(1):S1–S22. [PubMed] [Google Scholar]
- 11.Mao F, Leung W-Y, Xin X. Characterization of EvaGreen and the implication of its physicochemical properties for qPCR applications. BMC Biotechnol. 2007;7:76. doi: 10.1186/1472-6750-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Idaghdour Y, Czika W, Shianna KV, Lee SH, Visscher PM, Martin HC, et al. Geographical genomics of human leukocyte gene expression variation in southern Morocco. Nat Genet. 2010;42(1):62–7. doi: 10.1038/ng.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo W, Sarkar SK, Peddada SD. Controlling false discoveries in multidimensional directional decisions, with applications to gene expression data on ordered categories. Biometrics. 2010;66(2):485–92. doi: 10.1111/j.1541-0420.2009.01292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbosa-Morais NL, Dunning MJ, Samarajiwa SA, Darot JFJ, Ritchie ME, Lynch AG, et al. A re-annotation pipeline for Illumina BeadArrays: improving the interpretation of gene expression data. Nucleic Acids Res. 2010;38(3):e17. doi: 10.1093/nar/gkp942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank E, Hall M, Trigg L, Holmes G, Witten IH. Data mining in bioinformatics using Weka. Bioinformatics. 2004;20(15):2479–81. doi: 10.1093/bioinformatics/bth261. [DOI] [PubMed] [Google Scholar]
- 17.Villar J, Carroli G, Wojdyla D, Abalos E, Giordano D, Ba’aqeel H, et al. Preeclampsia, gestational hypertension and intrauterine growth restriction, related or independent conditions. Am J Obstet Gynecol. 2006;194(4):921–31. doi: 10.1016/j.ajog.2005.10.813. [DOI] [PubMed] [Google Scholar]
- 18.Loewenstein Y, Portugaly E, Fromer M, Linial M. Efficient algorithms for accurate hierarchical clustering of huge datasets: tackling the entire protein space. Bioinformatics. 2008;24(13):i41–9. doi: 10.1093/bioinformatics/btn174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yae K, Keng VW, Koike M, Yusa K, Kouno M, Uno Y, et al. Sleeping beauty transposon-based phenotypic analysis of mice: lack of Arpc3 results in defective trophoblast outgrowth. Mol Cell Biol. 2006;26(16):6185–96. doi: 10.1128/MCB.00018-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson NJ, Baker PN, Jones CJP, Aplin JD. A role for tissue transglutaminase in stabilization of membrane-cytoskeletal particles shed from the human placenta. Biol Reprod. 2007;77(4):648–57. doi: 10.1095/biolreprod.107.061747. [DOI] [PubMed] [Google Scholar]
- 21.Laflamme C, Assaker G, Ramel D, Dorn JF, She D, Maddox PS, et al. Evi5 promotes collective cell migration through its Rab-GAP activity. J Cell Biol. 2012;198(1):57–67. doi: 10.1083/jcb.201112114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu NQ, Kaplan AT, Lagishetty V, Ouyang YB, Ouyang Y, Simmons CF, et al. Vitamin D and the regulation of placental inflammation. J Immunol. 2011;186(10):5968–74. doi: 10.4049/jimmunol.1003332. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Liu Y, Ding M, Wang X. Reduced expression of indoleamine 2,3-dioxygenase participates in pathogenesis of preeclampsia via regulatory T cells. Mol Med Report. 2011;4(1):53–8. doi: 10.3892/mmr.2010.395. [DOI] [PubMed] [Google Scholar]
- 24.Yogi A, Callera GE, Antunes TT, Tostes RC, Touyz RM. Vascular biology of magnesium and its transporters in hypertension. Magnes Res. 2010;23(4):S207–15. doi: 10.1684/mrh.2010.0222. [DOI] [PubMed] [Google Scholar]
- 25.Li F-Y, Lenardo MJ, Chaigne-Delalande B. Loss of MAGT1 abrogates the Mg2+ flux required for T cell signaling and leads to a novel human primary immunodeficiency. Magnes Res. 2011;24(3):S109–14. doi: 10.1684/mrh.2011.0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arroyo JA, Brown LD, Galan HL. Placental mammalian target of rapamycin and related signaling pathways in an ovine model of intrauterine growth restriction. Am J Obstet Gynecol. 2009;201(6):616.e1–7. doi: 10.1016/j.ajog.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yung H-W, Calabrese S, Hynx D, Hemmings BA, Cetin I, Charnock-Jones DS, et al. Evidence of placental translation inhibition and endoplasmic reticulum stress in the etiology of human intrauterine growth restriction. Am J Pathol. 2008;173(2):451–62. doi: 10.2353/ajpath.2008.071193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roos S, Powell TL, Jansson T. Placental mTOR links maternal nutrient availability to fetal growth. Biochem Soc Trans. 2009;37(Pt 1):295–8. doi: 10.1042/BST0370295. [DOI] [PubMed] [Google Scholar]
- 29.Klammt J, Pfäffle R, Werner H, Kiess W. IGF signaling defects as causes of growth failure and IUGR. Trends Endocrinol Metab. 2008;19(6):197–205. doi: 10.1016/j.tem.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Netchine I, Azzi S, Le Bouc Y, Savage MO. IGF1 molecular anomalies demonstrate its critical role in fetal, postnatal growth and brain development. Best Pract Res Clin Endocrinol Metab. 2011;25(1):181–90. doi: 10.1016/j.beem.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 31.Chim SS, Shing TK, Hung EC, Leung TY, Lau TK, Chiu RW, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54(3):482–90. doi: 10.1373/clinchem.2007.097972. [DOI] [PubMed] [Google Scholar]
- 32.Le XF, Almeida MI, Mao W, Spizzo R, Rossi S, Nicoloso MS, et al. Modulation of MicroRNA-194 and cell migration by HER2-targeting trastuzumab in breast cancer. PloS One. 2012;7(7):e41170. doi: 10.1371/journal.pone.0041170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song Y, Zhao F, Wang Z, Liu Z, Chiang Y, Xu Y, et al. Inverse association between miR-194 expression and tumor invasion in gastric cancer. Ann Surg Oncol. 2012;19 (Suppl 3):S509–17. doi: 10.1245/s10434-011-1999-2. [DOI] [PubMed] [Google Scholar]
- 34.Hromadnikova I, Hromadnikova I, Hromadnikova I, Hromadnikova I, Hromadnikova I, Kotlabova K, et al. Absolute and relative quantification of placenta-specific micrornas in maternal circulation with placental insufficiency-related complications. J Mol Diagn. 2012;14(2):160–7. doi: 10.1016/j.jmoldx.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 35.Millar JG, Campbell SK, Albano JD, Higgins BR, Clark AD. Early prediction of pre-eclampsia by measurement of kallikrein and creatinine on a random urine sample. Br J Obstet Gynaecol. 1996;103(5):421–6. doi: 10.1111/j.1471-0528.1996.tb09767.x. [DOI] [PubMed] [Google Scholar]
- 36.Nishizawa H, Pryor-Koishi K, Suzuki M, Kato T, Kogo H, Sekiya T, et al. Increased levels of pregnancy-associated plasma protein-A2 in the serum of pre-eclamptic patients. Mol Hum Reprod. 2008;14(10):595–602. doi: 10.1093/molehr/gan054. [DOI] [PubMed] [Google Scholar]
- 37.Onodera T, Sakai T, Hsu JC, Matsumoto K, Chiorini JA, Yamada KM. Btbd7 regulates epithelial cell dynamics and branching morphogenesis. Science. 2010;329(5991):562–5. doi: 10.1126/science.1191880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anthony DF, Sin YY, Vadrevu S, Advant N, Day JP, Byrne AM, et al. β-Arrestin 1 inhibits the GTPase-activating protein function of ARHGAP21, promoting activation of RhoA following angiotensin II type 1A receptor stimulation. Mol Cell Biol. 2011;31(5):1066–75. doi: 10.1128/MCB.00883-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mishra N, Nugent WH, Mahavadi S, Walsh SW. Mechanisms of enhanced vascular reactivity in preeclampsia. Hypertension. 2011;58(5):867–73. doi: 10.1161/HYPERTENSIONAHA.111.176602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arroyo JA, Garcia-Jones P, Graham A, Teng CC, Battaglia FC, Galan HL. Placental TonEBP/NFAT5 osmolyte regulation in an ovine model of intrauterine growth restriction. Bio Reprod. 2012;86(3):94. doi: 10.1095/biolreprod.111.094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.