Abstract
Knowledge discovery from large and complex scientific data is a challenging task. With the ability to measure and simulate more processes at increasingly finer spatial and temporal scales, the growing number of data dimensions and data objects presents tremendous challenges for effective data analysis and data exploration methods and tools. The combination and close integration of methods from scientific visualization, information visualization, automated data analysis, and other enabling technologies —such as efficient data management— supports knowledge discovery from multi-dimensional scientific data. This paper surveys two distinct applications in developmental biology and accelerator physics, illustrating the effectiveness of the described approach.
Keywords: scientific visualization, information visualization, data analysis, multi-dimensional data, laser wakefield particle acceleration, 3D gene expression
1. Introduction
Knowledge discovery from large and complex collections of today’s scientific datasets is a challenging task. Due to advances in data acquisition and scientific computing, today’s datasets are becoming increasingly complex. With the ability to measure and simulate more processes at finer scales, the number of data dimensions and data objects has grown significantly in today’s scientific datasets, while the phenomena researchers are able to investigate become increasingly complex. Researchers are overwhelmed with data and standard tools are often insufficient to enable efficient data analysis and, hence, discovery of information and knowledge from the data.
We address these challenges via a combination of scientific visualization, information visualization, automated data analysis, and other enabling technologies. The tight coupling of different analysis methods and tools supports knowledge discovery from complex, multi-dimensional scientific data. To illustrate the effectiveness of this approach, we survey the processes and tools used to analyze: i) 3D gene expression data, and ii) laser wakefield particle acceleration data, demonstrating the applicability of the described basic concept to a large range of applications.
Analysis of 3D gene expression data is linked to the more general problem of understanding the control of embryo development, which is a fundamental question in biology. A cell’s unique fate is determined by specific combinations of developmental regulatory factors. These factors form part of complex genetic regulatory networks, which ultimately coordinate the expression of all genes. In order to study these complex systems, the BDTNP2 has developed so called PointCloud data, a novel type of spatial and temporal gene expression data. Single PointClouds are obtained via segmentation of two-photon microscopy images of whole Drosophila embryos and provide a quantitative representation of spatial gene expression levels of the Drosophila blastoderm at cellular resolution [1]. Multiple PointClouds representing a variety of genes at various developmental time points are registered into a single Atlas PointCloud describing the expression of about one hundred genes at multiple points in time [2]. Analysis of 3D gene expression data is challenging in particular due to the large number of data dimensions (genes) and the complex interactions between them.
Laser wakefield particle accelerators (LWFAs) [3] utilize an electron plasma wave to accelerate charged particles (e.g., electrons) to high energy levels over very short distances [4, 5]. Analysis, understanding, and control of the complex physical processes of plasma-based particle acceleration requires understanding of how particle beams are formed and accelerated. These processes are best understood by tracing the particles that form a beam over time and studying their temporal evolution [6, 7, 8, 9]. In laboratory experiments, however, it is impossible to record the complete evolution of a beam and much less to trace single particles within a plasma. Researchers from the LOASIS3 project perform simulation of LWFA experiments using VORPAL [10], in order to better understand the fundamental physics of plasma-based acceleration and the processes involved in experiments, as well as to improve experiments [11]. The datasets produced by LWFA simulations are (i) extremely large, (ii) of varying spatial and temporal resolution, (iii) heterogeneous, and (iv) high-dimensional, making analysis and knowledge discovery from complex LWFA simulation data a challenging task.
Section 2 introduces our general approach for knowledge discovery from multi-dimensional scientific data. We demonstrate the broad applicability of the described methodology in a survey of the analysis processes and tools used to analyze 3D gene expression (Section 3) and laser wakefield particle acceleration data (Section 4).
2. General Methodology
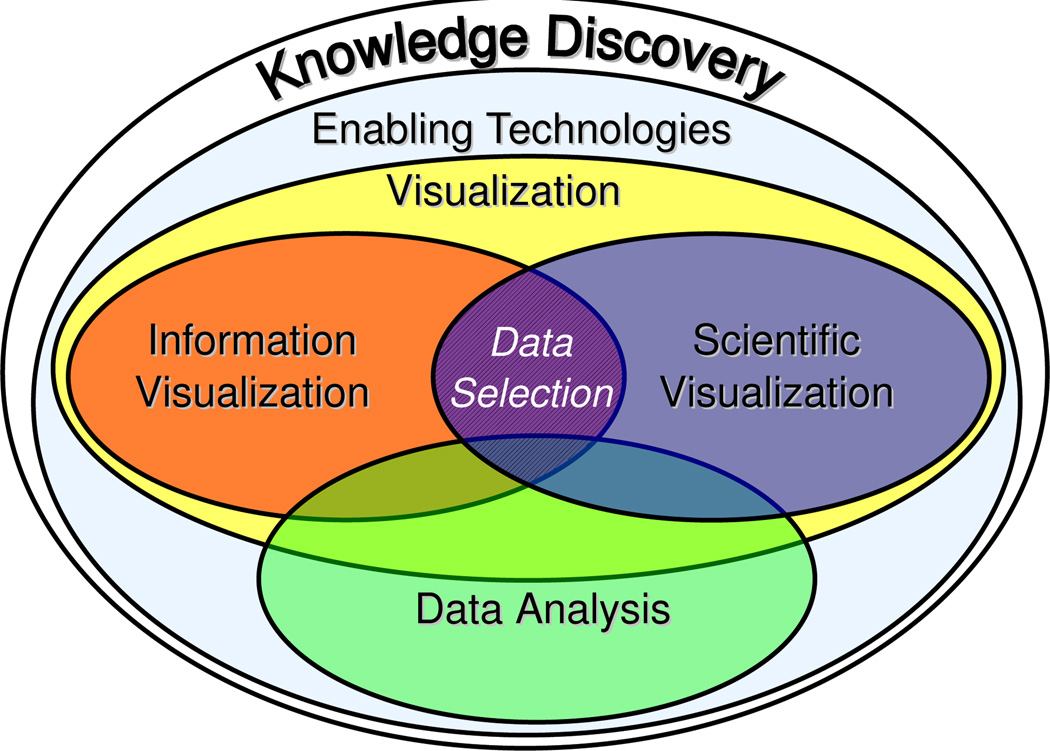
While the challenges in developmental biology and accelerator physics research are quite different, the same basic analytic methodology can be used for knowledge discovery from such complex data. The basic approach is based on the unique combination and close integration of: (i) enabling technologies, (ii) visualization, and (iii) data analysis (Figure 1). Enabling technologies are fundamental methods, needed for data analysis, that are not necessarily part of the analysis itself, e.g., methods for data retrieval, access, and management.
Figure 1.
Overview of the basic components we use in combination to enable knowledge discovery from multi-dimensional scientific data.
Visualization transforms data into readily comprehensible images and is an indispensable part of the scientific discovery process [12]. In particular in the context of multi-dimensional data, a single display is often not sufficient to reveal all aspects of the data. Scientific visualizations support detailed analysis of physical data characteristics, while information visualizations provide means for exploration of the variable space and identification of relationships between different data dimensions. We use multiple views —each highlighting different aspects of the data— linked via the concept of data selection (brushing) [13, 14, 15]. Selected data subsets can be highlighted in any view enabling detailed analysis and knowledge discovery.
While interactive data exploration based on linked multiple views is effective, it also has limitations. Manual data exploration can be time-consuming — hindering the analysis of large data collections — and visual detection of all fine and subtle data features is often impossible. Automated data analysis methods promise to overcome these limitations of visual data analysis by assisting in the most complex and time-consuming steps of the analysis pipeline, e.g., through automated feature detection [16].
In practice, interpretation of automated analysis results is often unintuitive and may lead to false interpretations, and proper definition of analysis parameters is often complicated. By linking automated data analysis and visualization, we overcome the difficulties with both visual and automated data analysis. Automating the detection of data features of interest enables, e.g., development of advanced visualizations that focus on the main data portions of interest, significantly reducing clutter and occlusion of important information. At the same time, visualization eases validation and interpretation of analysis results and definition of input parameters. Ultimately, it is the tight and meaningful integration of all these different methods that enables us to effectively discover new knowledge.
3. Application I: 3D Gene Expression Data
With the availability of 3D PointCloud gene expression data, new ways for analyzing the complex genetic regulatory networks controlling animal development are becoming possible. PointCloud data describes the output of these complex networks quantitatively. The information stored in a PointCloud can be represented as a table in which each row represents a single cell of the embryo containing information about: i) the location of the cell in physical space (x,y,z), ii) its neighbors, iii) the estimated cell-volume and surface normal, and iv) the recorded expression levels for currently up to ≈ 100 genes at multiple time intervals. PointCloud data effectively transforms hundreds of gigabytes of image data into a for computation easily accessible format, enabling analysis of the spatial patterns of gene expression, their temporal variation, and regulation.
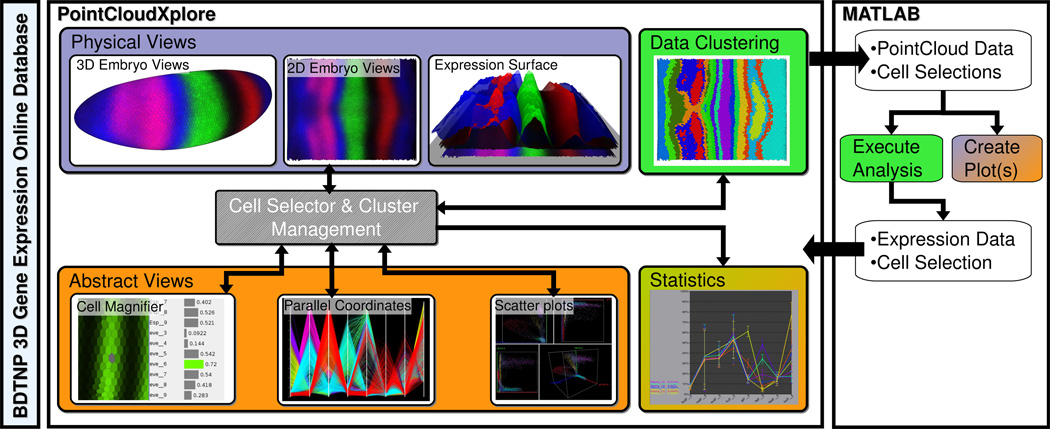
PointCloudXplore (PCX) [17] is a visualization and analysis system specifically developed for the analysis of 3D PointCloud data (Figure 2). PCX supports analysis of spatial gene expression patterns via dedicated 2D and 3D physical model representations of the embryo blastoderm [18]. The 3D embryo views show the morphology of the embryo blastoderm and allow biologists to study the spatial expression patterns of genes relative to the shape of the embryo. The 2D embryo views describe 2D projections of the embryo blastoderm and provide an overview of all blastoderm cells of the embryo. Dedicated information visualizations (abstract views) provide means for exploration of gene expression space and identification of relationships between genes. Parallel coordinates and scatter-plots are used for analysis and comparison of the expression of multiple genes in all cells of the embryo while the Cell Magnifier provides an overview of all expression values of a single cell via a bar-graph view. The concept of cell selection (brushing) allows the user to correlate the information shown in different views [19]. The user can select cells of interest in any view, e.g., via drawing on the embryo surface or via thresholding in parallel coordinates. This mechanism allows features of interest to be defined and highlighted in any view, making PointCloudXplore an effective tool for rapid data exploration (Figure 3).
Figure 2.
The system for knowledge discovery from 3D gene expression data. Color indicates the areas the different components of the system belong to, i.e., enabling technologies (light blue), scientific visualization (lilac), information visualization (orange), and data analysis (green).
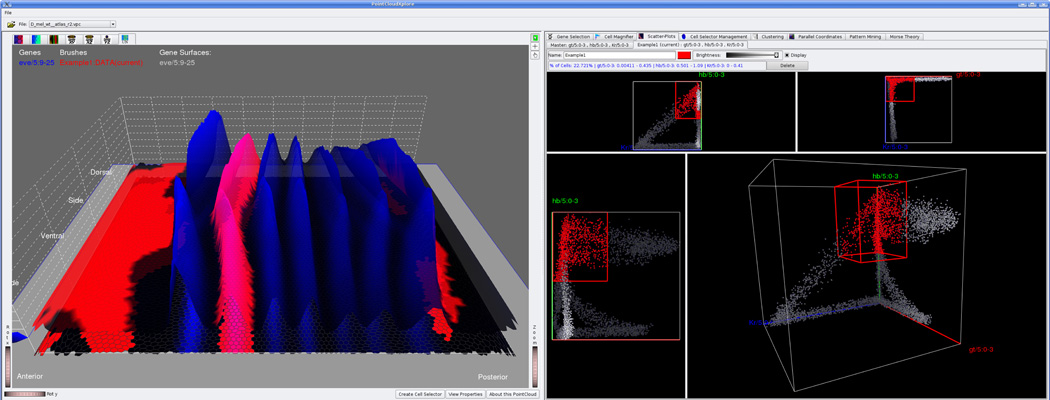
Figure 3.
Screenshot of PointCloudXplore showing an expression surface of eve (left) and 2D/3D scatter-plots of gt, hb, and Kr (right). The user selected a set of cells (red) via thresholding in the scatter-plots. The same cells are highlighted in the embryo view (left). A characteristic subset of the selected cells coincides with the second stripe of the pattern of eve, indicating that gt, hb, and Kr could potentially be involved in the regulation of eve stripe 2.
While visualization is a powerful approach for knowledge discovery from complex data sets, visual detection of all existing features is very difficult in this case due to the large number and subtlety of features and intricate nature of 3D gene expression data. A typical feature of interest defines, e.g., various groups of cells behaving similarly with respect to the expression of several genes or a single gene over time. In the context of conceptually simpler forms of expression data — such as microarray experiments — data clustering has already shown to be able to reveal details hidden in the data [20]. However, appropriately defining clustering parameters — such as the number of clusters — as well as validation and interpretation of clustering results, is still complicated.
PCX integrates data clustering directly with the visualization. Using a combination of visualization and dedicated algorithms for evaluating the quality of clustering results, the user can intuitively identify appropriate clustering parameters [21]. A cluster defines a selection of cells behaving similarly with respect to the expression of the set of genes used in the clustering process. Similar to user-defined cell selections, PCX can directly display and highlight automatically computed clusters in any view. The meaningful integration of data clustering with the visualization improves the visualization as well as the clustering process. Data clustering supports automatic detection and highlighting of data features in the visualization, enabling a more focused and accurate analysis process. Visualization provides effective means for accurate definition of clustering input parameters and allows intuitive validation and interpretation of clustering results.
In particular in the context of novel scientific data — such as PointCloud data — researchers need to be able to quickly develop new analysis functions. PCX addresses this need by providing an interface to MATLAB (The Math- Works Inc., Natick, MA, USA), allowing researchers to integrate custom analysis capabilities with PCX and providing biologists faster and more convenient access to advanced analysis functions [22]. With its interface to MATLAB, PCX supports fast prototyping and testing of new ideas, thus facilitating communication between bioinformatics researchers and experimental biologists. The close integration of MATLAB with the visualization improves the visualization by providing simple access to new, advanced analysis capabilities as well as the analysis implemented in MATLAB by providing efficient means for validation and exploration of analysis results.
While the different methods—i.e., scientific visualization, information visualization, and automated data analysis — are each useful in their own right, it is ultimately the combination and close integration of all these methods that allows us to effectively analyze PointCloud data. Automatic data analysis methods are commonly used for feature detection and to manipulate and summarize large amounts of information. Data visualization provides effective means for data exploration as well as analysis, validation, and control of the automatic data analysis.
4. Application II: Laser Wakefield Particle Acceleration Data
Analysis of and knowledge discovery from large, complex, multi-dimensional laser wakefield particle accelerator (LWFA) simulation data is a challenging task. Scientists of the LOASIS project model LWFA experiments computationally via particle-in-cell (PIC) simulations using VORPAL [10] to better understand nonlinear plasma response, beam trapping, self-consistent laser propagation, and beam acceleration—processes not accessible to analytic theory. In PIC simulations, collections of real charged particles are modeled as computational macro-particles while the electromagnetic field is spatially discretized via a computational grid. Particles are moved under Newton-Lorentz force obtained through interpolation from the fields. The current carried by the moving particles is then deposited onto the simulation-grid to solve Maxwell’s equations for the fields. For each dump —i.e., a snapshot of the simulation at a particular point in time — data about the particles, fields, and auxiliary state data is saved. For the analysis discussed here we mainly focus on the particle data. Each particle is represented as a vector describing the physical location (x, y, z), momentum (px, py, pz), identifier (id), and weight (wt) of the macro-particle. One main feature researchers are interested in are beams of high-energy particles formed during the course of LWFA simulations. To enable efficient and accurate data analysis, dedicated mechanisms for beam selection and detection are needed.
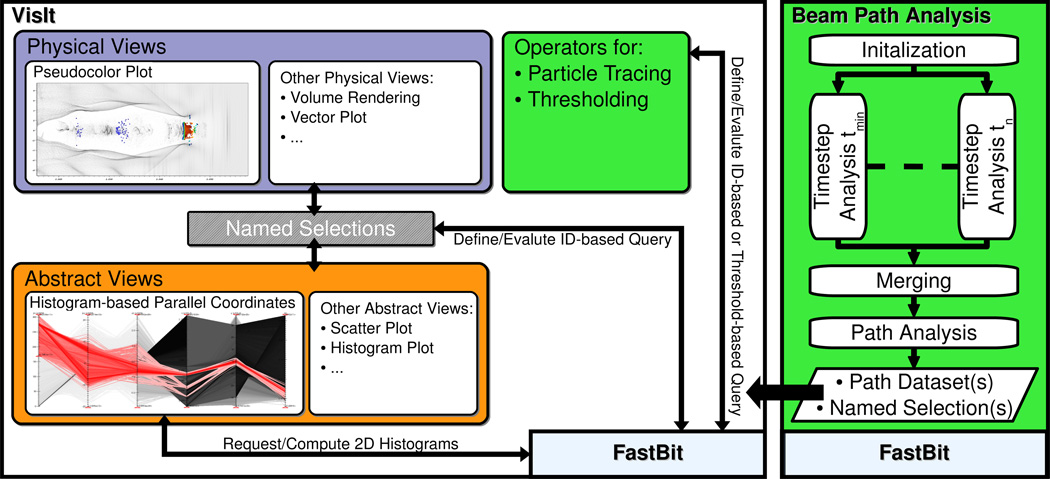
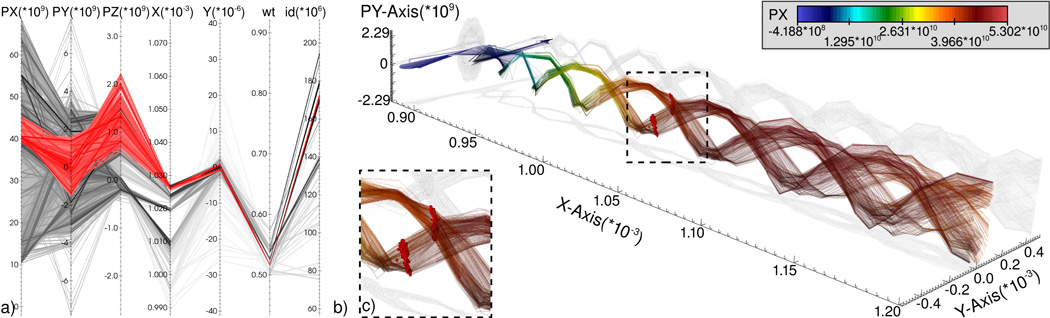
Figure 4 provides an overview of the system for knowledge discovery from LWFA simulations. The index/query system FastBit [23] serves as main interface to the data enabling fast computation of conditional histograms, threshold queries, ID-based queries, and particle tracing. The close integration of FastBit and the state-of-the-art visualization system VisIt [24] supports fast visual exploration of very large datasets [8]. VisIt implements an efficient rendering method for parallel coordinates based on 2D histograms, computed directly using FastBit. Histogram-based parallel coordinates serve as the main interface for defining multi-dimensional range queries used for selection of particle beams (Figure 5a). Once a subset of particles of interest has been identified, the particle IDs are saved as a named selection. Named selections can be applied to any plot in VisIt, enabling effective linking of multiple physical and abstract data views (Figure 5b, c). While parallel coordinates serve as the main interface for data selection, VisIt also supports creation of named selections based on a large range of other views allowing the user, e.g., to select particles in physical views using a bounding box. Common methods used for visualization of the particle data include point-based pseudocolor and scatter plot visualizations as well as density-based visualizations, such as histogram views and parallel coordinates. For visualization of the field data we commonly use vector plots, volume rendering, and iso-surface-based visualizations. The close integration of multiple views via the concept of named selections together with efficient data management supports interactive data exploration based on the iterative refinement and validation of data queries.
Figure 4.
The system for knowledge discovery from laser wakefield particle accelerator simulation data. Color indicates the areas the different components of the system belong to, i.e., enabling technologies (light blue), scientific visualization (lilac), information visualization (orange), and data analysis (green).
Figure 5.
a) Parallel coordinates of time step t = 86 of a particle dataset showing all particles with a momentum in x direction of px > 1e10 (gray) and a selected particle beam (red). b) Semi-transparent rendering of the paths of the selected particles over time, using time steps 0 through 105. Color indicates px (the momentum in the acceleration direction x) and height indicates momentum in y direction (py). The particles of the selected beam at time step t = 86 are shown in addition (red). The transition in color from blue to red along the particle paths shows that the selected particles are constantly accelerated over time. The cork-screw-like structure of the paths illustrates the oscillating motion of the particles in the wake. c) Close-up view of the region in figure b showing the selected particles at time step t = 86.
While interactive selection of particle beams is effective, it requires substantial manual input from the user and can be time-consuming. Automating the detection of particle beams supports a more focused and efficient analysis process [9, 25]. The beam path analysis algorithm [25] defines an efficient analysis pipeline that supports fast detection of particle beams (Figure 4 right). First, each time step is analyzed independently to detect individual particle bunches. The derived information is merged to define a single description for each main particle beam. Finally, the algorithm computes the different temporal phases of each beam — defining, e.g., the time frame when a beam was formed and accelerated — as well as two distance fields ds and dmdefining the distance of particles to the beam in physical and momentum space, respectively.
The automated beam detection is linked with the visualization in two ways. First, the beam path analysis automatically creates a set of named selections — one per detected beam — that can be applied to any plot in VisIt, enabling a fully automated beam analysis process. Second, a set of complementary files is created, containing additional information about the particle paths. These files can be visualized directly in the context of the simulated data and enable a more efficient manual data exploration process by providing information about: i) the temporal phases of a beam, ii) an appropriate reference time step for each beam, and iii) the beam distance fields ds and dmenabling a faster and more accurate selection of particle beams. Linking the automated beam detection with the visualization improves the visualization by enabling a more streamlined and focused analysis process as well as the automated analysis by providing effective means for investigation and validation of analysis results.
5. Conclusions
The increasing complexity and size of today’s scientific data poses tremendous challenges for data understanding and knowledge discovery. We have described an integrative approach for knowledge discovery from multidimensional scientific data based on the concept of linking visualization and data analysis. We have illustrated the effectiveness of this methodology by describing how it is applied in practice in the analysis of 3D gene expression and laser wakefield particle accelerator data.
The challenges we faced in both applications were quite different. First, LWFA simulation datasets are extremely large (several TBs) making computationally efficient data management methods, such as FastBit, indispensable. In direct comparison, PointCloud data sets consist of relatively few data objects —i.e., on the order of 6000 cells in the case of Drosophila PointCloud data compared to several million particles in LWFA simulation data— but contain information about many more data dimensions, i.e., gene expressions. Furthermore, the features researchers are interested in are quite different in the two applications. Particle beams are very small compared to the complete data. Effective means for fast data reduction are, therefore, essential for efficient analysis of LWFA simulation data. In the visual data exploration process, the user step-by-step refines advanced data queries while using multiple views for analysis and validation of query results. Similarly, the automatic beam path analysis is aimed at identifying specific small features of interest, i.e., particle beams. In contrast, the goal of the automatic data analysis in the context of 3D gene expression is usually to manipulate and summarize large amounts of information rather than extracting a specific small data subset. Data clustering is here often used to subdivide the complete data into meaningful groups (clusters), each defining a set of cells with similar expression behavior.
Despite the large differences between 3D PointCloud and LWFA simulation data, the basic concepts used to analyze these different types of data are similar. While different applications require different visual representations and analysis methods, it is the powerful combination and close integration of multiple different methods that enables effective knowledge discovery. The meaningful integration of visualization, data analysis, and enabling technologies (including efficient data storage, organization, and access methods) supports a more efficient, detailed, and focused analysis process than possible based solely on either visualization or data analysis methods alone.
Acknowledgment
The work concerning the analysis of 3D gene expression data was supported by the National Institutes of Health through grant GM70444 and by Lawrence Berkeley National Laboratory (LBNL) through the Laboratory Directed Research Development (LDRD) program. Research performed at LBNL was also supported by the Department of Energy under contract DE-AC02-05CH11231. In addition, we gratefully acknowledge the support of an International Research Training Group (IRTG 1131) grant provided by the German Research Foundation (DFG), awarded to the University of Kaiserslautern, Germany.
The work concerning the analysis of LWFA data was supported by the Director, Office of Science, Offices of High Energy Physics and Advanced Scientific Computing Research, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231 through the Scientific Discovery through Advanced Computing (SciDAC) program’s Visualization and Analytics Center for Enabling Technologies (VACET) and the COMPASS project. This research used resources of the National Energy Research Scientific Computing Center (NERSC), which is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
We thank the members of IDAV at UC Davis, the BDTNP and LOASIS at LBNL, the IRTG 1131, LBNL’s Visualization Group, and the VORPAL team.
Footnotes
Berkeley Drosophila Transcription Network Project (BDTNP): http://bdtnp.lbl.gov/Fly-Net/
Lasers, Optical Accelerator Systems Integrated Studies (LOASIS): http://loasis.lbl.gov/
References
- 1.Luengo Hendriks CL, Keränen SVE, Fowlkes CC, Simirenko L, Weber GH, DePace AH, Henriquez C, Kaszuba DW, Hamann B, Eisen MB, Malik J, Sudar D, Biggin MD, Knowles DW. Three-dimensional morphology and gene expression in the Drosophila blastoderm at cellular resolution I: Data acquisition pipeline. Genome Biology. 2006;7(12):R123. doi: 10.1186/gb-2006-7-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fowlkes CC, Luengo Hendriks CL, Keränen SVE, Weber GH, Rübel O, Huang M-Y, Chatoor S, DePace AH, Simirenko L, Henriquez C, Beaton A, Weiszmann R, Celniker S, Hamann B, Knowles DW, Biggin MD, Eisen MB, Malik J. A quantitative spatio-temporal atlas of gene expression in the drosophila blastoderm. Cell. 2008;133:364–374. doi: 10.1016/j.cell.2008.01.053. [DOI] [PubMed] [Google Scholar]
- 3.Tajima T, Dawson JM. Laser electron accelerator. Phys. Rev. Lett. 1979;43(4):267–270. [Google Scholar]
- 4.Geddes C, Toth C, van Tilborg J, Esarey E, Schroeder C, Bruhwiler D, Nieter C, Cary J, Leemans W. High-Quality Electron Beams from a Laser Wakefield Accelerator Using Plasma-Channel Guiding. Nature. 2004;438:538–541. doi: 10.1038/nature02900. [DOI] [PubMed] [Google Scholar]
- 5.Leemans WP, Nagler B, Gonsalves AJ, Toth C, Nakamura K, Geddes CGR, Esarey E, Schroeder CB, Hooker SM. GeV electron beams from a centimetre-scale accelerator. Nature Physics. 2006;2:696–699. [Google Scholar]
- 6.Geddes CGR. Ph.D. thesis. Berkeley: University of California; 2005. Plasma Channel Guided Laser Wakefield Accelerator. [Google Scholar]
- 7.Tsung FS, Narang R, Mori WB, Joshi C, Fonseca RA, Silva LO. Near-GeV-Energy Laser-Wakefield Acceleration of Self-Injected Electrons in a Centimeter-Scale Plasma Channel. Physical Review Letters (PRL) 93:185002. doi: 10.1103/PhysRevLett.93.185002. [DOI] [PubMed] [Google Scholar]
- 8.Rübel O, Prabhat, Wu K, Childs H, Meredith J, Geddes CGR, Cormier-Michel E, Ahern S, weber GH, Messmer P, Hagen H, Hamann B, Bethel EW. SuperComputing 2008 (SC08) Austin, Texas, USA: 2008. High Performance Multivariate Visual Data Exploration for Extemely Large Data. [Google Scholar]
- 9.Ushizima D, Rübel O, Prabhat, Weber G, Bethel EW, Aragon C, Geddes C, Cormier-Michel E, Hamann B, Messmer P, Hagen H. Proceedings of The Seventh International Conference on Machine Learning and Applications 2008 (ICMLA 08) Los Alamitos, CA, USA: IEEE Computer Society Press; 2008. Automated Analysis for Detecting Beams in Laser Wakefield Simulations; p. 382387. [Google Scholar]
- 10.Nieter C, Cary JR. VORPAL: A Versatile Plasma Simulation Code. J. Comput. Phys. 2004;196(2):448–473. [Google Scholar]
- 11.Geddes CGR, Bruhwiler DL, Cary JR, Mori WB, Vay J-L, Martins SF, Katsouleas T, Cormier-Michel E, Fawley WM, Huang C, Wang X, Cowan B, Decyk VK, Esarey E, Fonseca RA, Lu W, Messmer P, Mullowney P, Nakamura K, Paul K, Plateau GR, Schroeder CB, Silva LO, Toth C, Tsung FS, Tzoufras M, Antonsen T, Vieira J, Leemans WP. Computational Studies and Otimization of Wakefield Accelerators. Journal of Physics: Conference Series V. 2008;125:12002/1–12002/11. [Google Scholar]
- 12.Hansen CD, Johnson CR. The Visualization Handbook. Elsevier Academic Press; 2005. [Google Scholar]
- 13.Wang Baldonado MQ, Woodruff A, Kuchinsky A. AVI ’00: Proceedings of the working conference on Advanced visual interfaces. New York, NY, USA: ACM Press; 2000. Guidelines for using multiple views in information visualization; pp. 110–119. [Google Scholar]
- 14.Henze C. Feature detection in linked derived spaces. In: Ebert D, Rushmeier H, Hagen H, editors. Proceedings IEEE Visualization ’98. Los Alamitos, CA, USA: IEEE Computer Society Press; 1998. pp. 87–94. [Google Scholar]
- 15.Doleisch H, Gasser M, Hauser H. Interactive feature specification for focus+context visualization of complex simulation data. In: Bonneau G-P, Hahmann S, Hansen CD, editors. Data Visualization 2003 (Proceedings of the Eurographics/IEEE TCVG Symposim on Visualization) 2003. pp. 239–248. [Google Scholar]
- 16.Garth C, Tricoche X. Topology- and feature-based flow visualization: Methods and applications. SIAM Conference on Geometric Design and Computing. 2005 [Google Scholar]
- 17. PointCloudXplore is available from http://bdtnp.lbl.gov/Fly-Net/bioimaging.jsp?w=pcx.
- 18.Rübel O, Weber GH, Keränen SVE, Fowlkes CC, Luengo Hendriks CL, Simirenko L, Shah NY, Eisen MB, Biggin MD, Hagen H, Sudar D, Malik J, Knowles D, Hamann B. Pointcloudxplore: Visual analysis of 3d gene expression data using physical views and parallel coordinates. In: Santos BS, Ertl T, Joy K, editors. Data Visualization 2006 (Proceedings of EuroVis 2006) Aire-la-Ville, Switzerland: Eurographics Association; 2006. pp. 203–210. [Google Scholar]
- 19.Weber GH, Rübel O, Huang M-Y, DePace AH, Fowlkes CC, Keränen SV, Luengo Hendriks CL, Hagen H, Knowles DW, Malik J, Biggin MD, Hamann B. Visual exploration of three-dimensional gene expression using physical views and linked abstract views. IEEE Transactions on Computational Biology and Bioinformatics. 2009;6(2):296–309. doi: 10.1109/TCBB.2007.70249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rübel O, Weber GH, Huang M-Y, Bethel EW, Biggin MD, Fowlkes CC, Luengo Hendriks CL, Keränen SVE, Eisen MB, Knowles DW, Malik J, Hagen H, Hamann B. Integrating data clustering and visualization for the analysis of 3d gene expression data. IEEE Transactions on Computational Biology and Bioinformatics. 2010;7(1):64–79. doi: 10.1109/TCBB.2008.49. [DOI] [PubMed] [Google Scholar]
- 22.Rübel O, Keränen SVE, Biggin MD, Knowles DW, H.Weber G, Hagen H, Hamann B, Bethel EW. Mathematical Methods for Visualization in Medicine and Life Sciences. Springer Verlag; 2009. Linking Advanced Visualization and MATLAB for the Analysis of 3D Gene Expression Data. (submitted) [Google Scholar]
- 23. FastBit is available from https://codeforge.lbl.gov/projects/fastbit/.
- 24. VisIt is available from https://wci.llnl.gov/codes/visit/. [Google Scholar]
- 25.Rübel O, Geddes CGR, Cormier-Michel E, Wu K, Prabhat, Weber GH, Ushizima DM, Messmer P, Hagen H, Hamann B, Bethel W. Automatic beam path analysis of laser wakefield particle acceleration data. IOP Computational Science & Discovery 2 (015005) 2009:38. [Google Scholar]