Abstract
Kinesins are ATP-dependent molecular motors that carry cargos along microtubules, generally in an anterograde direction. They are classified into 14 distinct families with varying structural and functional characteristics. KIF17 is a member of the kinesin-2 family that is plus end-directed. It is a homodimer with a pair of head motor domains that bind microtubules, a coiled-coil stalk, and a tail domain that binds cargos. In neurons, KIF17 transports N-methyl-D-aspartate receptor NR2B subunit, kainate receptor GluR5, and potassium Kv4.2 channels from cell bodies exclusively to dendrites. These cargos are necessary for synaptic transmission, learning, memory, and other functions. KIF17’s interaction with NXF2 enables the transport of mRNA bidirectionally in dendrites. KIF17 or its homolog OSM-3 also mediates intraflagellar transport of cargos to the distal tips of flagella or cilia, thereby aiding in ciliogenesis. In many invertebrate and vertebrate sensory cells, KIF17 delivers cargos that contribute to chemosensory perception and signal transduction. In vertebrate photoreceptors, KIF17 is necessary for outer segment development and disc morphogenesis. In the testis, KIF17 (KIF17b) mediates microtubule-independent delivery of ACT from the nucleus to the cytoplasm and microtubule-dependent transport of Spatial-ε, both are presumably involved in spermatogenesis. KIF17 is also implicated in epithelial polarity and morphogenesis, placental transport and development, and the development of specific brain regions. The transcriptional regulation of KIF17 has recently been found to be mediated by nuclear respiratory factor 1 (NRF-1), which also regulates NR2B as well as energy metabolism in neurons. Dysfunctions of KIF17 are linked to a number of pathologies.
Keywords: ciliary transport, kinesin-2 family, microtubules, NR2B transport, transcriptional control
1. Kinesin superfamily proteins
The kinesin superfamily proteins (KIFs) are microtubule-based molecular motors that convert the chemical energy of ATP hydrolysis to the mechanical force of transporting cargos along microtubules. These ATPases were first identified and partially purified from squid giant axons and optic lobes, the bovine brain, the chick brain, and sea urchin eggs [1–3]. They form a high affinity complex with microtubules in the presence of a non-hydrolyzable ATP analog, and their kinetic property prompted the term “kinesin” [1] as the counterpart to the other microtubule-associated ATPase, dynein, described two decades earlier [4]. More than 98% of all KIFs (611 out of 623 KIF proteins) have been classified into 14 distinct families (kinesin-1 to -14) with many subfamily groupings [5, 6]. Their similarity resides principally in their conserved microtubule and ATP binding head domain, while their diversity in other structural features is associated with diverse functions, such as intracellular transport of vesicles, organelles, protein complexes, and mRNAs. KIFs also contribute to spindle and chromosomal stabilization and movement during mitosis and meiosis, engage in neuronal function and survival, and help in the regulation of microtubule dynamics and developmental patterning [6–9]. Unlike dynein, which moves cargos toward the minus-ends of microtubules [10], almost all kinesins (with the exception of KIF2a, kinesin-14, and perhaps kinesin-5 Cin8 when acting alone) move cargo toward the plus-ends of microtubules [6, 11]. In neurons, kinesins mediate primarily fast anterograde transport away from the cell body, as opposed to cytoplasmic dynein, which moves cargo toward the cell body [1, 12].
A typical kinesin, such as conventional kinesin 1, is composed of two heavy chains (each ~ 124 kDa) and two light chains (each ~ 64 kDa) [13]. Each heavy chain consists of three parts: (a) a structurally conserved globular head motor domain that contains a catalytic site for ATP hydrolysis and a binding site for microtubules; (b) an α-helical coiled-coil stalk domain that permits protein-protein interactions connected to the head domain via a family-specific flexible neck linker; and (c) a tail domain that binds cargos [6, 13, 14]. The light chains typically bind near the tail domain of the heavy chain [13] and is found, at least in kinesin-1, to promote cargo transport by suppressing the auto-inhibitory kinesin heavy chain tail-head and tail-microtubule interactions [15]. There are many exceptions to these organizing principles, including monomeric arrangement of the heavy chain (kinesin-3), the absence of a coiled-coil stalk domain (kinesin-13), a neck linker that is N-terminal to the catalytic core (kinesin-14), and the absence of a cargo binding light chain (kinesin-2) [6, 13, 14]. Members of the kinesin superfamily are found in protozoa, fungi, plants, invertebrates, and vertebrates; and at least 45 of them are encoded in the genomes of mice, rats, and humans [6]. The majority of KIFs have their motor domain or catalytic core at the N-terminus (formerly called N-kinesins); some, such as the kinesin-13 family, have it in the middle (previously known as M-kinesin) and may exhibit microtubule depolymerizing activity; whereas still others have the motor domain at the C-terminus, as exemplified by the kinesin-14 family members that are minus-end directed [6, 7, 16, 17]. Kinesin-14 Ncd also has the ability to bind microtubules via both its head and tail domains, thus enabling crosslinking and sliding of adjacent microtubules, such as in oocyte spindle assembly [18].
A large number of kinesin-associated proteins have also been identified. For example, the Miro/Milton is an adaptor protein complex that assists kinesin-1’s binding to its cargo, the mitochondrion [19], and Miro1 links mitochondria to KIF5 to be transported to postsynaptic sites [20]. The kinesin superfamily-associated protein 3 (KAP3) was originally purified as the 115 kDa subunit from sea urchin eggs in the heterotrimeric complex of kinesin-related protein KRP85/95/115 [21] and later from the mouse brain as a complex of 3 high molecular weight polypeptides (hence the name) that associate with KIF3A and KIF3B of the kinesin-2 family to mediate anterograde transport of membranous organelles [22]. This protein was first sequenced in sea urchin eggs [23] and cloned as well as characterized in the mouse brain [24]. KAP3 has splice variants with a single species in mouse testis, and the tissue-specific composition of KAP3 may determine functional diversity of the KIF3 complex in different cell types [22]. Furthermore, this heteromeric kinesin-2 motor is regarded as that canonical motor driving anterograde intraflagellar transport in virtually all cilia and flagella and is essential for ciliogenesis [25].
An excellent review of molecular motors, especially in neurons, appeared recently [26]. The present review focuses mainly on a key member of the kinesin-2 family, KIF17, and its diverse functions.
2. Phylogenetic relationships of kinesin-2 family members
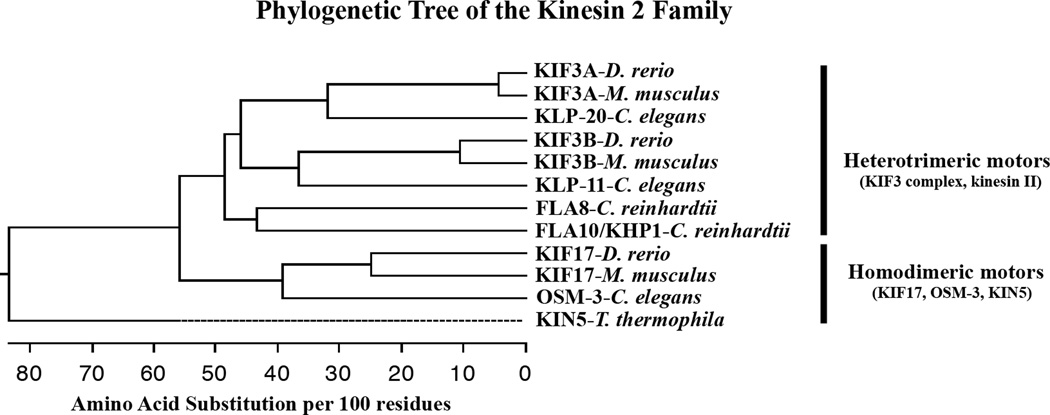
Phylogenetic analysis based on protein sequence has significantly simplified our understanding of the many kinesin gene families as well as the relationships of the many sequences from many species that are included in the kinesin-2 family (see [6] and associated website). As shown in Figure 1, which is a simplified kinesin 2 phylogenetic tree, the kinesin heavy chains in this family fall into two distinct classes. KIF17 and its homologues (OSM-3 and KIN5) form a class of related homodimeric motors in which two heavy chains associate through coiled-coil domains to form a functional motor with a C-terminal cargo binding domain. The second class of kinesin-2 motors is derived from closely related heavy chains that are referred to in vertebrates as KIF3A, KIF3B, and KIF3C [6, 22, 27–29]. As originally shown for proteins purified from sea urchin [21], these heavy chains or their homologues in other species, such as Danio rerio, Caenorhabditis elegans, and Chlamydomonas reinhardtii that are included in Figure 1 form heterotrimeric complexes consisting of two different heavy chains along with a C-terminal bound, cargo binding protein such as KAP3 [24]. The heterotrimer is often referred to as the KIF3 complex or simply as kinesin II [22, 29, 30]. The kinesin-2 family has not yet been found in fungi or higher plants, which are species that do not produce sperm or have cilia or flagella [6]. Although there are two evolutionarily conserved kinds of kinesin-2 family motors, the nomenclature can be challenging because of the plethora of gene/protein names and the frequent historical use of the term “kinesin II” for the heterotrimeric motor. From a functional point of view, the concept of a homodimeric motor compared to a heterotrimeric motor is most useful in distinguishing the two members of the kinesin-2 family (see Figure 1).
Figure 1.
Phylogenetic tree showing the relationship of kinesin 2 family heavy chains discussed in this review. The tree was constructed with the Clustal W method of multi-sequence alignment using DNASTAR software. A more complete analysis of all kinesin heavy chains including the kinesin 2 family can be seen in [6] and the associated website. The tree shows evolutionary distance measured as substitutions per 100 amino acids using the full-length sequences of each heavy chain. Included are the heterotrimeric heavy chains KIF3A and KIF3B from Danio rerio and Mus musculus. The comparable heavy chains, KLP-20 and KLP-11, from Caenorhabditis elegans, and FLA10/KHP1 and FLA8 from Chlamydomonas reinhardtii are included as well. FLA8 is the heavy chain binding partner of FLA10 [58]. The homodimeric heavy chains include KIF17 from D. rerio and M. musculus, OSM-3 from C. elegans, and KIN5 from Tetrahymena thermophila. The dashed line associated with KIN5 represents a negative branch length caused by averaging during the alignment. A homodimeric kinesin 2 motor has not been described in C. reinhardtii.
3. Kinesin superfamily protein KIF17
KIF17 is a 170 kDa protein cloned from the mouse brain that is highly expressed in the gray matter of the central nervous system, but not in the white matter. It was reportedly absent in other organs, such as the lung, heart, liver, spleen, kidney, and skeletal muscle [27] and was considered to be neuron-specific, although an earlier listing of the same protein claimed it to be testis-specific [31]. Now it is known that KIF17 is expressed in both neurons and testis, as well as in many other cell types with possible isoforms. KIF17 is a plus end-directed, homodimeric N-kinesin (N-4) belonging to the kinesin-2 family. KIF17 is the mammalian homolog of the molecular motor OSM-3 (osmotic avoidance abnormal protein 3; a dendritic motor for odorant receptors) in C. elegans and Kin5 in Tetrahymena thermophila, both mediating ciliary transport [32, 33]. The homodimeric KIF17 has a pair of head motor domains that bind to microtubules, a coiled-coil stalk, and a tail domain that binds cargos [6, 20, 22]. In the absence of cargo, KIF17 (or OSM-3) is thought to undergo auto-inhibition by folding about its central hinge to suppress motility [34, 35] (Figure 2). The folding enables the coiled-coil 2 (CC2) segment and the C-terminal tail domain of KIF17 to interact directly with the dimeric head motor domain. Two mechanisms have been proposed for autoinhibition: a) interaction of tail domain with the motor domain prevents the latter from binding to microtubules; and b) the CC2 segment interacts with the motor domain and inhibits processive motility [35]. Inhibition is relieved by the binding of cargos [34, 35]. The speed of KIF17 transport is calculated to be 0.8 to 1.2 µm/s, consistent with fast intracellular transport [27]. Kinesin-2 family members are involved in the transport of vesicles, melanosomes, intraflagellar proteins [6, 36], as well as many other functions. In intraflagellar transport, OSM-3 is thought to serve as an accessory motor interacting with the KIF3 complex for assembly of the cilium [37]. Those functions that pertain specifically to KIF17 (or OSM-3) are discussed in section 4.
Figure 2.
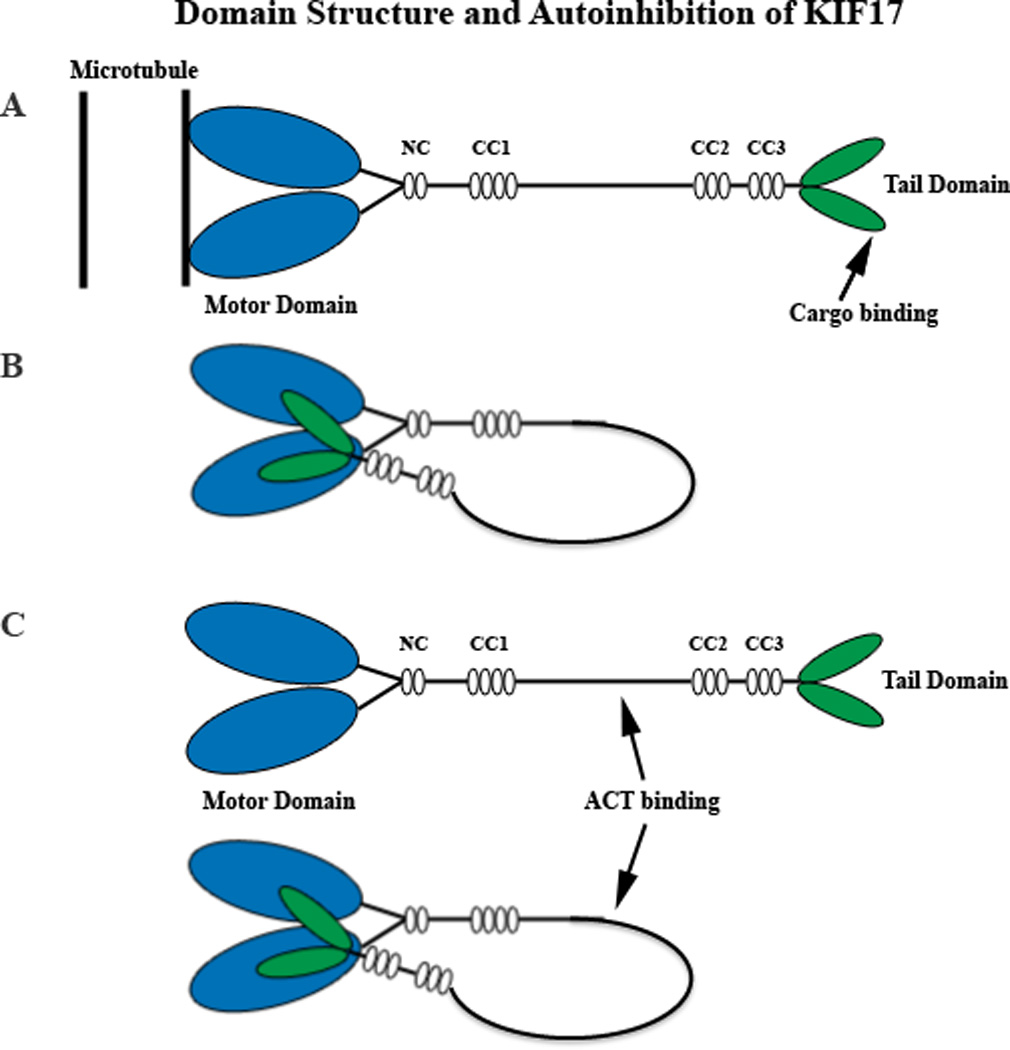
Diagrammatic representation of the KIF17 homodimer in its active (A) and auto-inhibited (B) configurations. In B, the cargo binding tail domain (green) associates with the microtubule binding ATPase head domain (blue). Cargo binding could involve intermediate adaptor complexes, as in the case of NR2B trafficking in dendrites or IFT trafficking in cilia. However, in the testis, ACT binds to an intermediate domain (C) within the stalk rather than the tail in a regulatory event that occurs independently of KIF17 motor activity and microtubules. The structural configuration of KIF17 for ACT binding is not known, but in theory could occur in either the extended or auto-inhibited configuration. Not drawn to scale. CC, coiled-coil domains 1, 2, and 3; NC, neck coil domain.
4. Diverse functions of KIF17
4.1 Dendritic transport of NR2B and synaptic transmission
When KIF17 was first discovered in the murine brain, it was found to selectively transport N-methyl-D-aspartate receptor subunit 2B (NR2B, GluN2B, or NRε2) in vesicles along microtubules from the cell body exclusively to dendrites of neurons [27]. Entry into axons is permissible for vesicles carried by some kinesins, such as KIF5, but is strictly prohibited for NR2B-vesicles carried by KIF17, indicating a selective filtering system at the initial segment of axons that directs cargo trafficking to specific neuronal compartments [38]. The velocity of movement along dendrites in brain slices is 0.7 µm/s [39]. The transport is via a specific binding of the tail domain of KIF17 to the C-terminal first PDZ (postsynaptic density-95/disc large/zona occludens-1) domain of the murine homolog of C. elegans Lin-10 within a large scaffolding protein complex that also includes the murine homologs of C. elegans Lin-2 and Lin-7 [27]. Lin-10 is comparable to the mammalian Mint 1/X11, a synaptic receptor sorting protein that contains two PDZ domains [40]. Lin-2 is similar to the mammalian CASK, a dlg/PSD95 homolog that interacts with neurexin [41]. CASK and another membrane-associated guanylate kinase (MAGUK) family of synaptic scaffold protein, SAP97, are required for the trafficking of NR1/NR2B-containing vesicles from somatic endoplasmic reticulum (ER) to dendritic ER and dendritic Golgi outposts, powered by the microtubule-dependent KIF17 [42]. Lin-7 is homologous to MALS/Velis, a mammalian or vertebrate LIN-7 protein [43, 44] with a single PDZ domain that binds to the C-terminus of NR2B [44]. Thus, the mLin complex links KIF17 to the NR2B (and possibly the ubiquitous NR1)-containing vesicles to be transported along microtubules to their dendritic destination [27], where NR2B forms an integral subunit of the postsynaptic glutamatergic NMDA receptor. The binding of KIF17 to the PDZ domain of mLin-10 is very precise, as disruption of either the first PDZ domain of mLin-10 or the C-terminus of KIF17 reduces or eliminates such interactions, and dendritic transport of NR2B is prohibited [27]. Such interaction is also regulated by a CaMKII (Ca2+/calmodulin-dependent protein kinase II)-dependent phosphorylation of KIF17’s tail region (at serine 1029), as phosphorylation disrupts the KIF17-Mint1 association and causes the release of cargo from its transporting microtubule [45]. A knockdown of KIF17 with chronic exposure to antisense oligonucleotides or a dominant negative construct down-regulates the expression of NR2B and mLin10, but up-regulates the expression of NR2A in hippocampal neurons [46]. Antisense knockdown of KIF17 in prefrontal cortical neurons reduces the basal whole cell NMDA receptor currents, and the application of a microtubule depolymerizing agent nocodazole, which normally decreases NMDA-evoked currents, has no effect on NMDA receptor currents in KIF-17 knockdown neurons [47]. Thus, NMDA receptor currents appear to be regulated by KIF17-mediated transport of NR2B along microtubules to dendrites. Up-regulating NR2B with an NMDA receptor antagonist D(-)-2-amino-5-phosphonopentanoic acid increases the expression of KIF17 [46]. These findings are consistent with a dual regulation of the cargo and its motor in neurons (see section 5).
4.2 Dendritic transport of kainate receptor subunit GluR5
Glutamate receptors can be broadly subdivided into NMDA and non-NMDA receptors. The latter can be further classified into AMPA, kainate, and metabotropic receptors [48–50]. Thus far, KIF17 has been linked primarily to the transport of NR2B in the CNS [27]. More recently, however, the kainate receptor GluR5 was found to be transported by KIF17 in a complex with other kainate receptor subunits GluR6 and KA2 from the soma to distal dendrites in cultured hippocampal neurons [51]. Whereas the transport of GluR5 to the distal dendrites is dependent on the presence of GluR6, KA2, as well as KIF17, its transport to the proximal dendrites is independent of all three [51], raising the possibility that other transport motors may be involved in the initial somato-dendritic transport of GluR5 and that KIF17 takes over when GluR5 is ready to complex with GluR6 and KA2 in distal dendrites.
4.3 Dendritic transport of potassium Kv4.2 channels
Besides NR2B, KIF17 also transports the voltage-gated K+ channel Kv4.2 from the cell body to dendrites of neurons [52]. Kv4.2 is found exclusively in dendrites and is a major regulator of dendritic excitability [52]. Although the extreme C-terminus of Kv4.2 (and not the dileucine dendritic targeting motif) is found to interact with KIF17, it is not known if the interaction is direct or indirect; and although KIF17 has been determined to be necessary for the localization of Kv4.2, by itself KIF17 does not specify dendritic localization of the channel protein [52]. These results suggest the participation of a linking protein, such as the Lin complex found for the transport of NR2B. Lin-7 (MALS) proteins, in particular, have been suspected to contribute to dendritic and postsynaptic targeting of neuronal transmembrane proteins [44].
4.4 Dendritic transport of mRNA
In vertebrate cells, the export of poly(A)+ RNA from the nucleus to the cytoplasm is mediated by a conserved family of proteins known as Tap/NXF (Tip-associated protein/nuclear RNA export factors) [53, 54]. Within the NXF family, NXF2 has been found to play an additional role. In the cytoplasm, it interacts with several motor proteins, such as KIF9, DyneinLC1-like protein, and KIF17, and thus may mediate cytoplasmic localization of mRNAs [55]. In cultured hippocampal neurons, the N-terminus of NXF2 interacts with the C-terminus of KIF17, and they form RNA granules that move bidirectionally along dendrites in a microtubule-dependent manner [55]. The movement, however, is much slower than that of vesicles transported by KIF17 (~ 0.13 µm/s versus 0.76 µm/s). NXF2 has also been found to bind FMRP (Fragile × mental retardation protein), a known RNA-binding protein [56], and this binding site coincides with that for KIF17 necessary for dendritic targeting of NXF2 [55]. However, the precise RNA species bound to NXF2 and/or FMRP and transported to dendrites is(are) as yet unknown.
4.5 Intraflagellar transport, ciliogenesis, and chemosensory transduction
Microtubules form the cytoskeletal backbone or axoneme of flagella (9 doublets plus 2 central microtubules), primary cilia (non-motile; 9 + 0), and motile cilia (9 + 2). These microtubules provide not only structural support, but are critical in the genesis, assembly, maintenance, and overall health of cilia and flagella. These processes are mediated, in large part, by intraflagellar transport (IFT) of axoneme precursors and membrane proteins via kinesin and dynein family motors along microtubules [25]. A variety of kinesins have been associated with cilia and flagella, the most prevalent ones are in the kinesin-2 family: KIF3A, KIF3B, and KIF17 or their homologs FLA10/FLA8, KLP-20/KLP-11 and OSM-3 (see Figure 1) that mediate IFT essential for the genesis and maintenance of cilia and flagella in diverse organisms [16, 25, 57, 58]. The discovery of IFT was made in the unicellular green alga, Clamydomonas reinhardtii [59], and it was subsequently found that a homolog of KIF3A known as KHP1 or FLA10 was important for IFT and flagella maintenance [60, 61]. These studies led to the finding that the heterotrimeric kinesin II motor is responsible for the anterograde transport of IFT protein complexes or particles within the flagellum [62]. This concept has now been generalized to include virtually all cilia and flagella [25, 57, 63].
In the chemosensory cilia of the nematode C. elegans, the heterotrimeric CeKinesin II and the homodimeric CeOSM-3, homologues of the KIF3 complex (or kinesin II) and KIF17, respectively, are both involved in the transport of ciliary components or IFT particles essential for the assembly of cilia in chemosensory neurons [25, 32]. The two motors function redundantly to transport IFT particles along doublet microtubules in forming the axoneme’s middle segment, but OSM-3 alone transports the particles along distal singlet microtubules to stabilize the distal segment of amphid channel cilia [64]. Interestingly, Cekinesin II and OSM-3 function completely redundantly in building the full-length of amphid wing cilia in C. elegans [65], and in yet other cilia, OSM-3 functions independently of Cekinesin II [66]. Thus, KIF17/OSM-3 in these specialized neurons takes part in the anterograde transport of axonemal components and membrane proteins in IFT particles, in ciliogenesis, and in chemosensory perception and signal transduction important in mating, egg laying, feeding, movement, and general development [32]. In Tetrahymena thermophila, KIN5 mediates intraciliary transport, just like its homologue OSM-3 [32], while in mammals, KIF17 is endogenously expressed in MDCK (Madin Darby canine kidney) epithelial cells, where it is required for the transport of exogenously expressed cyclic nucleotide-gated channels into the cilium [67].
Although kinesin-2 proteins are the key IFT motors in cilia and flagella, other factors are found to modulate their activities. In the cephalic male cilia of C. elegans, kinesin-3 KLP-6 modulates IFT dynamics and length by decreasing the velocity of OSM-3 and IFT [68]. A ciliary-cytoplasmic gradient of the small Ran GTPase (with high levels in the cilium) was recently proposed to regulate the entry of KIF17 into cilia, as disruption of this gradient prevents such admission [69]. It turns out that the entrance of kinesin motors into the cilium utilizes a mechanism that is very similar to that of protein entry into the nucleus. In the case of KIF17, the KRKK sequence in its tail domain serves as the ciliary localizing signal (CLS), similar to the nuclear localizing signal (NLS) of other proteins. Cytoplasmic interaction of KIF17 with a nuclear import protein importin-β2 is dependent on the CLS, and as the motor/importin complex crosses the ciliary barrier, it is confronted by a high level of RanGTP, which is proposed to dissociate the complex and release KIF17 to perform its function in the cilium [69].
4.6 Transport of cyclic nucleotide-gated channels to cilia of olfactory sensory neurons
KIF17 also exists in mammalian olfactory sensory neurons, where it is thought to play a role in the transport of chemosensory signaling cyclic nucleotide-gated (CNG) channels into the nonmotile cilia [67]. Although CNG channels are clustered in olfactory cilia [70], the data supporting a role for KIF17 are indirect in that they are based on the transport of exogenously expressed CNG channels in kidney epithelial cells. Both KIF3A (or the KIF3 complex) and KIF17 are implicated in chemosensory transduction in olfactory neurons [67]. The precise role of each, however, is not clear, as dominant negative suppression of KIF3A leads to a loss of cilia, while dominant negative KIF17 blocks ciliary targeting of CNG channels without disrupting cilium formation [67]. The C-terminal ciliary-targeting motif, RVxP, in the CNGB1b subunit is also critical, as mutation of this motif results in reduced or failed targeting of the channel to the cilia [67]. However, the mechanism by which the C-terminal domain of CNGB1b interacts with KIF17 and is transported into the cilium is not understood.
4.7 Photoreceptor outer segment development
The outer segment of photoreceptor cells is derived from a primary cilium that extends from the the inner segment of the cell body and remains in differentiated cells as a connecting cilium between the inner and outer segments [71]. This sensory cilium mediates phototransduction via visual pigments embedded in the outer segment discs. The central axoneme of the connecting cilium is made up of a ring-like 9 + 0 doublet microtubules that are linked to the adjacent plasma membrane in a manner similar to that occurring in the proximal transition zone common to virtually all cilia [71]. However, the axoneme extends into the outer segment and, in the most distal domain, the doublets lose their B-tubule to become singlets [72]. As with other sensory cilia, IFT mediated by the KIF3 complex (kinesin II) is essential for forming the outer segment [73, 74].
The organization of the photoreceptor sensory cilium resembles that of C. elegans sensory cilia in two ways. First, it forms from the distal region of the inner segment, which is structurally equivalent to the dendritic region of C. elegans sensory neurons. Second, distal segments of C. elegans sensory cilia often terminate in singlet microtubules. This structural similarity along with the finding that the KIF17 homologue, OSM-3, is required for building distal singlets in C. elegans [64] led directly to the idea that photoreceptor IFT would involve both the KIF3 complex (kinesin II) and KIF17 [75]. KIF17 is found to co-localize with IFT proteins (such as IFT88, IFT20, and IFT52) and mediate their transport in photoreceptor cilia [72, 76]. Immuno-EM analysis of zebrafish photoreceptors confirmed that both motors are associated with microtubule doublets of the connecting cilium and axonemal microtubules in the outer segments, as well as portions of inner segments that assemble IFT complex [72]. At the light microscopic level, KIF17 is found along the entire length of the axoneme [76]. Co-immunoprecipitation studies of mouse retinal extracts indicate that IFT88 interacts with both the KIF3 complex (kinesin II) and KIF17 [72]. Hence, the two family members may function in both cooperative and independent manner within the outer segment. Knocking down KIF17 in zebrafish with antisense morpholino has little effect on early embryogenesis or on the development of pronephric cilia, but the targeting of visual pigment proteins and the formation of outer segments are severely disrupted [75]. The severity of disruption is positively correlated with the degree of deficiency in KIF17, which is most prominent in central retina, where the outer segments almost completely fail to form [75]. To tease apart the respective roles played by the two members of the kinesin-2 family, dominant negative forms of KIF3B or KIF17 driven by a late-onset, cone-specific transducin promoter was generated in zebrafish cone photoreceptors [72, 77]. Dominant negative KIF3B disrupted the functions of both inner segments and synaptic terminals. This was characterized mainly by cone opsin mislocalization, accumulation of vacuoles and dense material in inner segments, and paucity of synaptic ribbons in cone pedicles, leading to cone cell death. Dominant negative KIF17, on the other hand, affected primarily outer segment assembly with disruption of outer segment discs and stunted growth of cone outer segments. Thus, the KIF3 complex and KIF17 have different roles in cone development and maintenance in the zebrafish, and KIF17 is critically important in outer segment development, a form of vertebrate ciliogenesis [72].
4.8 Spermatogenesis
When KIF17 was first listed but not characterized, it was considered “testis-specific” with Northern blotting [27]. Subsequent description of this motor protein in testis has referred to it as KIF17b [78–82]. KIF17b is highly expressed in male germ cells, where it associates with and directs the subcellular localization of a transcriptional coactivator ACT, an activator of cAMP-responsive element modulator or CREM, thereby regulating CREM-dependent transcription in the testis [78, 80, 83]. Interestingly, ACT binds to the central stalk rather than the C-terminus of KIF17b (see Figure 2), and the transport of ACT from the nucleus to the cytoplasm by KIF17b after the onset of spermatid elongation is independent of microtubules and the motor domain of the kinesin. This is a novel, microtubule-independent function of a kinesin; however, the transport is modulated by cyclic AMP-dependent protein kinase A-mediated phosphorylation of KIF17b [84]. ACT null mice are fertile, but have a markedly reduced amount of male sperm, and the remaining ones exhibit severe abnormalities [85]. Although this implies important roles for both ACT and KIF17b in spermatogenesis, loss of function studies have not yet been reported for KIF17b.
KIF17b also interacts with TB-RBP (testis brain RNA-binding protein), a murine orthologue of the human Translin) in the nucleocytoplasmic transport of CREM-regulated mRNAs in male germ cells [79]. When the mRNA cargo is released, it coincides with the commencement of mRNA translation [79]. Thus, it can be inferred that KIF17b is involved in the sequential regulation of transcription and transport, but not the translation, of CREM-dependent mRNAs in the male postmeiotic germ cells. These processes are vital for spermatogenesis.
KIF17b has also been localized to the chromatoid bodies of round spermatids, where it interacts with a testis-specific PIWI/Argonaute family member, MIWI, which is implicated in RNA metabolism as well as in the formation of chromatoid bodies [81]. Thus, KIF17b is implicated in the loading of haploid RNAs in the chromatoid bodies of spermatids.
Yet another cargo of KIF17b in the spermatids is Spatial (stromal protein associated with thymii and lymph-node), a putative nuclear factor highly expressed in thymus, brain, and testis [86]. The expression in testis is developmentally regulated, and appears only in adult mice commencing at 7 – 8 weeks of age, in step 2 – 10 spermatids during spermiogenesis [86]. The Spatial gene gives rise to five alternatively spliced variants (three short isoforms α, β, and γ, and two long isoforms δ and ε) with apparent tissue-specific distribution [87]. It is Spatial-ε that interacts with KIF17b in a microtubule-dependent manner in the manchette of elongating spermatids and in the principal piece of the spermatozoa tail, both of which have highly organized microtubules [82]. The function of KIF17b in these structures is unknown, but is postulated to involve intramanchette transport and intraflagellar transport. A distinct motif at the C-terminus of Spatial-ε is RVHP, which is reminiscent of the RVxP motif in CNGB1b essential for ciliary targeting (see section 4.6). However, definitive experiments have not yet been done to validate the mechanism of interaction between KIF17b and Spatial-ε, nor of their precise functions. Interestingly, Spatial-ε is also found in round spermatids, where KIF17b mediates the microtubule-independent, intracellular localization of ACT (see above), and the three (Spatial-ε, KIF17b, and ACT) can conceivably form a complex [82]. Thus, KIF17b is likely to play multifaceted roles in different stages of spermatogenesis.
4.9 Epithelial polarity and morphogenesis
Epithelial cells are often highly polarized, and this involves selective microtubule stabilization and reorganization. KIF17 has recently been found to contribute to such polarization [88]. The motor domain of KIF17 interacts with the C-terminal coiled-coil region of a plus-end binding protein (+TIP) known as EB1 (end-binding protein 1), and the latter targets KIF17 to the microtubule plus ends [88]. In addition, the coiled-coil region of KIF17 interacts with the N-terminal region of another +TIP protein called APC (adenomatous polyposis coli) and carries APC as a cargo to the plus ends of a subset of microtubules that converge in cell extensions. ATPase activity of KIF17 is necessary for this function and it may be regulated by EB1 [88]. As EB1 and APC are involved in the regulation of microtubule dynamics and stabilization in fibroblasts [89], the tripartite complex (KIF17, EB1, and APC) is hypothesized to play a critical role in stabilizing microtubules and in regulating microtubule dynamics in epithelial cells [88]. The stability of microtubules is enhanced by posttranslational acetylation, which appears to be influenced by KIF17, as a knockdown of KIF17 with shRNA reduces the level of acetylated microtubules [88]. Likewise, silencing KIF17 leads to a 30% faster growth of microtubules in epithelial cells, indicating that KIF17 is involved in the control of microtubule polymerization and dynamics in these cells [88]. By stabilizing specific sets of microtubules, perhaps via capping of the plus ends of microtubules, KIF17 also contributes to the apical-basolateral polarization of cells during epithelial morphogenesis [88]. However, the exact mechanisms by which KIF17 regulates microtubule polymerization and dynamics in epithelial cells remain unclear.
4.10 Placental transport and development
KIF17 has been localized to the vascular endothelium of early, normal and pathological term human placental villi, where vasculogenesis and angiogenesis occur [90]. Its presence in hematopoietic and Hofbauer cells of early but not term placentas raises the possibility that KIF17 may be involved in the development of hematopoietic cells and in placental vascular formation. The strong immunoreactivity of KIF17 in the endothelium of villi in preeclamptic and diabetic placentas implies that its activity is enhanced in these pathological conditions. However, the precise cargo is not known, and the role of KIF17 in placental vascular development and pathologies remains to be explored.
4.11 Development of specific brain regions
Spatial (see section 4.8) is also highly expressed in specific regions of the developing and adult murine central nervous system, such as the developing cerebellum, hippocampus, and the cortex, and its expression is temporally coincidental with the commencement of neuronal differentiation in some of these regions [91]. In primary cultures of hippocampal neurons, Spatial is colocalized with KIF17 in dendrites, and both Spatial-ε and Spatial-β physically interact with KIF17 in co-immunoprecipitation studies [91]. As Spatial-β expression overlaps with the beginning of granule and pyramidal cell differentiation in the hippocampus, it is tempting to suggest that Spatial-β complexes with Spatial-ε as they are being transported from the soma to the dendrites on microtubules powered by KIF17 [91]. However, the precise role of KIF17, if any, in neuronal morphogenesis during brain development awaits future investigation.
4.12 Neuronal plasticity and higher cognitive functions, including learning and memory
As discussed above, a major neuronal cargo of KIF17 is NR2B. It is an important subunit of the ionotropic, voltage-sensitive, heteromeric glutamatergic NMDA receptor, which is made up of the ubiquitous NR1 (GluN1) and various combinations of NR2A, 2B, 2C, and 2D (GluN2A-2D) subunits that differ with age and brain regions [50, 92]. NR2B binds to calcium/calmodulin-dependent protein kinase II (CamKII) at the postsynaptic density (PSD) sites [93], and this interaction is important for synapse formation and maturation and the induction of long-term potentiation (LTP), a cellular basis of learning and memory [94, 95]. NR2B binding is also found to cause an 11-fold increase in the affinity of CaMKII for ATP and confers an energy-efficient switch between phosphorylated and dephosphorylated states in the PSD that supports synaptic memory [96]. The involvement of NR2B in spatial memory performance and consolidation [97, 98] is strengthened by findings that adult transgenic mice with conditional knockout of NR2B in the cortex and CA1 region of the hippocampus exhibit impaired long-term depression, decreased dendritic spine density, and disrupted learning and memory [99]. On the other hand, over-expression of NR2B in the forebrain enhances learning and memory in transgenic mice [100].
The critical role of KIF17 in transporting NR2B subunit to postsynaptic dendrites [27] and the known involvement of NR2B in neuronal plasticity, learning and memory [97, 99] have prompted the question of whether the level of KIF17 directly affects such cognitive functions. Over-expression of KIF17 is found to enhance spatial and working memory in transgenic mice [39]. They learn the Morris water maze tasks more rapidly and they show significantly shorter escape latencies than their wild type littermates [39]. In these animals, the level of NR2B proteins in the cortex and hippocampus is up-regulated 1.5-fold and that of NR1 is also slightly increased. The level of NR2A proteins, on the other hand, is slightly reduced. By the same token, the level of NR2B mRNA is also up-regulated nearly 2-fold as compared to the wild type [39]. Consistent with these findings, recent loss-of-function studies in KIF17−/− mice [101] show reduced NR2B transport, impaired long-term potentiation, and impaired performance in tests of learning and memory.
4.13 Emotion and cognition
A link between KIF17 and cortical control of emotion and cognitive function can be gleaned from a cascade of events that mediate serotonergic regulation of NMDA receptors via a microtubule-dependent mechanism in prefrontal cortical neurons. It is known that one of the serotonergic receptors, 5-HT1A receptors, is highly expressed in the prefrontal cortex [102] and that it is up-regulated in schizophrenic patients [103]. On the other hand, 5-HT1A receptor null mice exhibit increased anxiety [104]. 5-HT1A receptors inhibit NMDA receptor-mediated ionic and synaptic currents by reducing microtubule stability, inhibiting the transport of NR2B by KIF17, and reducing the density of surface NR2B subunits on dendrites of prefrontal cortical neurons [105]. Cellular knock-down of KIF17 blocks the 5-HT1A effect on NMDA receptor-mediated currents [105]. Inhibition of CaMKII or MEK/ERK (mitogen-activated protein kinase kinase/extracellular signal-regulated kinase) also abolishes the 5-HT1A modulation of NMDA receptor-mediated currents [105], indicating that these kinases are involved in the transport of NR2B, perhaps by phosphorylating MAP2 (microtubule-associated protein 2), a known dendritic protein [106]. Chronic administration of methamphetamine for 2 weeks induces an increase in protein expression of both KIF17 and NR2B (NRε2) in the frontal cortex of mice, suggesting that both proteins may be involved in enhanced response to glutamate after repeated exposure to amphetamine [107].
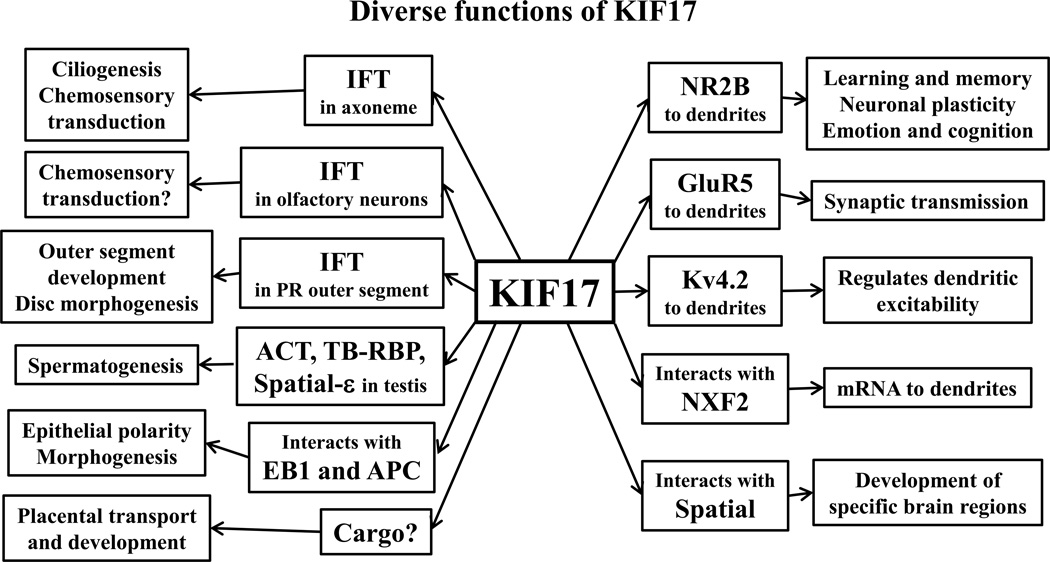
Thus, KIF17 performs a variety of functions. These are schematically summarized in Figure 3.
Figure 3.
Schematic diagram of diverse functions of KIF17. Radiating from KIF17 are cargos or interactions mediated by this molecular motor. Secondary extensions from the cargos and interactions signify proven or presumed functions resulting from KIF17 transport. See text for details.
5. Transcriptional regulation of KIF17
Although KIF17 in testis (KIF17b) is involved in the transcriptional regulation of target genes of CREM and in determining the subcellular localization of ACT [84], the transcriptional regulation of Kif17 gene itself, and indeed of many other Kif genes, has surprisingly received very little attention and is poorly understood.
Over-expression of KIF17 induced a 1.5-fold up-regulation of its cargo protein, NR2B, as well as a 2-fold increase of NR2B mRNA as compared to the wild type littermates, suggesting that KIF17 and NR2B mRNAs might be regulated by a “pretranscriptional process” in the transgenic mice [39]. The authors suggested that CREB may play such a role, as the level of phosphorylated CREB (but not total CREB) was increased in forebrain homogenates of these mice [39]. Likewise, Kif17 knockout leads to attenuated phosphorylation of CREB that is partially rescued by overexpression of NR2A, but not NR2B, presumably because NR2A contributes to CREB activation, but NR2B cannot reach the synaptic site for activation [101]. On the other hand, expression of KIF17 does rescue the CREB response in Kif17−/− neurons [101]. These experiments suggest an association between KIF delivery of NR2B and CREB activation, but they do not directly address a transcriptional link between KIF17 and NR2B.
Such a link was recently found to be mediated by a transcription factor known as nuclear respiratory factor 1 (NRF-1) [108]. NRF-1 plays an important role in mitochondrial energy metabolism, as it activates the transcription of nuclear genes encoding a number of subunits of the respiratory chain complexes, plus it regulates the expression of transcription factors A and B of mitochondria (TFAM, TFB1M, and TFB2M) responsible for the transcription and replication of mitochondrial DNA (reviewed in [109]). Of the five electron transport chain complexes, complex IV or cytochrome c oxidase (COX) has been proven to be a metabolic marker of neuronal activity, as energy in neurons is used primarily to repolarize membrane potentials after depolarizing activation, and neuronal activity and energy metabolism are tightly coupled processes [110]. COX is one of only four bigenomic enzymes, with three subunits encoded in the mitochondrial genome and ten in the nuclear DNA [111]. Transcriptional control of such a multisubunit, bigenomic, and multi-chromosomally-derived enzyme appeared daunting until the discovery that NRF-1 directly regulates the expression of all 10 nuclear-encoded COX subunit genes [112] in addition to its indirect regulation of the 3 mitochondrial-encoded COX subunit genes via its control of TFAM and TFBMs [109, 113]. Moreover, NRF-1 also directly regulates the expression of critical components of glutamatergic synapses, including NR1 and NR2B of NMDA receptors, GluR2 of AMPA receptors, and neuronal nitric oxide synthase, a downstream mediator of glutamatergic synaptic transmission [114–116]. Thus, NRF-1 coordinates the tight coupling between neuronal activity and energy metabolism at the transcriptional level.
By means of in vitro electrophoretic mobility shift and supershift assays, in vivo chromatin immunoprecipitation assays, promoter mutations, and real-time quantitative PCR, it was found that NRF-1 functionally regulates Kif17, but not Kif1a, gene in neurons [108]. NRF-1 binding sites on the promoter of Kif17 gene are highly conserved among rats, mice, and humans. Over-expressing NRF-1 rescues the suppressed expressions of Kif17 mRNA and proteins induced by tetrodotoxin-mediated impulse blockade, whereas silencing of NRF-1 with small hairpin RNA blocks the up-regulation of Kif17 transcripts and proteins induced by KCl-generated depolarizing stimulation [108]. Thus, NRF-1 co-regulates KIF17 and its cargo, NR2B in neurons [108, 114]. Moreover, by coupling the regulation of oxidative enzymes that generate energy and neurochemicals that consume energy during neuronal activity, NRF-1 ensures that energy produced matches energy utilized at the cellular and molecular levels in neurons. Whether NRF-1 acts alone or in combination with other transcription factors, such as CREB, in regulating KIF17 is as yet unknown. A schematic diagram of transcriptional co-regulation by NRF-1 is shown in Figure 4.
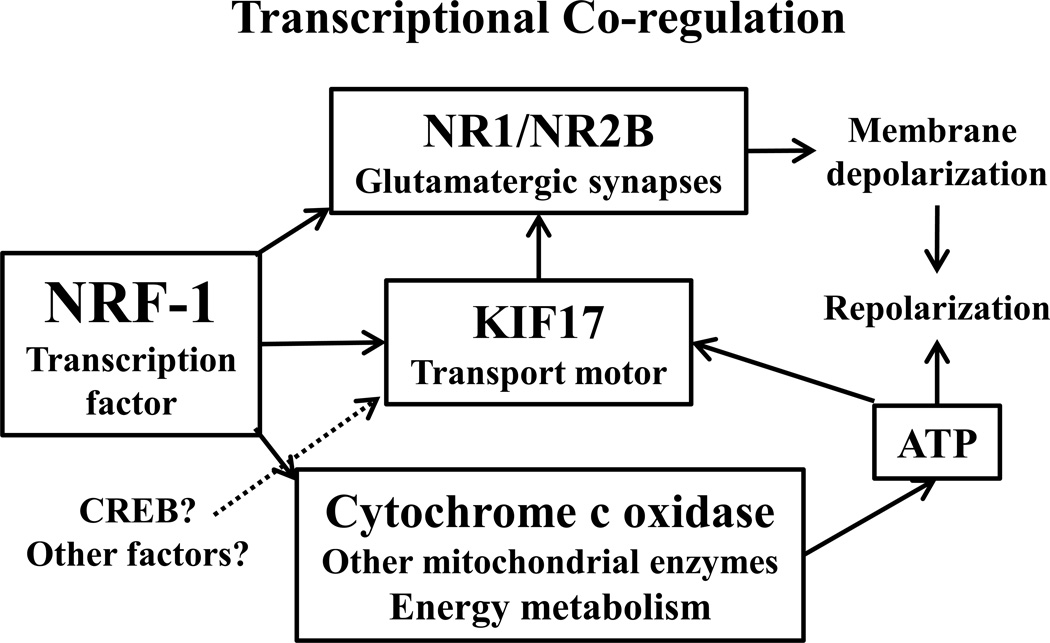
Figure 4.
Schematic rendition of transcriptional co-regulation by NRF-1. Not only does NRF-1 co-regulate glutamatergic neurochemicals, such as NR1/NR2B (and others not depicted here) and agents of energy metabolism, such as cytochrome c oxidase, it also regulates the motor that transports NR2B (and possibly NR1), i.e., KIF17. Membrane depolarization resulting from glutamatergic neurotransmission requires ATP generated by oxidative metabolism (of which cytochrome c oxidase is the terminal enzyme) for repolarization to enable reactivation. ATP is also used to fuel the transport motor KIF17. Thus, NRF-1 ensures that energy produced matches energy utilized in synaptic transmission by neurons. Other factors, such as CREB, may also contribute to this process, but it remains to be proven. See text for details.
6. Diseases and pathologies related to KIF17
6.1 Defect in cilia and flagella: ciliopathy
Structural and functional integrity of cilia and flagella are important to the health of the cell and of the organism. Defects in ciliogenesis and/or function can lead to a number of diseases, collectively known as ciliopathies [25, 117, 118]. By the same token, the kinesin-2 family members that mediate the assembly and maintenance of the cilia/flagella via the transport of IFT particles are vital motors in this regard [68]. Disruption of motile cilia that propel mucus across respiratory epithelia can lead to rhinitis, sinusitis, and bronchiectasis [119]. Defective assembly of non-motile cilia of kidney principal cells, such as those caused by an insertional mutation of the gene for IFT88 particles (Tg737 in mouse and human) may result in polycystic kidney disease [25, 119]. Mutation in the Tg737 gene in the murine retina causes developmental abnormality and eventual degeneration of rod outer segments [73]. A complete knockout of Tg737 in mice is embryonically lethal [120]. These embryos have neural tube defects, enlarged pericardial sac, and a prominent situs inversus, which results from a lack of nodal cilia that normally assist in left-right axis determination [120]. Mutations in genes for other IFT particles known to be transported by OSM-3 or KIF17, such as IFT80, IFT43, IGT121, IFT122, and IFT139, also cause multisystem ciliopathies in animals and humans, including Jeune asphyxiating thoracic dystrophy, cranioectodermal dysplasia or Sensenbrenner syndrome, and nephronophthisis [121–124]. Mutation or absent IFT140 leads to short flagella and possible defect in retrograde transport in trypanosomes [125]. Mutations in the osm-3 gene in C. elegans results in an inability to form normal ciliated chemosensory neurons and thus a defect in chemosensation [126]. In mammals, mutations or loss of KIF3A function in cilia results in severe hypoplasia and abnormal foliation of the cerebellum attributable to defects in granule cell proliferation [127]. Ciliopathies are also implicated in several human diseases involving cystic kidneys and retinal degeneration, such as Bardet-Biedl syndrome, Jeune syndrome, and Senior-Loken syndrome [25]. Nonetheless, at present a human ciliopathy has not been definitively linked to KIF17.
6.2 Defects in neuronal functioning, plasticity, and learning
Defective transport of receptor proteins and ion channels can lead to faulty signal transduction cascades, resulting in abnormal or failed neuronal functioning. Disruption of the Kif17 gene in mice inhibits the transport of NR2B, decreases Grin2b transcription, reduces the availability of synaptic NR2B, and attenuates NMDA receptor-mediated synaptic currents in hippocampal neurons [101]. Surprisingly, the level of NR2A is also reduced due to increased degradation via the ubiquitin-proteasome system [101]. As a result, both early and late long-term potentiation, long-term depression, and cAMP-response element binding protein (CREB)-dependent responses are also attenuated, leading to impairment in the acquisition and consolidation of memory in knockout mice [101].
6.2.1 Animal model of Down Syndrome
Down syndrome, or trisomy 21, is characterized by impaired learning and memory associated with synaptic dysfunction in the hippocampus, among other deficits [128]. In a Ts65Dn mouse model of Down syndrome that bears a triplicated segment of chromosome 16, which shares >50% homology with human chromosome 21, a significant decline in the level of KIF17 is found in the brain [129]. These animals exhibit a number of developmental, learning, and behavioral deficits found in Down syndrome [130, 131]. However, the true cause of learning deficits in these animals is not clear, as the levels of both NR2B and NR2A are reportedly unchanged [129]. Long-term potentiation, as induced by in vitro high-frequency stimulation of hippocampal neurons in adult Ts65Dn mice, is either reduced [132] or unchanged [133]. However, a significant reduction in LTP is observed with theta burst stimulation of the hippocampus in Ts65Dn mice as compared to euploid controls [133]. An increase in GABAA receptor-mediated inhibition may contribute to the defect, as bath application of picrotoxin, a GABAA receptor antagonist, normalizes NMDA receptor-mediated currents and restores LTP in the hippocampus [133, 134]. Decreased levels of KIF17 may play a role in reduced or abnormal trafficking of NR2B and/or GluR5 to the synapse, thereby directly or indirectly cause synaptic dysfunction. However, definitive proof is still lacking.
6.2.2 Schizophrenia
Abnormal glutamatergic neurotransmission has been implicated in schizophrenia, and the NMDA receptor hypofunction hypothesis has been proposed to mediate cognitive dysfunction [135]. This raises the intriguing question as to whether the expressions of NR2B and its transporter KIF17 would be altered in this illness. In a study of a group of elderly patients with schizophrenia, the levels of transcripts for a number of NR2B trafficking complex proteins, such as CASK, mLin7A, mLin7C, and APBA1 were increased in the prefrontal cortex, whereas those of proteins for CASK and mLin7C were decreased in the anterior cingulated cortex in postmortem schizophrenic brains as compared to a “comparison” group [136]. However, no change was observed for KIF17 or NR2B [136, 137]. On the other hand, chronic administration of antipsychotic drugs such as haloperidol in rats increases the expression of KIF17 and mLin7A in the frontal cortex, but does not affect that of CASK, mLin7C, and APBA1 [136]. Interpretations of these findings are made difficult for several reasons, including: a) natural heterogeneity among human subjects with respect to genetics, gender, medical history, and environmental factors; b) the precise neuronal type(s) that express(es) altered transcript and protein levels is(are) not known; c) altered levels of NR2B and/or KIF17 in dendrites may escape the detection of methods used; d) trafficking complex proteins such as CASK, mLin7, and APBA1 may have functions other than transporting NR2B and these functions may or may not be related to schizophrenia; e) postmortem delays may vary among the subjects, leading to varied outcome; and f) rodent models of diseases that affect higher cognitive functions are not always ideal, as these animals have relatively sparse association cortical areas compared to humans. Interestingly, the level of kainate receptor GluR5, a cargo of KIF17 (see section 4.2) is reportedly decreased in postmortem schizophrenic brains [138, 139]. It is possible that abnormal transporting of NR2B and GluR5 by KIF17 exists in schizophrenia via an as yet unknown mechanism.
Recently, the KIF17 gene has been proposed as one of the candidate gene for schizophrenia [140]. In a cohort of 188 patients with sporadic schizophrenia, a de novo nonsense truncating mutation in KIF17 (C1725A, Y575X) was found in one patient [140]. This results in a KIF17 protein that lacks the last 454 amino acids, which effectively removes the CamKII phosphorylation site and the C-terminal PDZ domain necessary for cargo binding and release (see above). Knocking down the orthologous Kif17 gene in the zebrafish with antisense morpholino oligonucleotides reportedly yields a truncated protein and developmental defects [140]. Despite the tantalizing possibility, the concept is based on a single patient and the effect of Kif17 knockdown on early zebrafish development contrasts sharply with prior studies showing that morpholino knockdown sufficient to suppress KIF17 protein and alter photoreceptor development had little or no effect on early embryogenesis [75]. Thus, the exact role of KIF17 in schizophrenia, if any, awaits future cumulative studies.
6.2.3 Response to oxidative and excitotoxic stress in cultured neurons with Cu/Zn SOD1 mutation related to amyotrophic lateral sclerosis (ALS)
Ten to twenty percent of familial ALS are caused by a gain of function mutation (G93A) of the Cu/Zn superoxide dismutase gene (SOD1) [141]. Progressive degeneration of motor neurons is thought to be due to oxidative stress or glutamate excitotoxicity, among many other plausible factors [142]. When cultured neurons harboring the mutant SOD1 gene were exposed to 50 µM H2O2 (an agent known to induce oxidative stress) for 6 hours, it caused a 39% reduction in neuronal survival as compared to controls [143]. At the same time, microarray analyses revealed that 59 transcripts were up-regulated and 104 were down-regulated by the H2O2 treatment in transgenic cortical neurons as compared to non-transgenic ones [143]. Among the up-regulated genes, KIF17 exhibited a 2.3-fold increase in transgenic versus non-transgenic neurons. However, the expression of this gene did not change when transgenic neurons were exposed to 2 mM NMDA treatment for 6 hours, a regimen known to induce excitotoxicity [143]. The complexicity of altered expressions of numerous transcripts and proteins in affected neurons exposed to neurotoxins suggests that multiple cellular mechanisms are involved.
7. Summary
The intracellular transport of proteins, nucleic acids, macromolecules, and organelles is a fundamental and vital function of all cells, and kinesin and dynein transport motors play critical roles in this process. The multitude of cargo species and cellular tasks during developmental and mature states necessitate the evolution of diverse forms of these motors. KIF17, though discovered only a dozen years ago, has its homolog OSM-3 in C. elegans and Kin5 in Tetrahymena, indicating a long evolutionary history. The main function of KIF17 is to deliver cargos in an ATP-dependent manner along and toward the plus ends of microtubules. The cargos vary in different cell types, but the overriding theme is that the delivery is to the ends of processes, such as to the tips of dendrites, cilia, or outer segments of photoreceptor cells. Of interest is the microtubule-independent transport of ACT from the nucleus to the cytoplasm in testis. Critical cellular functions depend on the transport ability of KIF17, whose dysfunction can lead to a multitude of diseases and pathologies. The transcriptional regulation of KIF17 is a relatively new focus, and the co-regulation of KIF17 and its cargo NR2B by the same transcription factor NRF-1, which also controls the expression of energy-generating enzyme cytochrome c oxidase, suggests a tight coupling at the molecular level between energy utilization by KIF17 and synaptic neurochemicals and energy production in neurons. Similar mechanisms may exist in other cell types.
8. Outlook
Much has been learned about KIF17 in recent years, but much remains to be explored. As more cell types, organs, brain regions, and species are probed, more cargos and more diverse functions of KIF17 will be uncovered. The list of kinesin-associated proteins is likely to grow, and their roles in identifying cargos and in assisting the transport mechanism will become clearer. Conditional knockouts of KIF17 will reveal the precise function of KIF17 in different cell types during different stages of development and in the adult. The basic mechanisms underlying various KIF17-related pathologies will undoubtedly be explored. Finally, the full transcriptional machinery in regulating KIF17 expression in various cell types will also be elucidated.
Supplementary Material
Highlights.
Some key unsolved issues are listed below. The list is by no means exhaustive.
What is the signal that regulates transport of various cargos by KIF17?
What is the signal that tells KIF17 to stop transporting or to cap the plus ends of microtubules?
What is the mechanism of microtubule-independent delivery of ACT in the testis and perhaps in other organs?
What prevents KIF17 from entering the axon?
What is the half-life of KIF17, and how is it regulated?
Besides CaMKII and protein kinase A, are there other post-translational modifications of KIF17 that would affect its function? If so, what are they?
Is abnormal trafficking of NR2B and other neurochemicals the primary underlying mechanism of diverse brain dysfunctions, or are there other factors involved? The same goes for other cargos and related pathologies.
Is CREB also a bona fide transcription factor of KIF17?
Does KIF17 send feedback signals to the NRF-1 transcriptional machinery?
Does NRF-1 cooperate with other transcription factors and co-activators in regulating KIF17, or do the factors function independently of each other?
Acknowledgements
It gives us great pleasure to thank all the investigators who have contributed to the discovery and understanding of KIF17. Unfortunately, space does not permit the citation of every paper published on this subject. In the Wong-Riley laboratory, we thank Dr. S. Dhar for her careful work on the transcriptional regulation of KIF17 in neurons. In the Besharse laboratory, we thank Christine Insinna and Jason Bader for their work on KIF17 in photoreceptors. Supported by the National Institutes of Health grants R01 EY018441 (to M. W-R) and R01 EY03222 (to J.C.B)
Non-standard abbreviations
- ACT
activator of cAMP-responsive element modulator
- APC
adenomatous polyposis coli
- ATP
adenosine triphosphate
- CaMKII
Ca2+/calmodulin-dependent protein kinase II
- CC2
coiled-coil 2
- CLS
ciliary localizing signal
- CNG
cyclic nucleotide-gated
- COX
cytochrome c oxidase
- CREB
cAMP-response element binding protein
- CREM
cAMP-response element modulator
- EB1
end-binding protein 1
- ERK
extracellular signal-regulated kinase
- FMRP
Fragile × mental retardation protein
- GABAA receptor
γ-amino butyric acid A receptor
- GluR5
kainate receptor 5
- Grin2b
gene for NR2B
- 5-HT1A receptor
5-hydroxytryptamine or serotonin 1A receptor
- IFT
intraflagellar transport
- KIF17
kinesin superfamily protein 17
- LTP
long-term potentiation
- MAGUK
membrane-associated guanylate kinase
- MAP2
microtubule-associated protein 2
- MDCK
Madin Darby canine kidney
- MEK
mitogen-activated protein kinase kinase
- NMDA
N-methyl-D-aspartate
- NR2A
NMDA receptor subunit 2A
- NR2B (GluN2B, NRε2)
NMDA receptor subunit 2B
- NRF-1
nuclear respiratory factor 1
- OSM-3
osmotic avoidance abnormal protein 3
- PDZ
postsynaptic density-95/disc large/zona occludens-1
- PSD
postsynaptic density
- SOD1
Cu/Zn superoxide dismutase gene
- Spatial
stromal protein associated with thymii and lymph-node
- Tap/NXF
Tip-associated protein/nuclear RNA export factors
- TB-RBP
testis brain RNA-binding protein
- TFAM
transcription factor A of mitochondria
- TFB1M/TFB2M
transcription factor B1 or B2 of mitochondria.
Footnotes
The authors have no conflict of financial and/or other interest.
References
- 1.Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brady ST. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature. 1985;317:73–75. doi: 10.1038/317073a0. [DOI] [PubMed] [Google Scholar]
- 3.Scholey JM, Porter ME, Grissom PM, McIntosh JR. Identification of kinesin in sea urchin eggs, and evidence for its localization in the mitotic spindle. Nature. 1985;318:483–486. doi: 10.1038/318483a0. [DOI] [PubMed] [Google Scholar]
- 4.Gibbons IR, Rowe AJ. Dynein: A Protein with Adenosine Triphosphatase Activity from Cilia. Science. 1965;149:424–426. doi: 10.1126/science.149.3682.424. [DOI] [PubMed] [Google Scholar]
- 5.Lawrence CJ, Dawe RK, Christie KR, Cleveland DW, Dawson SC, Endow SA, Goldstein LS, Goodson HV, Hirokawa N, Howard J, Malmberg RL, McIntosh JR, Miki H, Mitchison TJ, Okada Y, Reddy AS, Saxton WM, Schliwa M, Scholey JM, Vale RD, Walczak CE, Wordeman L. A standardized kinesin nomenclature. J Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Vale RD, Fletterick RJ. The design plan of kinesin motors. Annu Rev Cell Dev Biol. 1997;13:745–777. doi: 10.1146/annurev.cellbio.13.1.745. [DOI] [PubMed] [Google Scholar]
- 8.Sharp DJ, Rogers GC, Scholey JM. Microtubule motors in mitosis. Nature. 2000;407:41–47. doi: 10.1038/35024000. [DOI] [PubMed] [Google Scholar]
- 9.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 10.Schroer TA, Steuer ER, Sheetz MP. Cytoplasmic dynein is a minus end-directed motor for membranous organelles. Cell. 1989;56:937–946. doi: 10.1016/0092-8674(89)90627-2. [DOI] [PubMed] [Google Scholar]
- 11.Roostalu J, Hentrich C, Bieling P, Telley IA, Schiebel E, Surrey T. Directional switching of the kinesin Cin8 through motor coupling. Science. 2011;332:94–99. doi: 10.1126/science.1199945. [DOI] [PubMed] [Google Scholar]
- 12.Schnapp BJ, Reese TS. Dynein is the motor for retrograde axonal transport of organelles. Proc Natl Acad Sci U S A. 1989;86:1548–1552. doi: 10.1073/pnas.86.5.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bloom GS, Wagner MC, Pfister KK, Brady ST. Native structure and physical properties of bovine brain kinesin and identification of the ATP-binding subunit polypeptide. Biochemistry. 1988;27:3409–3416. doi: 10.1021/bi00409a043. [DOI] [PubMed] [Google Scholar]
- 14.Hirokawa N, Pfister KK, Yorifuji H, Wagner MC, Brady ST, Bloom GS. Submolecular domains of bovine brain kinesin identified by electron microscopy and monoclonal antibody decoration. Cell. 1989;56:867–878. doi: 10.1016/0092-8674(89)90691-0. [DOI] [PubMed] [Google Scholar]
- 15.Wong YL, Rice SE. Kinesin's light chains inhibit the head- and microtubule-binding activity of its tail. Proc Natl Acad Sci U S A. 2010;107:11781–11786. doi: 10.1073/pnas.1005854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Endow SA, Kull FJ, Liu H. Kinesins at a glance. J Cell Sci. 2010;123:3420–3424. doi: 10.1242/jcs.064113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirokawa N, Takemura R. Kinesin superfamily proteins and their various functions and dynamics. Exp Cell Res. 2004;301:50–59. doi: 10.1016/j.yexcr.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 18.Hallen MA, Endow SA. Anastral spindle assembly: a mathematical model. Biophys J. 2009;97:2191–2201. doi: 10.1016/j.bpj.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glater EE, Megeath LJ, Stowers RS, Schwarz TL. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006;173:545–557. doi: 10.1083/jcb.200601067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macaskill AF, Rinholm JE, Twelvetrees AE, Arancibia-Carcamo IL, Muir J, Fransson A, Aspenstrom P, Attwell D, Kittler JT. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron. 2009;61:541–555. doi: 10.1016/j.neuron.2009.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cole DG, Cande WZ, Baskin RJ, Skoufias DA, Hogan CJ, Scholey JM. Isolation of a sea urchin egg kinesin-related protein using peptide antibodies. J Cell Sci. 1992;101(Pt 2):291–301. doi: 10.1242/jcs.101.2.291. [DOI] [PubMed] [Google Scholar]
- 22.Yamazaki H, Nakata T, Okada Y, Hirokawa N. KIF3A/B: a heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. J Cell Biol. 1995;130:1387–1399. doi: 10.1083/jcb.130.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wedaman KP, Meyer DW, Rashid DJ, Cole DG, Scholey JM. Sequence and submolecular localization of the 115-kD accessory subunit of the heterotrimeric kinesin-II (KRP85/95) complex. J Cell Biol. 1996;132:371–380. doi: 10.1083/jcb.132.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamazaki H, Nakata T, Okada Y, Hirokawa N. Cloning and characterization of KAP3: a novel kinesin superfamily-associated protein of KIF3A/3B. Proc Natl Acad Sci U S A. 1996;93:8443–8448. doi: 10.1073/pnas.93.16.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 26.Hirokawa N, Niwa S, Tanaka Y. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 2010;68:610–638. doi: 10.1016/j.neuron.2010.09.039. [DOI] [PubMed] [Google Scholar]
- 27.Setou M, Nakagawa T, Seog DH, Hirokawa N. Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 2000;288:1796–1802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- 28.Kondo S, Sato-Yoshitake R, Noda Y, Aizawa H, Nakata T, Matsuura Y, Hirokawa N. KIF3A is a new microtubule-based anterograde motor in the nerve axon. J Cell Biol. 1994;125:1095–1107. doi: 10.1083/jcb.125.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Z, Roberts EA, Goldstein LS. Functional analysis of mouse kinesin motor Kif3C. Mol Cell Biol. 2001;21:5306–5311. doi: 10.1128/MCB.21.16.5306-5311.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cole DG, Chinn SW, Wedaman KP, Hall K, Vuong T, Scholey JM. Novel heterotrimeric kinesin-related protein purified from sea urchin eggs. Nature. 1993;366:268–270. doi: 10.1038/366268a0. [DOI] [PubMed] [Google Scholar]
- 31.Nakagawa T, Tanaka Y, Matsuoka E, Kondo S, Okada Y, Noda Y, Kanai Y, Hirokawa N. Identification and classification of 16 new kinesin superfamily (KIF) proteins in mouse genome. Proc Natl Acad Sci U S A. 1997;94:9654–9659. doi: 10.1073/pnas.94.18.9654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Signor D, Wedaman KP, Rose LS, Scholey JM. Two heteromeric kinesin complexes in chemosensory neurons and sensory cilia of Caenorhabditis elegans. Mol Biol Cell. 1999;10:345–360. doi: 10.1091/mbc.10.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Awan A, Bernstein M, Hamasaki T, Satir P. Cloning and characterization of Kin5, a novel Tetrahymena ciliary kinesin II. Cell Motil Cytoskeleton. 2004;58:1–9. doi: 10.1002/cm.10170. [DOI] [PubMed] [Google Scholar]
- 34.Imanishi M, Endres NF, Gennerich A, Vale RD. Autoinhibition regulates the motility of the C. elegans intraflagellar transport motor OSM-3. J Cell Biol. 2006;174:931–937. doi: 10.1083/jcb.200605179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hammond JW, Blasius TL, Soppina V, Cai D, Verhey KJ. Autoinhibition of the kinesin-2 motor KIF17 via dual intramolecular mechanisms. J Cell Biol. 2010;189:1013–1025. doi: 10.1083/jcb.201001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verhey KJ, Hammond JW. Traffic control: regulation of kinesin motors. Nat Rev Mol Cell Biol. 2009;10:765–777. doi: 10.1038/nrm2782. [DOI] [PubMed] [Google Scholar]
- 37.Scholey JM. Intraflagellar transport motors in cilia: moving along the cell's antenna. J Cell Biol. 2008;180:23–29. doi: 10.1083/jcb.200709133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Song AH, Wang D, Chen G, Li Y, Luo J, Duan S, Poo MM. A selective filter for cytoplasmic transport at the axon initial segment. Cell. 2009;136:1148–1160. doi: 10.1016/j.cell.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 39.Wong RW, Setou M, Teng J, Takei Y, Hirokawa N. Overexpression of motor protein KIF17 enhances spatial and working memory in transgenic mice. Proc Natl Acad Sci U S A. 2002;99:14500–14505. doi: 10.1073/pnas.222371099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okamoto M, Sudhof TC. Mints, Munc18-interacting proteins in synaptic vesicle exocytosis. J Biol Chem. 1997;272:31459–31464. doi: 10.1074/jbc.272.50.31459. [DOI] [PubMed] [Google Scholar]
- 41.Hata Y, Butz S, Sudhof TC. CASK: a novel dlg/PSD95 homolog with an N-terminal calmodulin-dependent protein kinase domain identified by interaction with neurexins. J Neurosci. 1996;16:2488–2494. doi: 10.1523/JNEUROSCI.16-08-02488.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeyifous O, Waites CL, Specht CG, Fujisawa S, Schubert M, Lin EI, Marshall J, Aoki C, de Silva T, Montgomery JM, Garner CC, Green WN. SAP97 and CASK mediate sorting of NMDA receptors through a previously unknown secretory pathway. Nat Neurosci. 2009;12:1011–1019. doi: 10.1038/nn.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Butz S, Okamoto M, Sudhof TC. A tripartite protein complex with the potential to couple synaptic vesicle exocytosis to cell adhesion in brain. Cell. 1998;94:773–782. doi: 10.1016/s0092-8674(00)81736-5. [DOI] [PubMed] [Google Scholar]
- 44.Jo K, Derin R, Li M, Bredt DS. Characterization of MALS/Velis-1, -2, and -3: a family of mammalian LIN-7 homologs enriched at brain synapses in association with the postsynaptic density-95/NMDA receptor postsynaptic complex. J Neurosci. 1999;19:4189–4199. doi: 10.1523/JNEUROSCI.19-11-04189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guillaud L, Wong R, Hirokawa N. Disruption of KIF17-Mint1 interaction by CaMKII-dependent phosphorylation: a molecular model of kinesin-cargo release. Nat Cell Biol. 2008;10:19–29. doi: 10.1038/ncb1665. [DOI] [PubMed] [Google Scholar]
- 46.Guillaud L, Setou M, Hirokawa N. KIF17 dynamics and regulation of NR2B trafficking in hippocampal neurons. J Neurosci. 2003;23:131–140. doi: 10.1523/JNEUROSCI.23-01-00131.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yuen EY, Jiang Q, Feng J, Yan Z. Microtubule regulation of N-methyl-D-aspartate receptor channels in neurons. J Biol Chem. 2005;280:29420–29427. doi: 10.1074/jbc.M504499200. [DOI] [PubMed] [Google Scholar]
- 48.Nakanishi S. Molecular diversity of glutamate receptors and implications for brain function. Science. 1992;258:597–603. doi: 10.1126/science.1329206. [DOI] [PubMed] [Google Scholar]
- 49.Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- 50.Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 51.Kayadjanian N, Lee HS, Pina-Crespo J, Heinemann SF. Localization of glutamate receptors to distal dendrites depends on subunit composition and the kinesin motor protein KIF17. Mol Cell Neurosci. 2007;34:219–230. doi: 10.1016/j.mcn.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chu PJ, Rivera JF, Arnold DB. A role for Kif17 in transport of Kv4.2. J Biol Chem. 2006;281:365–373. doi: 10.1074/jbc.M508897200. [DOI] [PubMed] [Google Scholar]
- 53.Gruter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber BK, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 54.Herold A, Suyama M, Rodrigues JP, Braun IC, Kutay U, Carmo-Fonseca M, Bork P, Izaurralde E. TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol Cell Biol. 2000;20:8996–9008. doi: 10.1128/mcb.20.23.8996-9008.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takano K, Miki T, Katahira J, Yoneda Y. NXF2 is involved in cytoplasmic mRNA dynamics through interactions with motor proteins. Nucleic Acids Res. 2007;35:2513–2521. doi: 10.1093/nar/gkm125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lai D, Sakkas D, Huang Y. The fragile X mental retardation protein interacts with a distinct mRNA nuclear export factor NXF2. Rna. 2006;12:1446–1449. doi: 10.1261/rna.94306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silverman MA, Leroux MR. Intraflagellar transport and the generation of dynamic, structurally and functionally diverse cilia. Trends Cell Biol. 2009;19:306–316. doi: 10.1016/j.tcb.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 58.Cole DG. Intraflagellar transport. In: Witman GB, editor. The Chlamydomonas Sourcebook: Cell Motility and Behavior. Oxford, U.K.: Academic Press; 2008. pp. 71–112. [Google Scholar]
- 59.Kozminski KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci U S A. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Walther Z, Vashishtha M, Hall JL. The Chlamydomonas FLA10 gene encodes a novel kinesin-homologous protein. J Cell Biol. 1994;126:175–188. doi: 10.1083/jcb.126.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kozminski KG, Beech PL, Rosenbaum JL. The Chlamydomonas kinesin-like protein FLA10 is involved in motility associated with the flagellar membrane. J Cell Biol. 1995;131:1517–1527. doi: 10.1083/jcb.131.6.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL. Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol. 1998;141:993–1008. doi: 10.1083/jcb.141.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baker SA, Freeman K, Luby-Phelps K, Pazour GJ, Besharse JC. IFT20 links kinesin II with a mammalian intraflagellar transport complex that is conserved in motile flagella and sensory cilia. J Biol Chem. 2003;278:34211–34218. doi: 10.1074/jbc.M300156200. [DOI] [PubMed] [Google Scholar]
- 64.Snow JJ, Ou G, Gunnarson AL, Walker MR, Zhou HM, Brust-Mascher I, Scholey JM. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat Cell Biol. 2004;6:1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- 65.Evans JE, Snow JJ, Gunnarson AL, Ou G, Stahlberg H, McDonald KL, Scholey JM. Functional modulation of IFT kinesins extends the sensory repertoire of ciliated neurons in Caenorhabditis elegans. J Cell Biol. 2006;172:663–669. doi: 10.1083/jcb.200509115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mukhopadhyay S, Lu Y, Qin H, Lanjuin A, Shaham S, Sengupta P. Distinct IFT mechanisms contribute to the generation of ciliary structural diversity in C. elegans. Embo J. 2007;26:2966–2980. doi: 10.1038/sj.emboj.7601717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jenkins PM, Hurd TW, Zhang L, McEwen DP, Brown RL, Margolis B, Verhey KJ, Martens JR. Ciliary targeting of olfactory CNG channels requires the CNGB1b subunit and the kinesin-2 motor protein, KIF17. Curr Biol. 2006;16:1211–1216. doi: 10.1016/j.cub.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 68.Morsci NS, Barr MM. Kinesin-3 KLP-6 Regulates Intraflagellar Transport in Male-Specific Cilia of Caenorhabditis elegans. Curr Biol. 2011;21:1239–1244. doi: 10.1016/j.cub.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dishinger JF, Kee HL, Jenkins PM, Fan S, Hurd TW, Hammond JW, Truong YN, Margolis B, Martens JR, Verhey KJ. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nat Cell Biol. 2010;12:703–710. doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flannery RJ, French DA, Kleene SJ. Clustering of cyclic-nucleotide-gated channels in olfactory cilia. Biophys J. 2006;91:179–188. doi: 10.1529/biophysj.105.079046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Besharse JC, Horst CJ. The photoreceptor connecting cilium. A model for the transition zone. In: Bloodgood RA, editor. Ciliary and Flagellar Membranes. New York: Plenum Publishing Corp; 1990. pp. 389–417. [Google Scholar]
- 72.Insinna C, Humby M, Sedmak T, Wolfrum U, Besharse JC. Different roles for KIF17 and kinesin II in photoreceptor development and maintenance. Dev Dyn. 2009;238:2211–2222. doi: 10.1002/dvdy.21956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pazour GJ, Baker SA, Deane JA, Cole DG, Dickert BL, Rosenbaum JL, Witman GB, Besharse JC. The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J Cell Biol. 2002;157:103–113. doi: 10.1083/jcb.200107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marszalek JR, Liu X, Roberts EA, Chui D, Marth JD, Williams DS, Goldstein LS. Genetic evidence for selective transport of opsin and arrestin by kinesin-II in mammalian photoreceptors. Cell. 2000;102:175–187. doi: 10.1016/s0092-8674(00)00023-4. [DOI] [PubMed] [Google Scholar]
- 75.Insinna C, Pathak N, Perkins B, Drummond I, Besharse JC. The homodimeric kinesin, Kif17, is essential for vertebrate photoreceptor sensory outer segment development. Dev Biol. 2008;316:160–170. doi: 10.1016/j.ydbio.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Insinna C, Besharse JC. Intraflagellar transport and the sensory outer segment of vertebrate photoreceptors. Dev Dyn. 2008;237:1982–1992. doi: 10.1002/dvdy.21554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Insinna C, Luby-Phelps K, Link BA, Besharse JC. Analysis of IFT kinesins in developing zebrafish cone photoreceptor sensory cilia. Methods Cell Biol. 2009;93:219–234. doi: 10.1016/S0091-679X(08)93012-0. [DOI] [PubMed] [Google Scholar]
- 78.Macho B, Brancorsini S, Fimia GM, Setou M, Hirokawa N, Sassone-Corsi P. CREM-dependent transcription in male germ cells controlled by a kinesin. Science. 2002;298:2388–2390. doi: 10.1126/science.1077265. [DOI] [PubMed] [Google Scholar]
- 79.Chennathukuzhi V, Morales CR, El-Alfy M, Hecht NB. The kinesin KIF17b and RNA-binding protein TB-RBP transport specific cAMP-responsive element modulator-regulated mRNAs in male germ cells. Proc Natl Acad Sci U S A. 2003;100:15566–15571. doi: 10.1073/pnas.2536695100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kimmins S, Kotaja N, Fienga G, Kolthur US, Brancorsini S, Hogeveen K, Monaco L, Sassone-Corsi P. A specific programme of gene transcription in male germ cells. Reprod Biomed Online. 2004;8:496–500. doi: 10.1016/s1472-6483(10)61094-2. [DOI] [PubMed] [Google Scholar]
- 81.Kotaja N, Lin H, Parvinen M, Sassone-Corsi P. Interplay of PIWI/Argonaute protein MIWI and kinesin KIF17b in chromatoid bodies of male germ cells. J Cell Sci. 2006;119:2819–2825. doi: 10.1242/jcs.03022. [DOI] [PubMed] [Google Scholar]
- 82.Saade M, Irla M, Govin J, Victorero G, Samson M, Nguyen C. Dynamic distribution of Spatial during mouse spermatogenesis and its interaction with the kinesin KIF17b. Exp Cell Res. 2007;313:614–626. doi: 10.1016/j.yexcr.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 83.Hogeveen KN, Sassone-Corsi P. Regulation of gene expression in post-meiotic male germ cells: CREM-signalling pathways and male fertility. Hum Fertil (Camb) 2006;9:73–79. doi: 10.1080/14647270500463400. [DOI] [PubMed] [Google Scholar]
- 84.Kotaja N, Macho B, Sassone-Corsi P. Microtubule-independent and protein kinase A-mediated function of kinesin KIF17b controls the intracellular transport of activator of CREM in testis (ACT) J Biol Chem. 2005;280:31739–31745. doi: 10.1074/jbc.M505971200. [DOI] [PubMed] [Google Scholar]
- 85.Kotaja N, De Cesare D, Macho B, Monaco L, Brancorsini S, Goossens E, Tournaye H, Gansmuller A, Sassone-Corsi P. Abnormal sperm in mice with targeted deletion of the act (activator of cAMP-responsive element modulator in testis) gene. Proc Natl Acad Sci U S A. 2004;101:10620–10625. doi: 10.1073/pnas.0401947101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Irla M, Puthier D, Le Goffic R, Victorero G, Freeman T, Naquet P, Samson M, Nguyen C. Spatial, a new nuclear factor tightly regulated during mouse spermatogenesis. Gene Expr Patterns. 2003;3:135–138. doi: 10.1016/s1567-133x(03)00024-3. [DOI] [PubMed] [Google Scholar]
- 87.Irla M, Puthier D, Granjeaud S, Saade M, Victorero G, Mattei MG, Nguyen C. Genomic organization and the tissue distribution of alternatively spliced isoforms of the mouse Spatial gene. BMC Genomics. 2004;5:41. doi: 10.1186/1471-2164-5-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jaulin F, Kreitzer G. KIF17 stabilizes microtubules and contributes to epithelial morphogenesis by acting at MT plus ends with EB1 and APC. J Cell Biol. 2010;190:443–460. doi: 10.1083/jcb.201006044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, Wallar BJ, Alberts AS, Gundersen GG. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- 90.Sati L, Seval-Celik Y, Unek G, Korgun ET, Demir R. The presence of kinesin superfamily motor proteins KIFC1 and KIF17 in normal and pathological human placenta. Placenta. 2009;30:848–854. doi: 10.1016/j.placenta.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 91.Irla M, Saade M, Fernandez C, Chasson L, Victorero G, Dahmane N, Chazal G, Nguyen C. Neuronal distribution of spatial in the developing cerebellum and hippocampus and its somatodendritic association with the kinesin motor KIF17. Exp Cell Res. 2007;313:4107–4119. doi: 10.1016/j.yexcr.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 92.Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 93.Strack S, Colbran RJ. Autophosphorylation-dependent targeting of calcium/ calmodulin-dependent protein kinase II by the NR2B subunit of the N-methyl- D-aspartate receptor. J Biol Chem. 1998;273:20689–20692. doi: 10.1074/jbc.273.33.20689. [DOI] [PubMed] [Google Scholar]
- 94.Barria A, Malinow R. NMDA receptor subunit composition controls synaptic plasticity by regulating binding to CaMKII. Neuron. 2005;48:289–301. doi: 10.1016/j.neuron.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 95.Gambrill AC, Barria A. NMDA receptor subunit composition controls synaptogenesis and synapse stabilization. Proc Natl Acad Sci U S A. 2011;108:5855–58560. doi: 10.1073/pnas.1012676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cheriyan J, Kumar P, Mayadevi M, Surolia A, Omkumar RV. Calcium/calmodulin dependent protein kinase II bound to NMDA receptor 2B subunit exhibits increased ATP affinity and attenuated dephosphorylation. PLoS One. 2011;6:e16495. doi: 10.1371/journal.pone.0016495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bannerman DM. Fractionating spatial memory with glutamate receptor subunit-knockout mice. Biochem Soc Trans. 2009;37:1323–1327. doi: 10.1042/BST0371323. [DOI] [PubMed] [Google Scholar]
- 98.Ge Y, Dong Z, Bagot RC, Howland JG, Phillips AG, Wong TP, Wang YT. Hippocampal long-term depression is required for the consolidation of spatial memory. Proc Natl Acad Sci U S A. 2010;107:16697–16702. doi: 10.1073/pnas.1008200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brigman JL, Wright T, Talani G, Prasad-Mulcare S, Jinde S, Seabold GK, Mathur P, Davis MI, Bock R, Gustin RM, Colbran RJ, Alvarez VA, Nakazawa K, Delpire E, Lovinger DM, Holmes A. Loss of GluN2B-containing NMDA receptors in CA1 hippocampus and cortex impairs long-term depression, reduces dendritic spine density, and disrupts learning. J Neurosci. 2010;30:4590–4600. doi: 10.1523/JNEUROSCI.0640-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tang YP, Shimizu E, Dube GR, Rampon C, Kerchner GA, Zhuo M, Liu G, Tsien JZ. Genetic enhancement of learning and memory in mice. Nature. 1999;401:63–69. doi: 10.1038/43432. [DOI] [PubMed] [Google Scholar]
- 101.Yin X, Takei Y, Kido MA, Hirokawa N. Molecular motor KIF17 is fundamental for memory and learning via differential support of synaptic NR2A/2B levels. Neuron. 2011;70:310–325. doi: 10.1016/j.neuron.2011.02.049. [DOI] [PubMed] [Google Scholar]
- 102.Kia HK, Miquel MC, Brisorgueil MJ, Daval G, Riad M, El Mestikawy S, Hamon M, Verge D. Immunocytochemical localization of serotonin1A receptors in the rat central nervous system. J Comp Neurol. 1996;365:289–305. doi: 10.1002/(SICI)1096-9861(19960205)365:2<289::AID-CNE7>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 103.Simpson MD, Lubman DI, Slater P, Deakin JF. Autoradiography with [3H]8-OH-DPAT reveals increases in 5-HT(1A) receptors in ventral prefrontal cortex in schizophrenia. Biol Psychiatry. 1996;39:919–928. doi: 10.1016/0006-3223(95)00026-7. [DOI] [PubMed] [Google Scholar]
- 104.Ramboz S, Oosting R, Amara DA, Kung HF, Blier P, Mendelsohn M, Mann JJ, Brunner D, Hen R. Serotonin receptor 1A knockout: an animal model of anxiety-related disorder. Proc Natl Acad Sci U S A. 1998;95:14476–14481. doi: 10.1073/pnas.95.24.14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yuen EY, Jiang Q, Chen P, Gu Z, Feng J, Yan Z. Serotonin 5-HT1A receptors regulate NMDA receptor channels through a microtubule-dependent mechanism. J Neurosci. 2005;25:5488–5501. doi: 10.1523/JNEUROSCI.1187-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bernhardt R, Matus A. Light and electron microscopic studies of the distribution of microtubule-associated protein 2 in rat brain: a difference between dendritic and axonal cytoskeletons. J Comp Neurol. 1984;226:203–221. doi: 10.1002/cne.902260205. [DOI] [PubMed] [Google Scholar]
- 107.Yamamoto H, Imai K, Kamegaya E, Takamatsu Y, Irago M, Hagino Y, Kasai S, Shimada K, Yamamoto T, Sora I, Koga H, Ikeda K. Repeated methamphetamine administration alters expression of the NMDA receptor channel epsilon2 subunit and kinesins in the mouse brain. Ann N Y Acad Sci. 2006;1074:97–103. doi: 10.1196/annals.1369.009. [DOI] [PubMed] [Google Scholar]
- 108.Dhar SS, Wong-Riley MT. The kinesin superfamily protein KIF17 is regulated by the same transcription factor (NRF-1) as its cargo NR2B in neurons. Biochim Biophys Acta. 2011;1813:403–411. doi: 10.1016/j.bbamcr.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 110.Wong-Riley MT. Cytochrome oxidase: an endogenous metabolic marker for neuronal activity. Trends Neurosci. 1989;12:94–101. doi: 10.1016/0166-2236(89)90165-3. [DOI] [PubMed] [Google Scholar]
- 111.Kadenbach B, Jarausch J, Hartmann R, Merle P. Separation of mammalian cytochrome c oxidase into 13 polypeptides by a sodium dodecyl sulfate-gel electrophoretic procedure. Anal Biochem. 1983;129:517–521. doi: 10.1016/0003-2697(83)90586-9. [DOI] [PubMed] [Google Scholar]
- 112.Dhar SS, Ongwijitwat S, Wong-Riley MT. Nuclear respiratory factor 1 regulates all ten nuclear-encoded subunits of cytochrome c oxidase in neurons. J Biol Chem. 2008;283:3120–3129. doi: 10.1074/jbc.M707587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gleyzer N, Vercauteren K, Scarpulla RC. Control of Mitochondrial Transcription Specificity Factors (TFB1M and TFB2M) by Nuclear Respiratory Factors (NRF-1 and NRF-2) and PGC-1 Family Coactivators. Mol Cell Biol. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dhar SS, Wong-Riley MT. Coupling of energy metabolism and synaptic transmission at the transcriptional level: role of nuclear respiratory factor 1 in regulating both cytochrome c oxidase and NMDA glutamate receptor subunit genes. J Neurosci. 2009;29:483–492. doi: 10.1523/JNEUROSCI.3704-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dhar SS, Liang HL, Wong-Riley MT. Nuclear respiratory factor 1 co-regulates AMPA glutamate receptor subunit 2 and cytochrome c oxidase: tight coupling of glutamatergic transmission and energy metabolism in neurons. J Neurochem. 2009;108:1595–1606. doi: 10.1111/j.1471-4159.2009.05929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dhar SS, Liang HL, Wong-Riley MT. Transcriptional coupling of synaptic transmission and energy metabolism: Role of nuclear respiratory factor 1 in co-regulating neuronal nitric oxide synthase and cytochrome c oxidase genes in neurons. Biochim Biophys Acta. 2009;1793:1604–1613. doi: 10.1016/j.bbamcr.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 118.Tobin JL, Beales PL. The nonmotile ciliopathies. Genet Med. 2009;11:386–402. doi: 10.1097/GIM.0b013e3181a02882. [DOI] [PubMed] [Google Scholar]
- 119.Eley L, Yates LM, Goodship JA. Cilia and disease. Curr Opin Genet Dev. 2005;15:308–314. doi: 10.1016/j.gde.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 120.Murcia NS, Richards WG, Yoder BK, Mucenski ML, Dunlap JR, Woychik RP. The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left-right axis determination. Development. 2000;127:2347–2355. doi: 10.1242/dev.127.11.2347. [DOI] [PubMed] [Google Scholar]
- 121.Beales PL, Bland E, Tobin JL, Bacchelli C, Tuysuz B, Hill J, Rix S, Pearson CG, Kai M, Hartley J, Johnson C, Irving M, Elcioglu N, Winey M, Tada M, Scambler PJ. IFT80, which encodes a conserved intraflagellar transport protein, is mutated in Jeune asphyxiating thoracic dystrophy. Nat Genet. 2007;39:727–729. doi: 10.1038/ng2038. [DOI] [PubMed] [Google Scholar]
- 122.Walczak-Sztulpa J, Eggenschwiler J, Osborn D, Brown DA, Emma F, Klingenberg C, Hennekam RC, Torre G, Garshasbi M, Tzschach A, Szczepanska M, Krawczynski M, Zachwieja J, Zwolinska D, Beales PL, Ropers HH, Latos-Bielenska A, Kuss AW. Cranioectodermal Dysplasia, Sensenbrenner syndrome, is a ciliopathy caused by mutations in the IFT122 gene. Am J Hum Genet. 2010;86:949–956. doi: 10.1016/j.ajhg.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Arts HH, Bongers EM, Mans DA, van Beersum SE, Oud MM, Bolat E, Spruijt L, Cornelissen EA, Schuurs-Hoeijmakers JH, de Leeuw N, Cormier-Daire V, Brunner HG, Knoers NV, Roepman R. C14ORF179 encoding IFT43 is mutated in Sensenbrenner syndrome. J Med Genet. 2011;48:390–395. doi: 10.1136/jmg.2011.088864. [DOI] [PubMed] [Google Scholar]
- 124.Davis EE, Zhang Q, Liu Q, Diplas BH, Davey LM, Hartley J, Stoetzel C, Szymanska K, Ramaswami G, Logan CV, Muzny DM, Young AC, Wheeler DA, Cruz P, Morgan M, Lewis LR, Cherukuri P, Maskeri B, Hansen NF, Mullikin JC, Blakesley RW, Bouffard GG, Gyapay G, Rieger S, Tonshoff B, Kern I, Soliman NA, Neuhaus TJ, Swoboda KJ, Kayserili H, Gallagher TE, Lewis RA, Bergmann C, Otto EA, Saunier S, Scambler PJ, Beales PL, Gleeson JG, Maher ER, Attie-Bitach T, Dollfus H, Johnson CA, Green ED, Gibbs RA, Hildebrandt F, Pierce EA, Katsanis N. TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat Genet. 2011;43:189–196. doi: 10.1038/ng.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Absalon S, Blisnick T, Kohl L, Toutirais G, Dore G, Julkowska D, Tavenet A, Bastin P. Intraflagellar transport and functional analysis of genes required for flagellum formation in trypanosomes. Mol Biol Cell. 2008;19:929–944. doi: 10.1091/mbc.E07-08-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- 127.Chizhikov VV, Davenport J, Zhang Q, Shih EK, Cabello OA, Fuchs JL, Yoder BK, Millen KJ. Cilia proteins control cerebellar morphogenesis by promoting expansion of the granule progenitor pool. J Neurosci. 2007;27:9780–9789. doi: 10.1523/JNEUROSCI.5586-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Epstein CJ. 2001 William Allan Award Address. From Down syndrome to the "human" in "human genetics". Am J Hum Genet. 2002;70:300–313. doi: 10.1086/338915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Roberson R, Toso L, Abebe D, Spong CY. Altered expression of KIF17, a kinesin motor protein associated with NR2B trafficking, may mediate learning deficits in a Down syndrome mouse model. Am J Obstet Gynecol. 2008;198:313, e1–e4. doi: 10.1016/j.ajog.2008.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Reeves RH, Irving NG, Moran TH, Wohn A, Kitt C, Sisodia SS, Schmidt C, Bronson RT, Davisson MT. A mouse model for Down syndrome exhibits learning and behaviour deficits. Nat Genet. 1995;11:177–184. doi: 10.1038/ng1095-177. [DOI] [PubMed] [Google Scholar]
- 131.Holtzman DM, Santucci D, Kilbridge J, Chua-Couzens J, Fontana DJ, Daniels SE, Johnson RM, Chen K, Sun Y, Carlson E, Alleva E, Epstein CJ, Mobley WC. Developmental abnormalities and age-related neurodegeneration in a mouse model of Down syndrome. Proc Natl Acad Sci U S A. 1996;93:13333–13338. doi: 10.1073/pnas.93.23.13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Siarey RJ, Stoll J, Rapoport SI, Galdzicki Z. Altered long-term potentiation in the young and old Ts65Dn mouse, a model for Down Syndrome. Neuropharmacology. 1997;36:1549–1554. doi: 10.1016/s0028-3908(97)00157-3. [DOI] [PubMed] [Google Scholar]
- 133.Costa AC, Grybko MJ. Deficits in hippocampal CA1 LTP induced by TBS but not HFS in the Ts65Dn mouse: a model of Down syndrome. Neurosci Lett. 2005;382:317–322. doi: 10.1016/j.neulet.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 134.Kleschevnikov AM, Belichenko PV, Villar AJ, Epstein CJ, Malenka RC, Mobley WC. Hippocampal long-term potentiation suppressed by increased inhibition in the Ts65Dn mouse, a genetic model of Down syndrome. J Neurosci. 2004;24:8153–8160. doi: 10.1523/JNEUROSCI.1766-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lindsley CW, Shipe WD, Wolkenberg SE, Theberge CR, Williams DL, Jr, Sur C, Kinney GG. Progress towards validating the NMDA receptor hypofunction hypothesis of schizophrenia. Curr Top Med Chem. 2006;6:771–785. doi: 10.2174/156802606777057599. [DOI] [PubMed] [Google Scholar]
- 136.Kristiansen LV, Bakir B, Haroutunian V, Meador-Woodruff JH. Expression of the NR2B-NMDA receptor trafficking complex in prefrontal cortex from a group of elderly patients with schizophrenia. Schizophr Res. 2010;119:198–209. doi: 10.1016/j.schres.2010.02.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kristiansen LV, Beneyto M, Haroutunian V, Meador-Woodruff JH. Changes in NMDA receptor subunits and interacting PSD proteins in dorsolateral prefrontal and anterior cingulate cortex indicate abnormal regional expression in schizophrenia. Mol Psychiatry. 2006;11:737–747. 705. doi: 10.1038/sj.mp.4001844. [DOI] [PubMed] [Google Scholar]
- 138.Scarr E, Beneyto M, Meador-Woodruff JH, Dean B. Cortical glutamatergic markers in schizophrenia. Neuropsychopharmacology. 2005;30:1521–1531. doi: 10.1038/sj.npp.1300758. [DOI] [PubMed] [Google Scholar]
- 139.Beneyto M, Kristiansen LV, Oni-Orisan A, McCullumsmith RE, Meador-Woodruff JH. Abnormal glutamate receptor expression in the medial temporal lobe in schizophrenia and mood disorders. Neuropsychopharmacology. 2007;32:1888–1902. doi: 10.1038/sj.npp.1301312. [DOI] [PubMed] [Google Scholar]
- 140.Tarabeux J, Champagne N, Brustein E, Hamdan FF, Gauthier J, Lapointe M, Maios C, Piton A, Spiegelman D, Henrion E, Millet B, Rapoport JL, Delisi LE, Joober R, Fathalli F, Fombonne E, Mottron L, Forget-Dubois N, Boivin M, Michaud JL, Lafreniere RG, Drapeau P, Krebs MO, Rouleau GA. De novo truncating mutation in Kinesin 17 associated with schizophrenia. Biol Psychiatry. 2010;68:649–656. doi: 10.1016/j.biopsych.2010.04.018. [DOI] [PubMed] [Google Scholar]
- 141.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 142.Morrison BM, Morrison JH. Amyotrophic lateral sclerosis associated with mutations in superoxide dismutase: a putative mechanism of degeneration. Brain Res Brain Res Rev. 1999;29:121–135. doi: 10.1016/s0165-0173(98)00049-6. [DOI] [PubMed] [Google Scholar]
- 143.Boutahar N, Wierinckx A, Camdessanche JP, Antoine JC, Reynaud E, Lassabliere F, Lachuer J, Borg J. Differential effect of oxidative or excitotoxic stress on the transcriptional profile of amyotrophic lateral sclerosis-linked mutant SOD1 cultured neurons. J Neurosci Res. 2011;89:1439–1450. doi: 10.1002/jnr.22672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.