Abstract
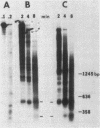
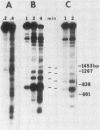
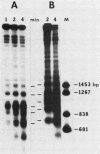
The functional relationship of nucleosome positioning and gene expression is not known. Using high-copy plasmids, containing the yeast phosphate-repressible acid phosphatase gene (PHO5) and the TRP1/ARS1 vector system, I have determined the nucleosomal structure of the 5' region of the PHO5 gene and demonstrated that the nucleosomal positioning of this region is independent of orientation or position in the various plasmid constructions utilized. However, deletion of a 278-base pair BamHI-ClaI fragment from the 5'-flanking sequences of the PHO5 gene causes the nucleosome positioning to become dependent on orientation or position in the plasmids tested. Use of PHO5-CYC1-lACZ fusions have demonstrated that this DNA fragment contains the sequences responsible for the transcriptional regulation of the PHO5 gene in response to the level of phosphate in the growth media. The nucleosome positioning in the 5' region of PHO5 may be determined by an interaction with the sequences or machinery responsible for transcriptional regulation of the gene.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bergman L. W., Kramer R. A. Modulation of chromatin structure associated with derepression of the acid phosphatase gene of Saccharomyces cerevisiae. J Biol Chem. 1983 Jun 10;258(11):7223–7227. [PubMed] [Google Scholar]
- Bergman L. W., Stranathan M. C., Preis L. H. Structure of the transcriptionally repressed phosphate-repressible acid phosphatase gene (PHO5) of Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jan;6(1):38–46. doi: 10.1128/mcb.6.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao M. V., Gralla J., Martinson H. G. DNA sequence directs placement of histone cores on restriction fragments during nucleosome formation. Biochemistry. 1979 Mar 20;18(6):1068–1074. doi: 10.1021/bi00573a021. [DOI] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Gottesfeld J. M., Bloomer L. S. Nonrandom alignment of nucleosomes on 5S RNA genes of X. laevis. Cell. 1980 Oct;21(3):751–760. doi: 10.1016/0092-8674(80)90438-9. [DOI] [PubMed] [Google Scholar]
- Guarente L., Ptashne M. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Lucchini G., Fink G. R. A synthetic HIS4 regulatory element confers general amino acid control on the cytochrome c gene (CYC1) of yeast. Proc Natl Acad Sci U S A. 1985 Jan;82(2):498–502. doi: 10.1073/pnas.82.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemire J. M., Willcocks T., Halvorson H. O., Bostian K. A. Regulation of repressible acid phosphatase gene transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1985 Aug;5(8):2131–2141. doi: 10.1128/mcb.5.8.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D. Organization of the GAL1-GAL10 intergenic control region chromatin. Nucleic Acids Res. 1984 Nov 26;12(22):8457–8474. doi: 10.1093/nar/12.22.8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D. The chromatin structure of an actively expressed, single copy yeast gene. Nucleic Acids Res. 1983 Oct 11;11(19):6755–6773. doi: 10.1093/nar/11.19.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis C., Schedl P., Samal B., Worcel A. Chromatin structure of the 5S RNA genes of D. melanogaster. Cell. 1980 Nov;22(2 Pt 2):387–392. doi: 10.1016/0092-8674(80)90349-9. [DOI] [PubMed] [Google Scholar]
- Lovett M. A., Guiney D. G., Helinski D. R. Relaxation complexes of plasmids ColE1 and ColE2: unique site of the nick in the open circular DNA of the relaxed complexes. Proc Natl Acad Sci U S A. 1974 Oct;71(10):3854–3857. doi: 10.1073/pnas.71.10.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. A. The regulation of yeast mating-type chromatin structure by SIR: an action at a distance affecting both transcription and transposition. Cell. 1982 Sep;30(2):567–578. doi: 10.1016/0092-8674(82)90253-7. [DOI] [PubMed] [Google Scholar]
- Rogers D. T., Lemire J. M., Bostian K. A. Acid phosphatase polypeptides in Saccharomyces cerevisiae are encoded by a differentially regulated multigene family. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2157–2161. doi: 10.1073/pnas.79.7.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin G. M. Three forms of the 5.8-S ribosomal RNA species in Saccharomyces cerevisiae. Eur J Biochem. 1974 Jan 3;41(1):197–202. doi: 10.1111/j.1432-1033.1974.tb03260.x. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Stafford D. W. Structural features of a phased nucleosome core particle. Proc Natl Acad Sci U S A. 1983 Jan;80(1):51–55. doi: 10.1073/pnas.80.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Thoma F., Simpson R. T. Local protein-DNA interactions may determine nucleosome positions on yeast plasmids. Nature. 1985 May 16;315(6016):250–252. doi: 10.1038/315250a0. [DOI] [PubMed] [Google Scholar]
- Toh-e A., Inouye S., Oshima Y. Structure and function of the PHO82-pho4 locus controlling the synthesis of repressible acid phosphatase of Saccharomyces cerevisiae. J Bacteriol. 1981 Jan;145(1):221–232. doi: 10.1128/jb.145.1.221-232.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]