Abstract
Cancer is driven by complex genetic and cellular mechanisms. Recently, the Drosophila community has become increasingly interested in exploring cancer issues. The Drosophila field has made seminal contributions to many of the mechanisms that are fundamental to the cancer process; several of these mechanisms have already been validated in vertebrates. Less well known are the Drosophila field's early direct contributions to the cancer field: some of the earliest tumor suppressors were identified in flies. In this review, we identify major contributions that Drosophila studies have made toward dissecting the pathways and mechanisms underlying tumor progression. We also highlight areas, such as drug discovery, where we expect Drosophila studies to make a major scientific impact in the future.
Keywords: cancer, metastasis, oncogene, tumor suppressor, Drosophila, animal models, drug discovery, therapeutics
INTRODUCTION
The fruit fly (or, vinegar fly) Drosophila has a long and impressive history as the genetic organism of choice for studying epithelial development. By layering a century of tool-building onto modern genetics, Drosophila offers the potential to study the detailed interactions between cells, tissues, and genes. With a few exceptions, only recently has the field begun to explore issues of disease. Cancer in particular has important unmet needs: cell line-based studies have proven disappointing as predictors of clinical relevance, and mouse models are still developing sophisticated genetic tools to recreate the full complexity of tumorigenesis. Here, we review what amounts to the first steps of several laboratories efforts to bring a fly approach to exploring cancer mechanisms and even therapeutics. These endeavors have primarily followed two strategies, which we describe as:
Drosophila to Mammals
Genetic screens and developmental studies have identified novel Drosophila oncogenes/tumor suppressors and related pathways independent of their known importance to mammalian tumorigenesis. In many cases, these same genes and pathways were subsequently implicated in human tumors.
Mammals to Drosophila to Mammals
A more recent approach for understanding cancer mechanisms has been to generate fly cancer models that reflect mammalian and tumor mutation data. Subsequent results from these fly models are then validated in mammals and humans. The advantage of this approach is that generating tumor specific mutations more accurately models deregulation of pathways that promote tumorigenesis.
MAMMALS TO DROSOPHILA TO MAMMALS
Single-hit Models
One of the difficulties of modeling cancer is its complexity. However, a small number of solid tumors are dependent on mutations in single loci, including tuberous sclerosis, neurofibromatosis, and Ret-based tumors such as multiple endocrine neoplasia type 2 (MEN2). Tuberous Sclerosis is an autosomal dominant disorder characterized by benign tumors in multiple organs. Most patients have inactivating mutations in the tsc1 or tsc2 genes, which encode a complex that mediates phosphatidylinositol-3 kinase (PI3K)/mammalian target of rapamycin (mTor) signaling (Sparagana and Roach, 2000). Since the discovery of the Drosophila orthologs in 1999, a large body of work has shown that Tsc-containing complexes integrate inputs from insulin signaling, nutrient sensing, as well as stress response pathways; together, these pathways are relevant for disease progression (Potter et al., 2001; Tapon et al., 2001; Pan et al., 2004; Table 1).
TABLE 1.
List of Genes and Pathways in Drosophila That Have Been Implicated in Human Tumors
| Cancer Model | Drosophila Genes | Mammalian Genes | Highlights/Mammalian Validation | |
|---|---|---|---|---|
| Mammals to Drosophila to Mammals | Tuberous Sclerosis | dTsc1, dTsc2 | TSC1, TSC2 | Tsc1/2 complex in PI3K pathway |
| Neurofibromatosis Type 1 | dNf1 | NF1 | Regulation of Ras Pathway | |
| Multiple Endocrine Neoplasia Type 2 | dRet, dSrc42/64, Btk29A, JNK | Ret, Src, JNK | Activation of pathways downstream, model or successful drug discovery | |
| Glioblastoma Multiforme | dEGFR, dPI3K | EGFR, PI3K | Pathway synergy | |
| Colon Cancer | dAPC, dMyc, Arm, dRb, dE2F | APC, c-Myc, RB, β-Catenin, E2F | First example of Rb as an oncogene | |
| Drosophila to Mammals | Junctional Tumor Suppressors | dlg, lgl, scrib, | lgl1/2, hDlg1, Scrb1, | Novel class of tumor suppressors, Dlg and Lgl, with a role in mammalian tumorigenesis |
| Growth Control (Hippo) | Hippo, Salvador, Warts, dMyc, dNF2/Merlin | Mst1/2, Sav1, Lats1/2, c-Myc, NF2 | Hippo in liver growth control, loss of function of NF2 in Neurofibromatosis II | |
| Tumor Invasion | dCsk, JNK, lgl, dlg, scrib, dRas, | Csk/Chk, Src, Lgl1/2, Dlg1, Serb1, Ras | Src, Actin Remodeling, MMP, Oncogene cooperation | |
| Metastasis | dRas, dlg, lgl, scrib, JNK, dCsk | Csk/Chk, Src, Lgl1/2, Dlg1, Scrb1, Ras | Oncogene cooperation, loss of polarity -> proliferation, Jnk | |
| Microenvironment | scrib, dRas, egr, Caspase | Scrb1, Ras, TNF, Caspase3 | Oncogene cooperation, non-autonmous modulation of tumorigenicity, role of immune system, cell competition, compensatory proliferation in Breast Cancer | |
| Chromatin Regulators | I(3)mbt, mi-2, PcG family | L3MBTL1, CHD3/4/5, PcG family | Single genes regulate multiple oncogenic pathways, CHD mutations in human tumors. Novel role of PcG's as tumor suppressors |
Neurofibromatosis 1 (NF1) is typically a childhood cancer syndrome characterized by benign brain tumors and tumors of the peripheral nervous system. Even though only a small fraction of these cancers develop into fatal metastatic disease, most children/adolescents suffer from debilitating skeletal defects and learning disabilities. Most patients show mutations in the NF1 gene, which encodes a large protein called neurofibromin (Ferner, 2007). Experiments performed in Drosophila provided some of the first evidence that NF1 regulates Ras pathway signaling (Williams et al., 2001; Table 1). Later mouse models showed that reduced NF1 activity can cooperate with other pathways including p19ARF and p53 to drive disease progression. This has led to efforts to treat neurofibromatosis patients with Ras pathway inhibitors (Barkan et al., 2006).
Perhaps the most focused Drosophila studies for single mutation tumor syndromes are the Ret-based cancers that primarily give rise to tumors of the thyroid. Activating mutations in the Ret (rearranged during transformation) receptor tyrosine kinase gene result in hyperproliferation of calcitonin producing C cells of the thyroid and a condition known as medullary thyroid carcinoma (MTC). Other tumors can emerge with lower penetrance including pheochromocytomas, parathyroid adenomas, and mucosal neuromas, and collectively this condition is referred to as MEN2 (Hazard et al., 1959; Donis-Keller et al., 1993; Carlson et al., 1994; Eng et al., 1996). MTCs can be treated by surgery if detected early but often—especially in patients with spontaneous disease—they are diagnosed late, when tumors have already metastasized to distant tissues including the brain, lung and liver (Jimenez et al., 2008; Bobinski et al., 2009). Little was known regarding the mechanisms underlying Ret transformation, nor had a useful chemotherapeutic been identified. Attempts to establish a useful mouse transgenic had met with mixed success (Smith-Hicks et al., 2000; Cranston and Ponder, 2003) and have not contributed to the search for a useful therapeutic.
The monogenic nature of MTC made it attractive to model in Drosophila. Targeting Drosophila Ret oncogenic isoforms (dRetMEN2) to the developing fly eye led to a rough eye phenotype characterized by excess proliferation, cell death, and developmental abnormalities. A genetic modifier screen identified 140 different loci that mediated oncogenic dRetMEN2's activity. These represented mediators of a broad array of signaling pathways including Ras, Src, Jnk, and Hedgehog (Read et al., 2005; Table 1). The broad spectrum of pathways was perhaps surprising, and pointed to the potential complexity of even a monogenic tumor.
A closer examination of the strongest genetic modifiers of dRetMEN2 has highlighted Ret's ability to direct many aspects of cancer. For example, our data indicated that dRetMEN2 signals through Src and Jnk to trigger the multiple steps required for metastasis-like behavior of cells, matrix metalloprotease production, and cell invasion (Read et al., 2005; Vidal et al., 2006). Ras pathway signaling, by contrast, appears to act primarily to promote proliferation (Read et al., 2005). Similar to other RTKs, Ret has a large number of docking sites, and presumably these account for its ability to activate many pathways (Cakir and Grossman, 2009). But these data pose a central question of which we have very little understanding: Ret is expressed broadly throughout our bodies, yet only C cells consistently respond by transforming in the presence of oncogenic Ret.
Monogenic models like dRetMEN2 hold considerable promise for the development of next generation therapeutics. After delineating the pathways relevant to tumor progression, rational strategies to inhibit important nodes of these pathways can be developed and tested in the whole animal. This approach has been successful for MEN2 therapeutics, a point that we discuss below.
Other Translational Models of Cancer
Efforts at developing cancer models with slightly more complexity have been successful, and recently a Drosophila model of human glioma, glioblastoma multiforme (GBM), was developed (Read et al., 2009). Human gliomas are deadly locally invading tumors that arise from glia and their precursor cells. Treatment options for gliomas are limited due to the nature of their location: chemotherapy, radiation, and if possible surgical resection are options. But most often the prognosis is poor with a mere 25% survival rate within 2 years of diagnosis (Kleihues et al., 1995; Maher et al., 2001; Furnari et al., 2007).
Gliomas commonly display constitutive activation of the epidermal growth factor receptor (EGFR) and PI3K pathways (Maher et al., 2001; Furnari et al., 2007). Activation of these two pathways in Drosophila embryonic glial cells resulted in massive over-proliferation of glial cells and a significantly larger larval brain. These brain-derived tumors grew and invaded into local tissues when transplanted into the abdomen of adult flies. It also led to activation of a network of genes linked to oncogenesis including Myc, Rb, Cyclins, and dTor (Read et al., 2009). Thus this model successfully showed that cooperative interaction between two pathways can induce an “oncogenic signature” that is distinct from the signatures that are observed by activation of either pathway alone, i.e., an emergent property (Table 1). Of interest, EGFR/PI3K pathway co-activation slightly earlier in the neuroblasts did not elicit glial hyper-proliferation. This suggested that the Drosophila larval glial cells have a “state” conducive to transformation that is distinct from the neuroblasts. Future studies will be required to identify differences between larval neuroblasts and committed glial precursors and establish how combinatorial signaling through PI3K and EGFR is kept under check in neuroblasts. This may prove helpful for identifying mechanisms that counter combinatorial activation of these pathways as a step toward targeted therapeutics.
A wing-based Drosophila genetic screen helped resolve a long-standing issue in another tumor type, colorectal cancer. The retinoblastoma gene (Rb) is a tumor suppressor that is mutated in a variety of cancers, yet Rb/E2F1 mutations are absent in colorectal tumors (Nevins, 2001). Most colorectal tumors require loss-of-function mutations in the adenomatous polyposis coli (Apc) gene; these mutations lead to stabilization of β-catenin and aberrant activation of the downstream effector cellular-myelocytomatosis (c-Myc; Kinzler and Vogelstein, 1996; Clevers, 2006). The Drosophila studies established E2F1 as a functional antagonist of β-catenin signaling. Using mouse studies and patient tumor analysis it was subsequently established that, in colon cancer, Rb represses E2F1 activity to allow aberrant β-catenin and c-Myc activity (Morris et al., 2008). Therefore, while Rb acts as a tumor suppressor in many tumors it acts as an oncogene in colorectal cancer. This study demonstrates the insights that Drosophila can provide into mechanisms driving human tumors (Table 1).
DROSOPHILA TO MAMMALS
Cell Polarity
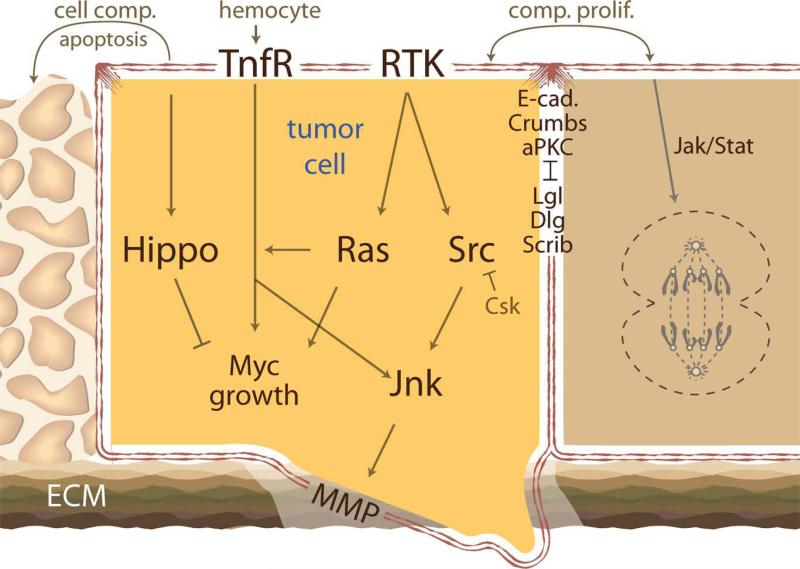
Most solid tumors arise from epithelial cells that have a defined apical-basal polarity and an early step of cell malignancy is the loss of this polarity. Pioneering studies by various groups in the 1970s identified the earliest polarity mutants and characterized the dramatic overgrowth and loss of polarity that occurred in tissues of these flies (Bryant and Schubiger, 1971; Gateff, 1978). When tissues mutant for the polarity genes discs-large (dlg) or lethal(2) giant larvae (lgl) were transplanted into wild-type host tissue, they invaded and colonized their surroundings, demonstrating some of the first in situ evidence for tumor suppressors (Woodhouse et al., 1994, 1998). Subsequent studies identified a host of other polarity genes and demonstrated an intimate regulatory network between all of them to promote cell polarization. In particular, Scribbled (Scrib), Lgl, and Dlg at the basolateral surface mutually antagonize two apical complexes also essential for polarity maintenance: atypical protein kinase C/Bazooka/PAR-6 and Crumbs/Stardust/Patj (Humbert et al., 2008; Fig. 2).
Fig. 2.
A schematic showing some major pathways implicated in cancer progression using Drosophila studies. Mechanisms of tumor cell behavior, like cell competition (cell comp.) and compensatory proliferation (comp. prolif.), were also first characterized in flies. Signal from the cell surface receptors to Hippo pathway depicted with an arrow.
The connection between loss of cell polarity and hyper-proliferation has gradually clarified in recent years. In general, it is context specific and depends on the mitotic status of the tissue (Grzeschik et al., 2007). In epithelial cells, Lgl localizes components of the Hippo pathway and affects Myc levels (Froldi et al., 2010; Grzeschik et al., 2010), pathways that are crucial regulators of growth and proliferation (see below). Studies of Scrib and Dlg have functionally classified specific structural domains with respect to the regulation of polarity versus growth (Brumby et al., 2003; Woods et al., 1996; Hough et al., 1997; Zeitler et al., 2004).
Human orthologs of Dlg display reduced protein levels as well as sub-cellular mislocalization in various gynecological malignancies (Gardiol et al., 2006). They interact with the colon cancer tumor suppressor Apc (Matsumine et al., 1996). Likewise human Lgl orthologs display lowered transcript/protein levels and lie within commonly deleted chromosomal regions in a variety of tumors (Grifoni et al., 2004; Korshunov et al., 2006; Lassmann et al., 2007; Table 1). While scrib mutations have not yet been identified in tumor sequencing studies, it shows reduced expression or mislocalization in tumor samples and its loss promotes tumor cell migration in vitro and in mouse models of breast cancer (Navarro et al., 2005; Gardiol et al., 2006; Dow et al., 2008; Zhan et al., 2008). aPKC has also been shown to be over-expressed in many cancer tissues and is associated with regional antagonism of human Lgl1 (Grifoni et al., 2007). As the early Drosophila screens predicted, cell polarity genes are important regulators of metastasis, a point further discussed below.
Growth and Control of Tissue Size
Deregulation of proliferation and growth control pathways is a defining feature of tumors. Genetic screens performed in Drosophila were instrumental in delineating a novel pathway that regulates cell growth and proliferation, the Hippo kinase cascade. Core kinase components Hippo (Hpo), Salvador (Sav), and Warts (Wts) work as tumor suppressors to negatively regulate the transcriptional co-activator Yorkie (Yki). Loss of hpo, sav, or wts, leads to excess Yorkie activity, transcriptional deregulation of its targets, and massive tissue overgrowth (Justice et al., 1995; Tapon et al., 2002; Udan et al., 2003; Huang et al., 2005; Pan, 2010). Loss of the Hippo pathway in imaginal discs leads to overgrowth; however, overall tissue structure and patterning remains unaffected suggesting that the pathway primarily regulates organ/tissue size. The Hippo pathway has been linked to apical basal polarity, planar cell polarity, and cell adhesion pathways. As such, there have been considerable efforts to understand more upstream components of the Hippo pathway, specifically how proteins at the membrane and associated adaptor proteins relay extracellular signals to the core cascade (Pan, 2010; Halder and Johnson, 2011).
An analogous Hippo kinase cascade exists in mammalian cells that similarly controls organ size. Over-expression of the mammalian Yki homolog YAP in the liver—or liver-specific knockout of the Hpo ortholog Mst1/2 or Sav ortholog Sav1—led to an enlarged liver and hepatocellular carcinoma (Dong et al., 2007; Lee et al., 2010; Lu et al., 2010). Interestingly, normal liver size can be restored following termination of Yap over-expression, suggesting that the Hippo cascade in mammals, like in flies, primarily regulates organ size control (Table 1; Fig. 2). However, just as in flies, the outcome of Hippo pathway deregulation seems to be context specific as deregulation of this pathway in other tissues leads to varying outcomes (Halder and Johnson, 2011).
Mutations of the mammalian Hippo pathway components are rare, the only described mutation being that of the tumor suppressor Merlin (Mer/NF2) that causes Neurofibromatosis 2 (Rouleau et al., 1993; Trofatter et al., 1993). But recent studies have shown that altered activity of this pathway is fairly common in many cancers (Pan, 2010). Hypermethylation and decreased expression of Mst1/2 (Minoo et al., 2007; Seidel et al., 2007) have been observed in sarcomas and colorectal/prostate cancer respectively. Likewise functional down-regulation of the Wts ortholog Lats1/2 through hypermethylation and by micro-RNA mediated silencing has been observed in testicular cancer (Takahashi et al., 2005; Jiang et al., 2006). Thus, genetic studies in Drosophila were instrumental in not only establishing the Hippo pathway as regulator of tissue size and growth but also in drawing attention to its possible role in human cancer. Future studies should establish to what extent the transforming ability of Hippo pathway components is dose sensitive as well as context dependent, similar to its role during development. Its utility as a therapeutic target is also an open question.
Invasion
One of the key stages of tumor progression is the ability of tumor cells to invade into surrounding tissues, and components of the c-Jun N-terminal kinase (JNK) pathway are key mediators of this process. The JNK pathway has long been implicated in tumori-genesis, having both pro-tumorigenic and anti-tumorigenic properties (Wagner and Nebreda, 2009). These contrasting features of JNK activity have been teased out in several developmental and translational studies in Drosophila. These in situ studies have proven a useful complement to work done in vertebrates and in many cases are identifying novel features that remain to be investigated in more complex organisms.
Early work in developmental models identified a critical role for JNK in epithelial morphogenesis. A prototypical example of this is embryonic dorsal closure, a process with molecular and morphological similarities to vertebrate wound healing (Grose and Martin, 1999; Stronach and Perrimon, 1999; Jacinto et al., 2000; Kockel et al., 2001). Transcriptional profiling of JNK-defective embryos undergoing dorsal closure identified several putative transcriptional targets with known roles in cytoskeletal remodeling or cell adhesion. Subsequent characterization of one such hit, the ortholog for Profilin, demonstrated its important role as an effector of this process (Jasper et al., 2001). Time-lapse imaging showed that JNK activity resulted in increased Actin cable tension and elevated integrin levels at the leading edge of the contralateral epithelial sheets, both of which were necessary to promote “zippering” of the tissue at the dorsal mid-line (Homsy et al., 2006).
This suspicion was borne out in more translational studies where the role of JNK in “oncogene cooperativity” was addressed using GFP-marked clones in the eye. These studies showed that loss of the polarity regulator Scrib resulted in transient proliferation, though eventually these cells were eliminated through JNK-dependent apoptosis. However, pairing genotypically scrib–/– null clones with Ras pathway activation led to dramatic tissue overgrowth, invasion into adjacent tissues, and organismal lethality (Brumby and Richardson, 2003). This finding was consistent with studies which showed that Ras pathway activation could inhibit the function of pro-apoptotic factor hid, thereby unleashing JNK pathway's ability to promote invasion (Bergmann et al., 1998; Kurada and White, 1998). Thus the genetic context in which JNK pathway is activated determines whether cells undergo apoptosis or invade (Brumby and Richardson, 2003; Igaki et al., 2006; Uhlirova and Bohmann, 2006). Additional experiments indicated that JNK simultaneously mediates the apoptosis of the scrib–/– cells as well as “compensatory proliferation” of the adjacent tissue (see below; Table 1; Uhlirova et al., 2005). More intriguingly the matrix metalloprotease ortholog Mmp1, a component essential for tumor invasiveness, was shown to be a direct transcriptional target of the JNK pathway. Reducing JNK activity suppressed Mmp1 expression and invasiveness into the adjacent tissue (Uhlirova and Bohmann, 2006) suggesting that JNK plays an important role in tumor invasion.
More recent work has expanded the role of JNK as a mediator of tumorigenesis, showing that it can link the Ras and Rho signaling axes. A genetic screen carried out to identify enhancers of Ras-mediated hyperplasia identified several members of the Rho family of GTPases; again, synergy between the two pathways depended on JNK (Brumby et al., 2011). Finally, the complexity of JNK pathway signaling is becoming apparent from recent findings which show that JNK can promote proliferation through the Hippo pathway (Sun and Irvine, 2011).
Fly Models of Metastasis
Most cancer work has focused on the primary tumor. However, most cancer patients die of metastases, the derivative secondary tumors found at distant sites. Recent sequencing data have indicated that these secondary tumors are genetically similar to sub-regions of primary tumors (Campbell et al., 2010; Yachida et al., 2010). Nevertheless, they likely develop significant differences as they remake themselves to invade the bone marrow, liver, brain, etc. The process of metastasis is complicated and likely depends in part on the details of the mutations that drive the primary tumor as well as the specific invaded site. From a mechanistic standpoint, Drosophila has much to offer in understanding these complex processes, and the field of metastasis will have perhaps the greatest growth in the fly cancer field in the next few years.
Metastasis screens
Our current understanding of tumor progression has progressed significantly since the “two-hit” model of tumorigenesis was proposed (Knudson, 1971). We know that multiple mutations/genetic alterations that affect multiple pathways often cooperate to drive tumorigenesis (Nowell, 1976; Fearon and Vogelstein, 1990). Tissue culture and mouse models have been developed over the past decade to more accurately study the effect of cooperating genetic mutations. A pioneering genetic screen made use of the genetic tractability of Drosophila to identify second-loci mutations that cooperated with oncogenic RasV12 mutation. Clones of cells with mutations in cell polarity genes dlg, lgl, or scrib are poorly invasive and often eliminated from the eye epithelia. But in the presence of RasV12 mutation these dlg, lgl, or scrib mutant eye tissues over-proliferated and colonized distant tissues (Pagliarini and Xu, 2003; Table 1). Over 30% of human tumors screened have activating mutations in one of the three Ras orthologs (Fernandez-Medarde and Santos, 2011) and, as mentioned previously, both Dlg and Lgl have been shown to be deregulated in several human tumors. Whether the vertebrate Ras mutations actually cooperate with the Dlg/Lgl axis to drive tumor progression and metastasis remains to be shown. Nevertheless, the ability to perform such unbiased genetic screens to identify tumor promoting genetic combinations has made Drosophila an immensely useful system to model cancer.
Src and metastasis
Our laboratory has taken a somewhat different approach to studying metastasis, and it stems from our work on dRetMEN2. Src and its negative regulator Csk were identified as genetic modifiers of dRetMEN2 (Read et al., 2005). Our subsequent work on Src/Csk pointed to its role in metastasis, a view supported by a large body of work in the mammalian world. Our contribution has been to explore elevated Src in the context of local cell–cell interactions, where Src plays a central role in promoting migration, invasion, and metastasis-like cell behavior.
Knockdown of Csk with the patched promoter led to activation of Src in a stripe across the anterior/posterior mid-line of the developing wing disc. Cells within this stripe exhibited increased proliferation, a well-known outcome of increased Src activity. Of interest, cells at the border of the patched stripe shifted basally and then migrated away from the original patched domain. Migration of these cells was triggered by a pathway that included E-cadherin, Rho1, Jnk, and matrix metallopro-teases. These basal cells also contained high levels of cleaved caspases and were ultimately eliminated by apoptosis. Interestingly, removing one functional copy of E-cadherin blocked migration, suggesting it is required to mediate part of the Src signal (Vidal et al., 2006). Our laboratory has begun to more carefully explore the mechanisms by which the Src-Cadherin-Rho-Jnk axis directs migration (Table 1). We are also exploring how other oncogenes such as Ras synergize with Src in this paradigm (Vidal et al., 2007; Fig. 2).
Tumor Microenvironment
Since Hanahan and Weinberg proposed six “hallmarks of cancer” that focused on the emerging tumor (Hanahan and Weinberg, 2000), over a decade of research has shed considerable light on the importance of the tumor cell environment in regulating tumorigenesis. Cancer-associated fibroblasts (CAFs) and pericytes are cell types often associated with human tumor cells (Kalluri and Zeisberg, 2006; Ostman and Augsten, 2009). These “nontransformed” cells have been shown to regulate tumor progression by secreting growth promoting signals and anti-apoptotic survival signals, promoting angiogenesis, and contributing to tissue invasion and metastasis (Pietras and Ostman, 2010; Hanahan and Weinberg, 2011).
A key “hallmark” of tumor cells is their ability to avoid apoptotic death. The mammalian tumor necrosis factor (TNF)—a cytokine regulating inflammation, immunity, and cell architecture—has a dual context-dependent role as a tumor suppressor as well as an oncogene. On one hand, several studies show that TNF can promote apoptotic death in both normal and malignant cells while other studies show that TNF can promote inflammation and metastasis (Mocellin and Nitti, 2008). Recent Drosophila studies have added to this knowledge showing that the Drosophila TNF ortholog Eiger (Egr) can promote as well as suppress tumorigenesis depending on the genetic context of the tumor. scrib–/– cells of the eye and wing are eliminated by egr-JNK mediated apoptotic signaling from endocytic vesicles. Removal of egr from scrib mutant cells (scrib–/–; egr–/–) resulted in suppression of death and enhanced tumor proliferation, suggesting egr has a tumor suppressor-like role in the context of scrib mutant tissue (Igaki et al., 2009). Conversely, removal of egr from cells with a different genetic background, scrib–/–; RasV12;egr–/–, suppressed invasive tumorigenesis suggesting egr may function as a tumor promoter in the context of activated Ras and pointing to the importance of genetic context. Of interest, latter studies showed that removal of egr function from circulating hemocytes (Drosophila immune cells) also suppressed tumor progression of scrib–/–; RasV12 cells, indicating a nonautonomous role of the egr pathway in promoting Ras tumor progression (Cordero et al., 2010). Thus, the tumor-microenvironment appears to be a site of immune cell enrichment (Fig. 1A–C) that, through egr/TNF signaling, controls apoptotic death or invasion and regulates tumor progression (Table 1; Fig. 2).
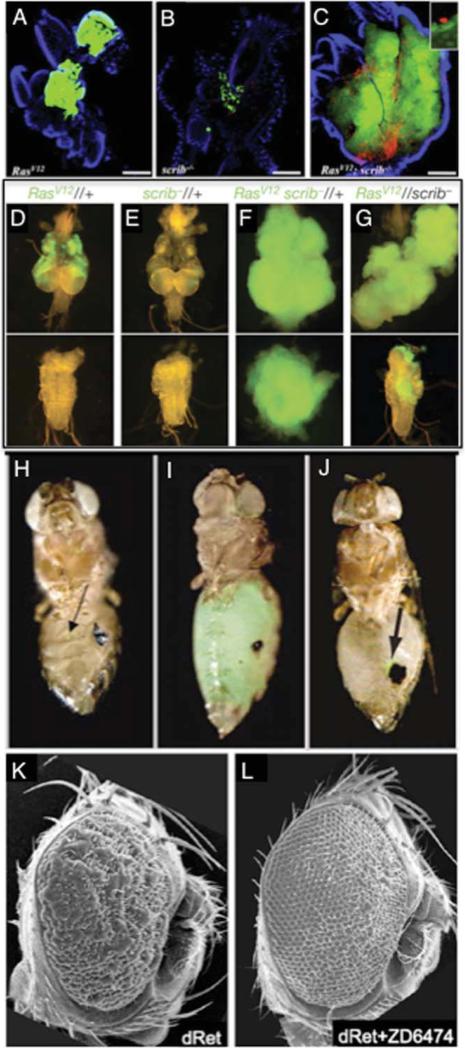
Fig. 1.
Teaching an old fly new tricks. A–C: Immune cells known as hemocytes were labeled with red fluorescent protein (RFP) and transplanted into flies expressing oncogenic Ras and/or lacking the tumor suppressor scrib. These hemocytes were only recruited to site of the transgene expression in RasV12;scrib tumors (Cordero et al., 2010). D–G: Oncogenic RasV12 and scrib loss of function synergize within a given clone to produce malignant over-growth and invasion into adjacent tissues. However, they may also synergize when present in adjacent clones (Wu et al., 2010). H–J: Wild-type (left) and l(3)mbt mutant (middle) tissue labeled with green fluorescent protein (GFP) were serially transplanted into the abdomen of adult hosts. Comparison of expression profiles showed l(3)mbt tumors were enriched for mRNA's of genes required for germline maintenance. Removal of some germline genes, such as piwi (right), abrogated tumor overgrowth (Janic et al., 2010). K,L: Overexpression of dRetMEN2 in the fly eye results in dysplastic growth, reminiscent of tumors from the MEN2 cancer syndrome. Administration of a kinase inhibitor ZD6474 restores these eyes to a wild-type pattern (Vidal et al., 2005). This therapeutic has been validated for MEN2 patients.
Studies in Drosophila have shown that genetic differences between tumor cells and their microenvironment cooperate to promote tumorigenesis. When clonal patches of cells expressing different levels of the oncoprotein Myc are juxtaposed, clones expressing lower levels of Myc were eliminated by apoptosis (de la Cova et al., 2004; Moreno and Basler, 2004). This process of “cell competition” has recently been observed in mammalian cell line studies using activated Ras and Src, and in transgenic mouse studies with p53 (Hogan et al., 2009; Bondar and Medzhitov, 2010; Kajita et al., 2010). Our Csk knockdown studies showed that the invasive ability of tumor cells could be enhanced when juxtaposed to wild-type cells, suggesting that tumor cells at the periphery of tumors are more invasive. But what pathways in the neighboring wild-type cells might mediate this process? A recent study showed that genetic alterations of the tumor microenvironment could itself alter tumor progression. Juxtaposing patches of cells mutant for scrib–/– with patches of RasV12 expressing tumor cells led to a significant enhancement of tumorigenesis, almost equivalent to having both genetic changes in the same cell (Fig. 1D–G). JNK and the JAK-STAT pathways in adjacent cell populations were involved in providing the “communication” through which these mutations promoted neoplastic overgrowth (Wu et al., 2010). Taken together, these findings may lead us to broaden the multi-hit model of tumor progression, indicating that multiple genetic changes need not be in the same cell (Table 1; Fig. 2).
Several Drosophila studies have shown that physical and genetic abuse of cells that lead to Caspase-dependent apoptotic death can stimulate neighboring cells to grow (Huh et al., 2004; Perez-Garijo et al., 2004; Ryoo et al., 2004). This phenomenon of “compensatory proliferation” allows tissues to restore overall organ size even when a significant number of cells are initially lost. Caspase activity has been intricately connected to tumor progression, leading to the idea that Caspase-induced “compensatory proliferation” might be involved in human tumors (Gallant, 2005). Consistent with this view, recent evidence using transgenic mouse and cancer cell lines have shown Caspase-dependent “compensatory proliferation” to be operative during tumor progression. Irradiated feeder 4T1 breast cancer cells activated Caspase3 and, through prostaglandin signaling, promoted proliferation of overlying nonirradiated 4T1 cells in vitro and in xenografts (Huang et al., 2011; Table 1). This finding may pose a cautionary note for treatment of tumors. Ineffective killing of tumor cells (or even treatment-induced Caspase3 activation in normal cells) could spur non-autonomous proliferation of a few surviving tumor cells and lead to rapid re-establishment of tumors.
Chromatin Regulators in Tumorigenesis
Genetic alterations that affect chromatin regulators and simultaneously deregulate multiple pathways are being uncovered as mediators of tumorigenesis. By virtue of controlling multiple pathways relevant to tumorigenesis—proliferation, invasion, differentiation, death, etc.—chromatin regulators may prove to be ideal targets for deregulation during cancer progression. Through serial tumor transplantations in the Drosophila abdomen, loss of function mutations in the chromatin repressor lethal(3) malignant brain tumor [l(3)mbt] led to aggressive brain-derived tumors (Fig. 1H–J). Whole genome expression profiling of these serial tumors revealed loss of somatic cell fate and up-regulation of a cohort of germline specific genes (Janic et al., 2010). Interestingly, acquisition of soma-to-germline characteristics was previously observed in Caenorhabditis elegans mutants of Rb, a global chromatin regulator and transcription factor (Wang et al., 2005; Strome and Lehmann, 2007; Curran et al., 2009). Up-regulation of germline-specific genes has also been documented in several cancer types, leading to the hypothesis that acquisition of germline fate aids tumor fitness and survival (Simpson et al., 2005). Serial tumor transplantation strategy coupled with expression profiling in Drosophila could become a useful tool to model metastatic cancers and identify deregulated pathways and “gene signatures” that drive metastasis.
But how commonly are chromatin regulators actually involved in vertebrate tumorigenesis? Mouse models and patient tumor analysis recently showed that down-regulation and loss of histone variant macroH2A (mH2A) can lead to metastatic melanoma. mH2A is a histone variant that regulates a large cohort of genes involved in a large number of developmental programs. Reduction of mH2A promoted tumorigenesis partly through CDK8 activation (Kapoor et al., 2010). Polycomb group (PcG) proteins are an evolutionarily conserved family of epigenetic chromatin repressors that control expression of genes regulating cell cycle and differentiation. Vertebrate PcG protein, proto-oncogene BmI1, negatively regulates key inhibitors of cell cycle progression, INK4A and ARF. Other PcG proteins show significant up-regulation in a variety of human cancers, indicating these proteins are important regulators of neoplastic progression (Sparmann and van Lohuizen, 2006; Schuettengruber et al., 2007). Genetic studies in Drosophila showed that loss of function of certain PcG genes could actually drive neoplastic progression in developing discs, through up-regulation of JAK/STAT and Notch signaling pathways (Classen et al., 2009; Martinez et al., 2009). Thus the fly studies uncovered novel complex context-dependent roles of chromatin regulators, like the PcG proteins, both as oncogenes as well as tumor suppressors. Our own studies identified mi-2/CHD3, a chromodomain helicase gene, as a genetic modifier of oncogenic RetMEN2 in the Drosophila eye. As an important parallel, we showed that there was frequent loss of heterozygosity (LOH) of the CHD3 locus in Ret dependent pheochromocytomas, tumors of the adrenal gland (Read et al., 2005). CHD3 is closely related to its paralog CHD5, a gene whose deficiency in transgenic mouse models down-regulated the p19Arf/p53 pathway predisposing cells to oncogenic transformation. The CHD5 locus is also frequently lost in several neural, epithelial, and hematopoietic tumors (Bagchi et al., 2007; Bagchi and Mills, 2008; Fujita et al., 2008; Mulero-Navarro and Esteller, 2008; Wang et al., 2009; Table 1). The exact mechanism by which other members of the CHD3/4/5 subfamily of chromodomain genes can contribute to tumor progression remains to be shown. Therefore, multiple lines of evidence including Drosophila studies have documented several cancer paradigms where deregulation of chromatin regulators promotes tumor progression.
Drug Discovery and Therapeutics
The current emphasis on cell- and luciferase-based drug screening is giving way to a more nuanced view of drug discovery: while target-based screens will remain dominant, whole animal phenotype-based screening is gathering interest. Whole animal model systems hold promise as important contributors to this new (really old) approach as they provide an accurate readout for “therapeutic index” of treatment.
To date, Drosophila has been used sparingly in drug discovery. The reasons are primarily cultural: as a field we have been primarily concerned with issues in development and cell biology, and we prefer to explore mechanism vs. more directly practical issues of therapeutics. This view is slowly changing as interest in therapeutics and pressure for practical outcomes increases. This is one field where Drosophila is playing catch-up, but the fly holds some unique advantages. Drosophila's famously powerful genetic tools are readily adapted to building sophisticated disease models, the important first step for useful drug discovery. Our laboratory and others are developing technology for screening these models in a whole animal setting.
A small number of Drosophila drug screening efforts have been reported to date. Early models of neuronal degeneration have explored candidate compounds that slow neuronal loss (Min and Benzer, 1999; Zhang et al., 2005). A pioneering study used a well-designed fly model of Fragile-X by establishing that loss of the fmr1 gene leads to lethality in the presence of excess glutamate (Chang et al., 2008). Screening flies in a 96-well format led to the identification of multiple compounds that rescued lethality, including chemical modulators of the γ-aminobutyric acid-ergic (GABAergic) pathway. These results demonstrate some of the promise for whole animal screening presented by Drosophila.
To our knowledge the sole example of a cancer drug reported to suppress disease in Drosophila that has been subsequently approved for clinical treatment is our MEN2 model, described above. Recall that targeting oncogenic dRet-MEN2 to the developing eye led to a rough eye phenotype due to over-proliferation, cell fate defects, and compensatory apoptosis (Read et al., 2005). Screening a limited set of compounds including clinically relevant drugs led to the identification of ZD6474/Vandetanib, an anilinoquinazoline-class kinase inhibitor that acts as a broad RTK inhibitor (Wedge et al., 2002). Cell culture and xenograft studies demonstrated its utility to target Ret-based growth (Carlo-magno et al., 2002). When fed orally to our fly MEN2 model Vandetanib was strongly effective in suppressing effects of the dRetMEN2 at concentrations far lower than those required to demonstrate whole animal toxicity (Vidal et al., 2005; Fig. 1K,L). The latter point is key: other compounds can also rescue the rough eye phenotype but most show reduced animal viability, the sort of information only gained from whole animal screening. Subsequent clinical trials demonstrated Vandetanib's utility as a first-line therapeutic in patients, and the FDA recently approved it as the first chemotherapeutic for medullary thyroid carcinoma (Deshpande et al., 2010; Table 1).
Vandetanib suppresses the activity of a broad range of RTKs (Karaman et al., 2008) so why was it successful in clinics? One view is that its activity against Ret is the only relevant activity. Our experiments suggest otherwise: we have not observed a clear correlation between activity against Ret and an overall useful therapeutic index in flies. This is surprising given that transformation is strictly due to introduction of dRetMEN2 in our fly models and due to activated Ret iso-forms in patients. In fact, we expect that its “off-target” effects are an important part of Vandetanib's usefulness. Suppressing VEGFR, EGFR, and other kinases may contribute to its utility, and not suppressing Ret activity too efficiently may reduce its toxicity. Importantly, these considerations are only accounted for with whole animal screening: reporter-based assays would reject Vandetanib as too inefficient to the target and too low in its specificity. Of note, most successful drugs have off-target activities that are important for their utility (Petrelli and Giordano, 2008).
Novel inhibitors identified using whole animal models will not be immediately available to patients/clinics due to the long drug approval process. So there is an enormous effort at NIH to provide immediate treatment options to patients through combinations of approved drugs. Rational combinations that target different aspects of tumor signaling could be useful to suppress cancer progression and also delay emergence of drug resistance (Dancey and Chen, 2006). Again, combinatorial screens using cell lines are not predictive of clinical success while combinatorial screens with mouse models are inordinately expensive. Large number of approved drug-dose combinations could be rapidly screened in fly models and useful combinations easily validated in mice and cell lines (Dancey and Chen, 2006; Das and Cagan, 2010).
PERSPECTIVES/FUTURE DIRECTIONS
At the start of this review we described two basic approaches used by Drosophila in the exploration of cancer mechanism and therapeutics. Both approaches will continue to contribute to our understanding of tumor progression, albeit in different ways.
Mammals to Drosophila to Mammals
The Drosophila models of MEN2 and Ras/Csk metastasis have been successful in elucidating novel disease mechanisms and even in identifying novel therapeutics that have succeeded in clinical trials. The development of more cancer models guided by patient tumor data will be extremely useful to accurately model different types of tumors. These models will also be amenable to drug screening to identify novel therapeutics. Beyond simple single-hit models, the sophisticated genetic tools in flies will allow for the development of more complex multigenic models of cancer. The presence of multiple genetic alterations driving tumor progression is the rule in most major types of cancer including colon, pancreatic, lung, breast, etc. Informed by patient tumor data and disease stratification strategies, Drosophila models should continue to improve in complexity, accuracy, and usefulness. Modeling multigenic disease holds the promise of stratifying patient populations into functional subgroups and finding tailored therapeutic strategies most appropriate for each group, that is, personalized medicine.
Complex cancer progression paradigms, e.g., colon cancer, also require the precise temporal accumulation of genetic/epigenetic changes (Fearon and Vogelstein, 1990; de Lau et al., 2007). To accurately reflect this in our multigenic models we foresee a need for technical improvements in the Drosophila genetic toolkit. The GAL4/UAS system is currently the only widely used, spatially controlled over-expression system available in flies. Development or better use of orthogonal, regulatable expression systems—e.g., TetR ON/OFF system (Stebbins et al., 2001; Stebbins and Yin, 2001)—pairwise with the GAL4/UAS system will allow for different transgenes to be activated in a precise temporal sequence, more accurately modeling the sequential deregulation of different oncogenic pathways.
Drosophila to Mammals
The identification of cell polarity mutants and their subsequent implication in human tumors is a classic example of how basic Drosophila research has contributed to our understanding of cancer. Several mechanistic insights into oncogenic pathways driven by Ras, Src, JNK, Egr, etc., have come from flies. Fundamental conceptual insights into tumor cell behavior including “compensatory proliferation” and “cell competition” have also been discovered using flies, which specialize in high-resolution examination of local cellular events (Fig. 2). Discovery of such complex cellular behavior is made possible by the sophisticated genetic tools available in flies, and has helped challenge vertebrate researchers to continue exploring the full complexity of tumors in the context of the larger environment. Future developmental, cell-biological, and molecular studies in flies should continue to inform us about tumor biology in known as well as unexpected ways.
ACKNOWLEDGMENTS
V.R. is supported by T32 grant from NIH. R.C. is supported by NIH grants. T.D. is supported by ACS fellowship grant.
REFERENCES
- Bagchi A, Mills AA. The quest for the 1p36 tumor suppressor. Cancer Res. 2008;68:2551–2556. doi: 10.1158/0008-5472.CAN-07-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagchi A, Papazoglu C, Wu Y, Capurso D, Brodt M, Francis D, Bredel M, Vogel H, Mills AA. CHD5 is a tumor suppressor at human 1p36. Cell. 2007;128:459–475. doi: 10.1016/j.cell.2006.11.052. [DOI] [PubMed] [Google Scholar]
- Barkan B, Starinsky S, Friedman E, Stein R, Kloog Y. The Ras inhibitor farnesylthiosalicylic acid as a potential therapy for neurofibromatosis type 1. Clin Cancer Res. 2006;12:5533–5542. doi: 10.1158/1078-0432.CCR-06-0792. [DOI] [PubMed] [Google Scholar]
- Bergmann A, Agapite J, McCall K, Steller H. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell. 1998;95:331–341. doi: 10.1016/s0092-8674(00)81765-1. [DOI] [PubMed] [Google Scholar]
- Bobinski M, Greco CM, Schrot RJ. Giant intracranial medullary thyroid carcinoma metastasis presenting as apoplexy. Skull Base. 2009;19:359–362. doi: 10.1055/s-0029-1220202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondar T, Medzhitov R. p53-mediated hematopoietic stem and progenitor cell competition. Cell Stem Cell. 2010;6:309–322. doi: 10.1016/j.stem.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumby AM, Richardson HE. scribble mutants cooperate with oncogenic Ras or Notch to cause neoplastic over-growth in Drosophila. EMBO J. 2003;22:5769–5779. doi: 10.1093/emboj/cdg548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumby AM, Goulding KR, Schlosser T, Loi S, Galea R, Khoo P, Bolden JE, Aigaki T, Humbert PO, Richardson HE. Identification of novel Ras-cooperating oncogenes in Drosophila melanogaster: a RhoGEF/Rho-family/JNK pathway is a central driver of tumorigenesis. Genetics. 2011;188:105–125. doi: 10.1534/genetics.111.127910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant PJ, Schubiger G. Giant and duplicated imaginal discs in a new lethal mutant of Drosophila melanogaster. Dev Biol. 1971;24:233–263. doi: 10.1016/0012-1606(71)90097-2. [DOI] [PubMed] [Google Scholar]
- Cakir M, Grossman AB. Medullary thyroid cancer: molecular biology and novel molecular therapies. Neuroendocrinology. 2009;90:323–348. doi: 10.1159/000220827. [DOI] [PubMed] [Google Scholar]
- Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, McBride DJ, Varela I, Nik-Zainal SA, Leroy C, Jia M, Menzies A, Butler AP, Teague JW, Griffin CA, Burton J, Swerdlow H, Quail MA, Stratton MR, Iacobuzio-Donahue C, Futreal PA. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlomagno F, Vitagliano D, Guida T, Ciardiello F, Tortora G, Vecchio G, Ryan AJ, Fontanini G, Fusco A, Santoro M. ZD6474, an orally available inhibitor of KDR tyrosine kinase activity, efficiently blocks oncogenic RET kinases. Cancer Res. 2002;62:7284–7290. [PubMed] [Google Scholar]
- Carlson KM, Dou S, Chi D, Scavarda N, Toshima K, Jackson CE, Wells SA, Jr, Goodfellow PJ, Donis-Keller H. Single missense mutation in the tyro-sine kinase catalytic domain of the RET protooncogene is associated with multiple endocrine neoplasia type 2B. Proc Natl Acad Sci U S A. 1994;91:1579–1583. doi: 10.1073/pnas.91.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Bray SM, Li Z, Zarnescu DC, He C, Jin P, Warren ST. Identification of small molecules rescuing fragile X syndrome phenotypes in Drosophila. Nat Chem Biol. 2008;4:256–263. doi: 10.1038/nchembio.78. [DOI] [PubMed] [Google Scholar]
- Classen AK, Bunker BD, Harvey KF, Vaccari T, Bilder D. A tumor suppressor activity of Drosophila Polycomb genes mediated by JAK-STAT signaling. Nat Genet. 2009;41:1150–1155. doi: 10.1038/ng.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Cordero JB, Macagno JP, Stefanatos RK, Strathdee KE, Cagan RL, Vidal M. Oncogenic Ras diverts a host TNF tumor suppressor activity into tumor promoter. Dev Cell. 2010;18:999–1011. doi: 10.1016/j.devcel.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cranston AN, Ponder BA. Modulation of medullary thyroid carcinoma penetrance suggests the presence of modifier genes in a RET transgenic mouse model. Cancer Res. 2003;63:4777–4780. [PubMed] [Google Scholar]
- Curran SP, Wu X, Riedel CG, Ruvkun G. A soma-to-germline transformation in long-lived Caenorhabditis elegans mutants. Nature. 2009;459:1079–1084. doi: 10.1038/nature08106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancey JE, Chen HX. Strategies for optimizing combinations of molecularly targeted anticancer agents. Nat Rev Drug Discov. 2006;5:649–659. doi: 10.1038/nrd2089. [DOI] [PubMed] [Google Scholar]
- Das T, Cagan R. Drosophila as a novel therapeutic discovery tool for thyroid cancer. Thyroid. 2010;20:689–695. doi: 10.1089/thy.2010.1637. [DOI] [PubMed] [Google Scholar]
- de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA. Drosophila myc regulates organ size by inducing cell competition. Cell. 2004;117:107–116. doi: 10.1016/s0092-8674(04)00214-4. [DOI] [PubMed] [Google Scholar]
- de Lau W, Barker N, Clevers H. WNT signaling in the normal intestine and colorectal cancer. Front Biosci. 2007;12:471–491. doi: 10.2741/2076. [DOI] [PubMed] [Google Scholar]
- Deshpande H, Roman S, Thumar J, Sosa JA. Vandetanib (ZD6474) in the Treatment of Medullary Thyroid Cancer. Clin Med Insights Oncol. 2010;5:213–221. doi: 10.4137/CMO.S6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis–Keller H, Dou S, Chi D, Carlson KM, Toshima K, Lairmore TC, Howe JR, Moley JF, Goodfellow P, Wells SA., Jr Mutations in the RET protooncogene are associated with MEN 2A and FMTC. Hum Mol Genet. 1993;2:851–856. doi: 10.1093/hmg/2.7.851. [DOI] [PubMed] [Google Scholar]
- Dow LE, Elsum IA, King CL, Kinross KM, Richardson HE, Humbert PO. Loss of human Scribble cooperates with H-Ras to promote cell invasion through deregulation of MAPK signalling. Oncogene. 2008;27:5988–6001. doi: 10.1038/onc.2008.219. [DOI] [PubMed] [Google Scholar]
- Eng C, Clayton D, Schuffenecker I, Lenoir G, Cote G, Gagel RF, van Amstel HK, Lips CJ, Nishisho I, Takai SI, Marsh DJ, Robinson BG, Frank-Raue K, Raue F, Xue F, Noll WW, Romei C, Pacini F, Fink M, Niederle B, Zedenius J, Nordenskjold M, Komminoth P, Hendy GN, Mulligan LM, et al. The relationship between specific RET proto-onco-gene mutations and disease phenotype in multiple endocrine neoplasia type 2. International RET mutation consortium analysis. JAMA. 1996;276:1575–1579. [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Fernandez-Medarde A, Santos E. Ras in cancer and developmental diseases. Genes Cancer. 2011;2:344–358. doi: 10.1177/1947601911411084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferner RE. Neurofibromatosis 1 and neurofibromatosis 2: a twenty first century perspective. Lancet Neurol. 2007;6:340–351. doi: 10.1016/S1474-4422(07)70075-3. [DOI] [PubMed] [Google Scholar]
- Froldi F, Ziosi M, Garoia F, Pession A, Grzeschik NA, Bellosta P, Strand D, Richardson HE, Grifoni D. The lethal giant larvae tumour suppressor mutation requires dMyc oncoprotein to promote clonal malignancy. BMC Biol. 2010;8:33. doi: 10.1186/1741-7007-8-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T, Igarashi J, Okawa ER, Gotoh T, Manne J, Kolla V, Kim J, Zhao H, Pawel BR, London WB, Maris JM, White PS, Brodeur GM. CHD5, a tumor suppressor gene deleted from 1p36.31 in neuroblastomas. J Natl Cancer Inst. 2008;100:940–949. doi: 10.1093/jnci/djn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, Chin L, DePinho RA, Cavenee WK. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–2710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- Gallant P. Myc, cell competition, and compensatory proliferation. Cancer Res. 2005;65:6485–6487. doi: 10.1158/0008-5472.CAN-05-1101. [DOI] [PubMed] [Google Scholar]
- Gardiol D, Zacchi A, Petrera F, Stanta G, Banks L. Human discs large and scrib are localized at the same regions in colon mucosa and changes in their expression patterns are correlated with loss of tissue architecture during malignant progression. Int J Cancer. 2006;119:1285–1290. doi: 10.1002/ijc.21982. [DOI] [PubMed] [Google Scholar]
- Gateff E. Malignant neoplasms of genetic origin in Drosophila melanogaster. Science. 1978;200:1448–1459. doi: 10.1126/science.96525. [DOI] [PubMed] [Google Scholar]
- Grifoni D, Garoia F, Schimanski CC, Schmitz G, Laurenti E, Galle PR, Pession A, Cavicchi S, Strand D. The human protein Hugl-1 substitutes for Drosophila lethal giant larvae tumour suppressor function in vivo. Oncogene. 2004;23:8688–8694. doi: 10.1038/sj.onc.1208023. [DOI] [PubMed] [Google Scholar]
- Grifoni D, Garoia F, Bellosta P, Parisi F, De Biase D, Collina G, Strand D, Cavicchi S, Pession A. aPKCzeta cortical loading is associated with Lgl cytoplasmic release and tumor growth in Drosophila and human epithelia. Oncogene. 2007;26:5960–5965. doi: 10.1038/sj.onc.1210389. [DOI] [PubMed] [Google Scholar]
- Grose R, Martin P. Parallels between wound repair and morphogenesis in the embryo. Semin Cell Dev Biol. 1999;10:395–404. doi: 10.1006/scdb.1999.0326. [DOI] [PubMed] [Google Scholar]
- Grzeschik NA, Amin N, Secombe J, Brumby AM, Richardson HE. Abnormalities in cell proliferation and apico-basal cell polarity are separable in Drosophila lgl mutant clones in the developing eye. Dev Biol. 2007;311:106–123. doi: 10.1016/j.ydbio.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzeschik NA, Parsons LM, Allott ML, Harvey KF, Richardson HE. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hazard JB, Hawk WA, Crile G., Jr Medullary (solid) carcinoma of the thyroid; a clinicopathologic entity. J Clin Endocrinol Metab. 1959;19:152–161. doi: 10.1210/jcem-19-1-152. [DOI] [PubMed] [Google Scholar]
- Hogan C, Dupre-Crochet S, Norman M, Kajita M, Zimmermann C, Pelling AE, Piddini E, Baena-Lopez LA, Vincent JP, Itoh Y, Hosoya H, Pichaud F, Fujita Y. Characterization of the interface between normal and transformed epithelial cells. Nat Cell Biol. 2009;11:460–467. doi: 10.1038/ncb1853. [DOI] [PubMed] [Google Scholar]
- Homsy JG, Jasper H, Peralta XG, Wu H, Kiehart DP, Bohmann D. JNK signaling coordinates integrin and actin functions during Drosophila embryo-genesis. Dev Dyn. 2006;235:427–434. doi: 10.1002/dvdy.20649. [DOI] [PubMed] [Google Scholar]
- Hough CD, Woods DF, Park S, Bryant PJ. Organizing a functional junctional complex requires specific domains of the Drosophila MAGUK Discs large. Genes Dev. 1997;11:3242–3253. doi: 10.1101/gad.11.23.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, Pan D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Huang Q, Li F, Liu X, Li W, Shi W, Liu FF, O'sullivan B, He Z, Peng Y, Tan AC, Zhou L, Shen J, Han G, Wang XJ, Thorburn J, Thorburn A, Jimeno A, Raben D, Bedford JS, Li CY. Caspase 3-mediated stimulation of tumor cell repopulation during cancer radio-therapy. Nat Med. 2011;17:860–866. doi: 10.1038/nm.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh JR, Guo M, Hay BA. Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr Biol. 2004;14:1262–1266. doi: 10.1016/j.cub.2004.06.015. [DOI] [PubMed] [Google Scholar]
- Humbert PO, Grzeschik NA, Brumby AM, Galea R, Elsum I, Richardson HE. Control of tumourigenesis by the Scribble/Dlg/Lgl polarity module. Oncogene. 2008;27:6888–6907. doi: 10.1038/onc.2008.341. [DOI] [PubMed] [Google Scholar]
- Igaki T, Pagliarini RA, Xu T. Loss of cell polarity drives tumor growth and invasion through JNK activation in Drosophila. Curr Biol. 2006;16:1139–1146. doi: 10.1016/j.cub.2006.04.042. [DOI] [PubMed] [Google Scholar]
- Igaki T, Pastor-Pareja JC, Aonuma H, Miura M, Xu T. Intrinsic tumor suppression and epithelial maintenance by endocytic activation of Eiger/TNF signaling in Drosophila. Dev Cell. 2009;16:458–465. doi: 10.1016/j.devcel.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto A, Wood W, Balayo T, Turmaine M, Martinez-Arias A, Martin P. Dynamic actin-based epithelial adhesion and cell matching during Drosophila dorsal closure. Curr Biol. 2000;10:1420–1426. doi: 10.1016/s0960-9822(00)00796-x. [DOI] [PubMed] [Google Scholar]
- Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science. 2010;330:1824–1827. doi: 10.1126/science.1195481. [DOI] [PubMed] [Google Scholar]
- Jasper H, Benes V, Schwager C, Sauer S, Clauder-Munster S, Ansorge W, Boh-mann D. The genomic response of the Drosophila embryo to JNK signaling. Dev Cell. 2001;1:579–586. doi: 10.1016/s1534-5807(01)00045-4. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Li X, Hu J, Zhou W, Jiang Y, Li G, Lu D. Promoter hypermethylation-mediated down-regulation of LATS1 and LATS2 in human astrocytoma. Neurosci Res. 2006;56:450–458. doi: 10.1016/j.neures.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Jimenez C, Hu MI, Gagel RF. Management of medullary thyroid carcinoma. Endocrinol Metab Clin North Am. 2008;37:481–496. x–xi. doi: 10.1016/j.ecl.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Justice RW, Zilian O, Woods DF, Noll M, Bryant PJ. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- Kajita M, Hogan C, Harris AR, Dupre-Crochet S, Itasaki N, Kawakami K, Charras G, Tada M, Fujita Y. Interaction with surrounding normal epithelial cells influences signalling pathways and behaviour of Src-transformed cells. J Cell Sci. 2010;123:171–180. doi: 10.1242/jcs.057976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- Kapoor A, Goldberg MS, Cumberland LK, Ratnakumar K, Segura MF, Emanuel PO, Menendez S, Vardabasso C, Leroy G, Vidal CI, Polsky D, Osman I, Garcia BA, Hernando E, Bernstein E. The his-tone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468:1105–1109. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaman MW, Herrgard S, Treiber DK, Gallant P, Atteridge CE, Campbell BT, Chan KW, Ciceri P, Davis MI, Edeen PT, Faraoni R, Floyd M, Hunt JP, Lockhart DJ, Milanov ZV, Morrison MJ, Pal-lares G, Patel HK, Pritchard S, Wodicka LM, Zarrinkar PP. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–132. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- Kleihues P, Aguzzi A, Ohgaki H. Genetic and environmental factors in the etiology of human brain tumors. Toxicol Lett. 1995;82–83:601–605. doi: 10.1016/0378-4274(95)03503-6. [DOI] [PubMed] [Google Scholar]
- Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kockel L, Homsy JG, Bohmann D. Drosophila AP-1: lessons from an invertebrate. Oncogene. 2001;20:2347–2364. doi: 10.1038/sj.onc.1204300. [DOI] [PubMed] [Google Scholar]
- Korshunov A, Sycheva R, Golanov A. Genetically distinct and clinically relevant subtypes of glioblastoma defined by array-based comparative genomic hybridization (array-CGH). Acta Neuropathol. 2006;111:465–474. doi: 10.1007/s00401-006-0057-9. [DOI] [PubMed] [Google Scholar]
- Kurada P, White K. Ras promotes cell survival in Drosophila by downregulating hid expression. Cell. 1998;95:319–329. doi: 10.1016/s0092-8674(00)81764-x. [DOI] [PubMed] [Google Scholar]
- Lassmann S, Weis R, Makowiec F, Roth J, Danciu M, Hopt U, Werner M. Array CGH identifies distinct DNA copy number profiles of oncogenes and tumor suppressor genes in chromosomal- and microsatellite-unstable sporadic colorectal carcinomas. J Mol Med (Berl) 2007;85:293–304. doi: 10.1007/s00109-006-0126-5. [DOI] [PubMed] [Google Scholar]
- Lee KP, Lee JH, Kim TS, Kim TH, Park HD, Byun JS, Kim MC, Jeong WI, Calvisi DF, Kim JM, Lim DS. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc Natl Acad Sci U S A. 2010;107:8248–8253. doi: 10.1073/pnas.0912203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, Wang Y, Halder G, Finegold MJ, Lee JS, Johnson RL. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci U S A. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher EA, Furnari FB, Bachoo RM, Row-itch DH, Louis DN, Cavenee WK, DePinho RA. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- Martinez AM, Schuettengruber B, Sakr S, Janic A, Gonzalez C, Cavalli G. Polyhomeotic has a tumor suppressor activity mediated by repression of Notch signaling. Nat Genet. 2009;41:1076–1082. doi: 10.1038/ng.414. [DOI] [PubMed] [Google Scholar]
- Matsumine A, Ogai A, Senda T, Okumura N, Satoh K, Baeg GH, Kawahara T, Kobayashi S, Okada M, Toyoshima K, Akiyama T. Binding of APC to the human homolog of the Drosophila discs large tumor suppressor protein. Science. 1996;272:1020–1023. doi: 10.1126/science.272.5264.1020. [DOI] [PubMed] [Google Scholar]
- Min KT, Benzer S. Preventing neurodegeneration in the Drosophila mutant bubblegum. Science. 1999;284:1985–1988. doi: 10.1126/science.284.5422.1985. [DOI] [PubMed] [Google Scholar]
- Minoo P, Zlobec I, Baker K, Tornillo L, Terracciano L, Jass JR, Lugli A. Prognostic significance of mammalian sterile20-like kinase 1 in colorectal cancer. Mod Pathol. 2007;20:331–338. doi: 10.1038/modpathol.3800740. [DOI] [PubMed] [Google Scholar]
- Mocellin S, Nitti D. TNF and cancer: the two sides of the coin. Front Biosci. 2008;13:2774–2783. doi: 10.2741/2884. [DOI] [PubMed] [Google Scholar]
- Moreno E, Basler K. dMyc transforms cells into super-competitors. Cell. 2004;117:117–129. doi: 10.1016/s0092-8674(04)00262-4. [DOI] [PubMed] [Google Scholar]
- Morris EJ, Ji JY, Yang F, Di Stefano L, Herr A, Moon NS, Kwon EJ, Haigis KM, Naar AM, Dyson NJ. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455:552–556. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero-Navarro S, Esteller M. Chromatin remodeling factor CHD5 is silenced by promoter CpG island hyper-methylation in human cancer. Epigenetics. 2008;3:210–215. doi: 10.4161/epi.3.4.6610. [DOI] [PubMed] [Google Scholar]
- Navarro C, Nola S, Audebert S, Santoni MJ, Arsanto JP, Ginestier C, Marchetto S, Jacquemier J, Isnardon D, Le Bivic A, Birnbaum D, Borg JP. Junctional recruitment of mammalian Scribble relies on E-cadherin engagement. Oncogene. 2005;24:4330–4339. doi: 10.1038/sj.onc.1208632. [DOI] [PubMed] [Google Scholar]
- Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- Ostman A, Augsten M. Cancer-associated fibroblasts and tumor growth--bystanders turning into key players. Curr Opin Genet Dev. 2009;19:67–73. doi: 10.1016/j.gde.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Pagliarini RA, Xu T. A genetic screen in Drosophila for metastatic behavior. Science. 2003;302:1227–1231. doi: 10.1126/science.1088474. [DOI] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Dong J, Zhang Y, Gao X. Tuberous sclerosis complex: from Drosophila to human disease. Trends Cell Biol. 2004;14:78–85. doi: 10.1016/j.tcb.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Perez-Garijo A, Martin FA, Morata G. Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila. Development. 2004;131:5591–5598. doi: 10.1242/dev.01432. [DOI] [PubMed] [Google Scholar]
- Petrelli A, Giordano S. From single-to multi-target drugs in cancer therapy: when aspecificity becomes an advantage. Curr Med Chem. 2008;15:422–432. doi: 10.2174/092986708783503212. [DOI] [PubMed] [Google Scholar]
- Pietras K, Ostman A. Hallmarks of cancer: interactions with the tumor stroma. Exp Cell Res. 2010;316:1324–1331. doi: 10.1016/j.yexcr.2010.02.045. [DOI] [PubMed] [Google Scholar]
- Potter CJ, Huang H, Xu T. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell. 2001;105:357–368. doi: 10.1016/s0092-8674(01)00333-6. [DOI] [PubMed] [Google Scholar]
- Read RD, Goodfellow PJ, Mardis ER, Novak N, Armstrong JR, Cagan RL. A Drosophila model of multiple endocrine neoplasia type 2. Genetics. 2005;171:1057–1081. doi: 10.1534/genetics.104.038018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RD, Cavenee WK, Furnari FB, Thomas JB. A drosophila model for EGFR-Ras and PI3K-dependent human glioma. PLoS Genet. 2009;5:e1000374. doi: 10.1371/journal.pgen.1000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouleau GA, Merel P, Lutchman M, Sanson M, Zucman J, Marineau C, Hoang-Xuan K, Demczuk S, Desmaze C, Plougastel B, et al. Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature. 1993;363:515–521. doi: 10.1038/363515a0. [DOI] [PubMed] [Google Scholar]
- Ryoo HD, Gorenc T, Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev Cell. 2004;7:491–501. doi: 10.1016/j.devcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Seidel C, Schagdarsurengin U, Blumke K, Wurl P, Pfeifer GP, Hauptmann S, Tau-bert H, Dammann R. Frequent hypermethylation of MST1 and MST2 in soft tissue sarcoma. Mol Carcinog. 2007;46:865–871. doi: 10.1002/mc.20317. [DOI] [PubMed] [Google Scholar]
- Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5:615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- Smith-Hicks CL, Sizer KC, Powers JF, Tischler AS, Costantini F. C-cell hyperplasia, pheochromocytoma and sympathoadrenal malformation in a mouse model of multiple endocrine neoplasia type 2B. EMBO J. 2000;19:612–622. doi: 10.1093/emboj/19.4.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparagana SP, Roach ES. Tuberous sclerosis complex. Curr Opin Neurol. 2000;13:115–119. doi: 10.1097/00019052-200004000-00001. [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Stebbins MJ, Yin JC. Adaptable doxycycline-regulated gene expression systems for Drosophila. Gene. 2001;270:103–111. doi: 10.1016/s0378-1119(01)00447-4. [DOI] [PubMed] [Google Scholar]
- Stebbins MJ, Urlinger S, Byrne G, Bello B, Hillen W, Yin JC. Tetracyclineinducible systems for Drosophila. Proc Natl Acad Sci U S A. 2001;98:10775–10780. doi: 10.1073/pnas.121186498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Lehmann R. Germ versus soma decisions: lessons from flies and worms. Science. 2007;316:392–393. doi: 10.1126/science.1140846. [DOI] [PubMed] [Google Scholar]
- Stronach BE, Perrimon N. Stress signaling in Drosophila. Oncogene. 1999;18:6172–6182. doi: 10.1038/sj.onc.1203125. [DOI] [PubMed] [Google Scholar]
- Sun G, Irvine KD. Regulation of Hippo signaling by Jun kinase signaling during compensatory cell proliferation and regeneration, and in neoplastic tumors. Dev Biol. 2011;350:139–151. doi: 10.1016/j.ydbio.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Miyoshi Y, Takahata C, Irahara N, Taguchi T, Tamaki Y, Noguchi S. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin Cancer Res. 2005;11:1380–1385. doi: 10.1158/1078-0432.CCR-04-1773. [DOI] [PubMed] [Google Scholar]
- Tapon N, Ito N, Dickson BJ, Treisman JE, Hariharan IK. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell. 2001;105:345–355. doi: 10.1016/s0092-8674(01)00332-4. [DOI] [PubMed] [Google Scholar]
- Tapon N, Harvey KF, Bell DW, Wahrer DC, Schiripo TA, Haber DA, Hariharan IK. salvador Promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Trofatter JA, MacCollin MM, Rutter JL, Murrell JR, Duyao MP, Parry DM, Eldridge R, Kley N, Menon AG, Pulaski K, et al. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell. 1993;72:791–800. doi: 10.1016/0092-8674(93)90406-g. [DOI] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Uhlirova M, Bohmann D. JNK-and Fos-regulated Mmp1 expression cooperates with Ras to induce invasive tumors in Drosophila. EMBO J. 2006;25:5294–5304. doi: 10.1038/sj.emboj.7601401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlirova M, Jasper H, Bohmann D. Non-cell-autonomous induction of tissue overgrowth by JNK/Ras cooperation in a Drosophila tumor model. Proc Natl Acad Sci U S A. 2005;102:13123–13128. doi: 10.1073/pnas.0504170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Wells S, Ryan A, Cagan R. ZD6474 suppresses oncogenic RET iso-forms in a Drosophila model for type 2 multiple endocrine neoplasia syndromes and papillary thyroid carcinoma. Cancer Res. 2005;65:3538–3541. doi: 10.1158/0008-5472.CAN-04-4561. [DOI] [PubMed] [Google Scholar]
- Vidal M, Larson DE, Cagan RL. Csk-deficient boundary cells are eliminated from normal Drosophila epithelia by exclusion, migration, and apoptosis. Dev Cell. 2006;10:33–44. doi: 10.1016/j.devcel.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Vidal M, Warner S, Read R, Cagan RL. Differing Src signaling levels have distinct outcomes in Drosophila. Cancer Res. 2007;67:10278–10285. doi: 10.1158/0008-5472.CAN-07-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- Wang D, Kennedy S, Conte D, Jr, Kim JK, Gabel HW, Kamath RS, Mello CC, Ruvkun G. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature. 2005;436:593–597. doi: 10.1038/nature04010. [DOI] [PubMed] [Google Scholar]
- Wang X, Lau KK, So LK, Lam YW. CHD5 is down-regulated through promoter hypermethylation in gastric cancer. J Biomed Sci. 2009;16:95. doi: 10.1186/1423-0127-16-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedge SR, Ogilvie DJ, Dukes M, Ken-drew J, Chester R, Jackson JA, Boffey SJ, Valentine PJ, Curwen JO, Mus-grove HL, Graham GA, Hughes GD, Thomas AP, Stokes ES, Curry B, Richmond GH, Wadsworth PF, Bigley AL, Hennequin LF. ZD6474 inhibits vascular endothelial growth factor signaling, angiogenesis, and tumor growth following oral administration. Cancer Res. 2002;62:4645–4655. [PubMed] [Google Scholar]
- Williams JA, Su HS, Bernards A, Field J, Sehgal A. A circadian output in Drosophila mediated by neurofibromatosis-1 and Ras/MAPK. Science. 2001;293:2251–2256. doi: 10.1126/science.1063097. [DOI] [PubMed] [Google Scholar]
- Woodhouse E, Hersperger E, Stetler-Stevenson WG, Liotta LA, Shearn A. Increased type IV collagenase in lgl-induced invasive tumors of Drosophila. Cell Growth Differ. 1994;5:151–159. [PubMed] [Google Scholar]
- Woodhouse E, Hersperger E, Shearn A. Growth, metastasis, and invasiveness of Drosophila tumors caused by mutations in specific tumor suppressor genes. Dev Genes Evol. 1998;207:542–550. doi: 10.1007/s004270050145. [DOI] [PubMed] [Google Scholar]
- Woods DF, Hough C, Peel D, Callaini G, Bryant PJ. Dlg protein is required for junction structure, cell polarity, and proliferation control in Drosophila epithelia. J Cell Biol. 1996;134:1469–1482. doi: 10.1083/jcb.134.6.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Pastor-Pareja JC, Xu T. Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion. Nature. 2010;463:545–548. doi: 10.1038/nature08702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, Velculescu VE, Kinzler KW, Vogelstein B, Iacobuzio-Donahue CA. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitler J, Hsu CP, Dionne H, Bilder D. Domains controlling cell polarity and proliferation in the Drosophila tumor suppressor Scribble. J Cell Biol. 2004;167:1137–1146. doi: 10.1083/jcb.200407158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan L, Rosenberg A, Bergami KC, Yu M, Xuan Z, Jaffe AB, Allred C, Muthuswamy SK. Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell. 2008;135:865–878. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Smith DL, Meriin AB, Engemann S, Russel DE, Roark M, Washington SL, Maxwell MM, Marsh JL, Thompson LM, Wanker EE, Young AB, Housman DE, Bates GP, Sherman MY, Kazantsev AG. A potent small molecule inhibits polyglutamine aggregation in Huntington's disease neurons and suppresses neurodegeneration in vivo. Proc Natl Acad Sci U S A. 2005;102:892–897. doi: 10.1073/pnas.0408936102. [DOI] [PMC free article] [PubMed] [Google Scholar]