SYNOPSIS
Silencing of GATA5 gene expression as a result of promoter hypermethylation has been observed in lung, gastrointestinal, and ovarian cancers. However, the regulation of GATA5 gene expression has been poorly understood. Here, we demonstrate that an enhancer (E)-box in the GATA5 promoter (bp −118 to −113 in mouse; bp −164 to −159 in the human) positively regulates GATA5 transcription by binding upstream stimulatory factor 1 (USF1). Using site-directed mutagenesis, EMSA, and affinity chromatography, we found that USF1 specifically binds to the E-box sequence (5′-CACGTG-3′), but not to a mutated E-box. CpG methylation of this E-box significantly diminished its binding of transcription factors. Mutation of the E-box within a GATA5 promoter fragment significantly decreased promoter activity in a luciferase reporter assay. Chromatin immunoprecipitation identified that USF1 physiologically interacts with the GATA5 promoter E-box in mouse intestinal mucosa, which has the highest GATA5 gene expression in mouse. Co-transfection with USF1 expression plasmid significantly increased GATA5 promoter-driven luciferase transcription. Furthermore, real-time and RT-PCR analyses confirmed that overexpression of USF1 activates endogenous GATA5 gene expression in human bronchial epithelial cells. This study presents the first evidence that USF1 activates GATA5 gene expression through E-box motif, and suggests a potential mechanism (disruption of the E-box) by which GATA5 promoter methylation reduces GATA5 expression in cancer.
Keywords: differentiation, transcription factor, cancer
INTRODUCTION
GATA5 is a member of the GATA transcription factor family whose six members contain a highly conserved DNA binding domain consisting of two zinc fingers. While GATA1, 2, and 3 are expressed in the hematopoietic system, GATA4, 5, and 6 are expressed within multiple mesoderm- and endoderm-derived tissues, and are involved in tissue-specific transcriptional regulation during cell differentiation and embryogenesis. Silencing of the genes encoding GATA4 and GATA5, or of their downstream activation targets trefoil factor 1 (TFF1), TFF2, TFF3 and inhibin-α, impairs proper maturation of endoderm-derived epithelial cells [1-4]. Loss of GATA5 gene expression due to promoter hypermethylation has been observed in malignancy in various organs, including lung [5-8], pancreas [9], ovarian [10], breast [11], esophageal [12], stomach [13-15] , and colon [16-18]. Despite its apparent importance in differentiation and development, very little is known about how GATA5 transcription is regulated normally, or about specifically how hypermethylation reduces its expression. In this study, we address the transcriptional regulation of GATA5 gene expression.
In many genes, an enhancer (E)-box lies within the promoter region to provide a binding site for members of the basic helix-loop-helix (bHLH) transcription factor family and enhance transcription of the downstream gene. In the 5′-CANNTG-3′ E-box core sequence, the two central nucleotides (NN) are in most cases either GC or CG, and altering the spacing between the CA and TG abrogates nuclear factor binding [19]. Upstream stimulating factor (USF) 1 is a member of the eukaryotic evolutionary conserved basic-Helix-Loop-Helix-Leucine Zipper transcription factor family, and exhibits higher binding affinity for the 5′-CACGTG-3′ core motif than the 5′-CATGTG-3′ motif. In this study, we identified an E-box sequence (5′-CACGTG-3′) in the promoter regions of both the human and the murine GATA5 genes, and found that USF1 activates GATA5 gene transcription by specially binding to the E-box motif of GATA5 promoter. Furthermore, experimental methylation of the E-box prevented USF1 binding, suggesting a mechanism by which GATA5 promoter hypermethylation might reduce GATA5 expression in cancers.
EXPERIMENTAL
Cell culture and animal tissue
16HBE14o-, CMT-93, HEK 293, HCT116, and NIH 3T3 cells (ATCC, Manassas, VA) were maintained in Dulbecco′s Modified Essential Medium (DMEM)/F12 supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin at 37°C in a 5% CO2 humidified incubator. Mouse tissue was used in accordance with institutional and National Institutes of Health guidelines and was approved by the University of Chicago Institutional Animal Care and Use Committee.
Plasmids
The pGL3-basic vector (Promega, Madison, WI) was used to construct reporter plasmids to measure GATA5 promoter activity as described previously [20]. A series of DNA fragments comprising bp −3053, −1706, −1289, −984, −731, −367, −317, −258, −150 or −71 to +311 (relative to the known GATA5 major transcription start site, TSS; Ensembl Gene ID ENSMUSG00000015627) of the mouse GATA5 gene were generated by PCR amplification using primers listed in Supplementary Table 1 and inserted into MluI/SpeI sites upstream of the firefly luciferase gene. Plasmids in which luciferase expression was driven by GATA5 bp −258 to +85 harboring either of two double-point mutations (−108/−107 CG>TA [Mut −108/−107] or −115/−114 GT>AC [Mut −115/−114]) were generated similarly. PCR amplifications were performed with LA Taq™ polymerase and GC buffer (Takara, Madison, WI). The mammalian USF1 expression vector (psv-USF1), a dominant-negative mutant AUSF (psv-AUSF), and their control vector (pSG424) were all previously described [21] and generously provided by Dr. Nanyue Chen (University of Texas M. D. Anderson Cancer Center). All constructs were verified by DNA sequence analysis and prepared using an EndoFree Plasmid Maxi Kit (Qiagen, Valencia, CA).
Transient transfection and luciferase assay
The dual-luciferase reporter assay system was used to determine promoter activity in HEK 293 cells. Transient transfection was performed in 12-well plates at 60,000 cells/well with 1 μg of pGL3-basic or equal molar amounts of pGL3-basic-derived plasmids containing mouse GATA5 promoter constructs, and 3 ng of pKT-null vector containing the Renilla luciferase reporter gene as an internal control for transfection efficiency. pGL3-basic or pGL3-basic derived plasmids containing the GATA5 promoter region −258 to +85 (wild type, Mut −115/−114, Mut −108/−107) were co-transfected with 1 μg of expression vectors pGS424, psv-USF1, or psv-AUSF. All mammalian cells were transfected with Qiafect Reagent (Qiagen) according to the manufacturer’s instructions. At 48 hrs post-transfection, cells were harvested and reporter activity was measured using a dual luciferase assay system (Promega). Promoter activity was calculated by normalizing luciferase to Renilla and was expressed as average ± SD from at least three independent experiments, each performed in triplicate wells.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts from CMT-93 and 16HBE14o- cells were prepared using NE-PER Nuclear and Cytoplasmic Extraction Kit (Pierce, Rockford, IL) and used to identify protein-DNA interaction in mouse and in human, respectively.
The oligonucleotide probe with mouse GATA5 promoter sequence was from bp −127 to −100 (WT −127 to −100). Modified oligonucleotide probes contained the methylated mCpG dinucleotide at a single site on each strand (Met −116) or two double-point mutations (Mut −108/−107 or Mut −115/−114). Another oligonucleotide probe corresponding to bp −173 to −146 of the human GATA5 promoter (hWT −173 to −146) was generated. Modified probes contained methyl-CpG dinucleotide (hMet −162) at single site on each strand or a double-point mutation at −162/−161 CG>AA (hMut −162/−161) (Supplementary Table 1). All oligonucleotides were synthesized from IDT (Coralville, IA), labeled with [γ-32P]-ATP using T4 polynucleotide kinase and purified by chromatography through ProbeQuant™ G-50 micro columns (GE Healthcare, Piscataway, NJ).
5 μg of nuclear proteins or 2.5 μg of nuclear extracts depleted of USF1 with anti-USF1 antibody (H-86 X, Santa Cruz Biotechnology) were incubated with [γ-32P]-ATP labeled probe (20,000 cpm) at room temperature for 20 min in a total of 20 μl of a reaction mixture containing (final concentrations): 10 mM Tris/HCl, pH 7.5, 50 mM NaCl, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 4% (v/v) glycerol and 1 μg of poly(dI-dC) as non-specific competitor. For competition EMSA, 100 pmole or a serial dilution of unlabeled probe was added in the reaction mixture and pre-incubated at room temperature for 10 min to verify the specificity of protein-DNA interactions. Supershift assays were performed using polyclonal antibodies directed against Sp1, AP2, Oct-1, USF1, or USF2 (Santa Cruz Biotechnology, Santa Cruz, CA). The resulting protein-DNA complexes were analyzed by electrophoresis on a 5% polyacrylamide gel followed by autoradiography and densitometry analysis.
Bisulfite sequencing to identify DNA methylation
Genomic DNA was extracted from wild-type C57Bl/6J mouse intestinal mucosa or from cultured HCT116 human colon carcinoma cells using Puregene DNA Purification System (Gentra Systems, Minneapolis, MN) and treated with sodium bisulfite using EZ DNA Methylation Kit (Zymo Research, Irvine, CA) by following the manufacturer′s instructions. The DNA fragment containing the GATA5 promoter E-box was amplified by PCR using the primers in Supplementary Table 1 and cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, CA). Five clones for each sample were sequenced with M13 reverse primer via automated sequencing system (University of Chicago Sequencing Core Facility).
Biotin affinity purification and western analysis of DNA binding proteins
Biotin labeled sense strand and unlabeled antisense oligonucleotides, including WT −127 to −100, Mut −115/−114, Mut −108/−107, WT −215 to −173, and WT −610 to −575 (Supplementary Table 1), were purchased (IDT, Coralville, IA). Biotinylated double–stranded oligonucleotides were prepared by annealing 2.5 nmoles of each single-stranded oligonucleotide resuspended in 100 μl annealing buffer (10 mM Tris-HCL, pH 7.5, 50 mM NaCl, 10 mM MgCl2), heated for 3 min at 80°C, and cooled to room temperature.
Dynabeads MyOne Streptavidin C1 (Invitrogen) were washed three times for 10 min each in washing buffer (10 mM Tris-HCL, pH 7.5, 100 mM NaCl, 2 mM DTT, 0.1% bovine serum albumin). 100 pmoles of biotinylated double-stranded DNA in 50 μl of coupling buffer (10 mM Tris-HCL, pH 8.0, 1 M NaCl, 1 mM EDTA, 2 mM DTT, 0.1% bovine serum albumin) was mixed with 1 mg of streptavidin C1 coated Dynabeads and incubated at room temperature for 30 min on a rotating wheel. After incubation, the Dynabeads coated with biotinylated double-stranded DNA were washed three times in coupling buffer and resuspended in 100 μl of TNE buffer (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 1 mM EDTA) supplemented with 2 mM DTT and 20 μg/ml insulin [22].
100 μl of DNA-coupled Dynabeads were washed four times with 500 μl of binding buffer (20 mM Tris-HCL, pH 8.0, 10% glycerol, 2 mM EDTA, 0.01% Triton-X-100, 150 mM NaCl, 1 mM DTT, 10 μg/ml insulin, and 0.5 mM Pefabloc), resuspended in 50μl of 2X binding buffer, and incubated with equal volume of CMT-93 nuclear extract (30 μg protein) for 15 min at room temperature. After magnetic separation, supernatants were removed and reserved for subsequent electrophoresis analysis. The beads containing the protein/biotinylated fragment/streptavidin complex was washed three times with the binding buffer and boiled in Laemmli sample buffer. The bound proteins were resolved on 12% SDS-PAGE along with the corresponding supernatant described above. Proteins were electroblotted to nitrocellulose membranes according to standard protocols. Each membrane was blocked with 2% BSA in TBS (10 mM Tris–HCl, pH8.0, 150 mM NaCl) for 1 h at room temperature. The membrane was then incubated overnight at 4 °C with rabbit polyclonal anti-USF1 or Sp1 antibody at 1/200 dilution in TBS with 2% BSA. After five washes in TBS with 0.1% Tween 20, the membrane was incubated with peroxidase-conjugated secondary antibody at 1/10,000 dilution in TBS with 5% milk for 1 h at room temperature. The membrane was washed three times with TBST and the specific protein bands were visualized using the SuperSignal Chemiluminescence system (Pierce).
ChIP assay
ChIP assay was performed on mouse intestinal mucosa using a ChIP Assay Kit (Upstate, Temecula, CA). Briefly, the cleaned intestinal mucosa from C57B/6J mice was removed by scratching with a glass slide and fixed at room temperature for 10 min in the presence of DMEM/F12 culture medium with 1% formaldehyde following by quenching with 125 mM glycine for 5min. After two washes with cold PBS containing protease inhibitor cocktail (Roche Diagnostic), the pellet of intestinal mucosa was homogenized in SDS lysis buffer (1% SDS, 10 mM EDTA, 30 mM Tris-Cl) plus proteinase inhibitor on ice for 10 min and then sonicated to shear the DNA into 200 to 1000 bp fragments using a Branson Sonifier Cell Disruptor 185 (Danbury, CT). Immunoprecipitation was conducted with rabbit anti-USF1 or USF2 antibody (Santa Cruz Biotechnology) or without any antibody by incubation at 4°C overnight. The chromatin protein complex was immobilized with protein A beads and washed to remove unbound or non-specific DNA. After cross-links were reversed and the protein was digested with proteinase K (40 μg/ml), DNA was recovered and purified using a DNeasy Tissue Kit (Qiagen) and resuspended in 20 μl elution buffer. The input control DNA and immunoprecipitated DNA were amplified and PCR products were separated by a 1.5% agarose gel electrophoresis. Quantitative PCR analysis also was performed in duplicate for each of three independent CHIP experiments and the fold enrichment of the GATA5 E-box promoter region from USF1 and USF2 antibody-bound chromatin DNA over a non-promoter region of GATA5 gene was calculated and normalized to non-immune serum control, essentially as described [23, 24]. All PCR primer sequences are given in Supplementary Table 1.
Construction of standard curves and real-time PCR assay
To quantitatively measure human GATA5 gene expression activated by overexpression of USF1, real-time PCR standard curves for the GATA5 gene and for a housekeeping control gene μ-actin were generated by 10-fold serial dilution of each modified pCR 2.1-TOPO plasmid (Invitrogen) which was inserted the target sequence and its flanking sequence of the human GATA5 or μ-actin genes.
Total RNA was isolated from 16HBE14o- cells transfected with pSG424, psv-USF1, or psv-AUSF. RNA samples were treated with DNase I (Roche) to remove any contaminating DNA and cDNAs were synthesized from 0.5 μg RNA using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA). A fragment of human GATA5 (192 bp) and μ-actin (159 bp) were amplified during real-time PCR using gene specific primer pairs listed in Supplementary Table 1. All reactions (standard and unknown samples) were run in triplicate for each condition on the same 96-well plate. Thermal cycling conditions were 94°C for 2 min followed by 40 cycles of 94°C for 30 sec, 57°C for 30 sec, and 72°C for 30 sec in a MyiQ™ Single Color Real-time PCR detection System (Bio-Rad Laboratories). Results reported here are averages of the triplicates. Human GATA5 gene expression level was normalized by μ-actin expression level. Reverse-transcription (RT)-PCR was also performed using gene specific RT-PCR primer pairs (Supplementary Table 1). The RT-PCR amplification protocol was 94°C for 3 min; 30 cycles of 94°C for 30 sec, 60°C for 30 sec, and 72°C for 30 sec, followed by extension at 72°C for 5 min in an Applied Biosystems Thermocycler (Foster City, CA). The PCR products of human GATA5 (336 bp) and μ-actin (159 bp) were visualized after electrophoresis on 2% agarose gels.
Statistical analysis
All experiments were preformed independently at least three times. Data are expressed as mean ± standard deviation (SD). The statistical significance of differences of the means was determined by paired t test. P ≤ 0.05 was considered statistically significant.
RESULTS
Bp −150 to −71 of the mouse GATA5 gene contains positive regulatory elements
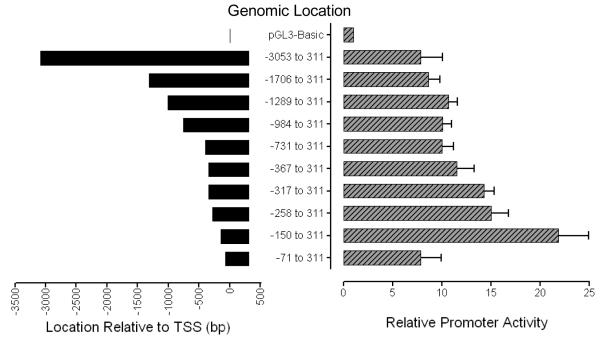
To determine the location of putative regulatory elements of the mouse GATA5 promoter, the promoter activity of the cloned 5′ end of GATA5 gene was studied by transient expression assays with the luciferase gene as a reporter. A genomic DNA fragment containing 3053 bp of 5′ flanking sequence and 311 bp of GATA5 exon 1 was inserted into MluI/SpeI sites upstream of the firefly luciferase gene in pGL3-basic vector. This construct or equal molar amounts of a series of deletion constructs were transfected into HEK 293 cells and the luciferase activity in cell extracts was analyzed after 48 hrs. The construct containing bp −150 to 311 exhibited the greatest promoter activity. Further 5′ truncation from −150 to −71 led to a remarkable decrease in luciferase activity, indicating the presence of positive regulatory elements in this region (Fig. 1). Similar results were observed in canine tracheal smooth muscle cells (data not shown). These findings indicate that the upstream sequence −150 to −71 contains strongly positive regulatory element(s) that regulate mouse GATA5 gene expression.
Figure 1.
Function analysis of mouse GATA5 promoter in HEK 293 cells. Promoter regions containing progressive 5′ truncations of the GATA5 5′-flanking sequence, ranging from bp −3053 through −71 relative to the major GATA5 transcriptional start site and extending to +311 bp (within exon 1 untranslated region), were cloned upstream of the firefly luciferase reporter gene in pGL3-basic. Each of the constructs was transiently co-transfected into HEK 293 cells with a pKT-null vector containing the Renilla luciferase reporter gene as an internal control. The Renilla-normalized firefly luciferase activity of each plasmid is shown relative to that of promoter-less pGL3-basic. The region between bp −150 to −71 contains strongly positive regulatory elements. The region between bp −71 to +311 exhibits core promoter activity. Data represent the mean ± SD for 3 experiments in each of which each datum is the average of results from 3 wells.
The GATA5 promoter contains an E-box motif which is a critical binding site
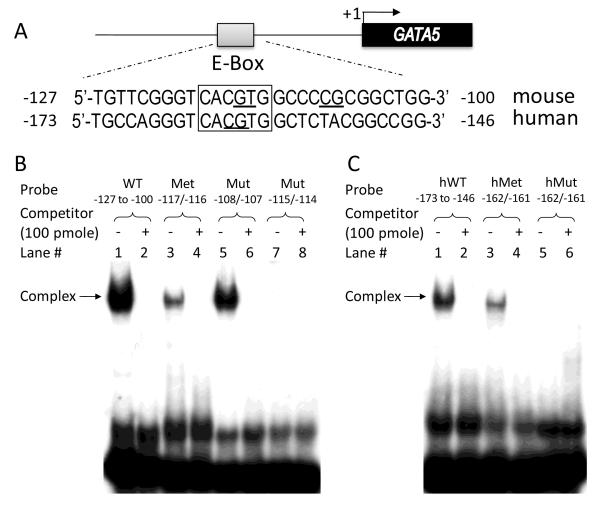
To understand how these positive regulatory elements regulate GATA5 gene expression, we analyzed the sequence of the 500-bp region upstream from the human and the mouse GATA5 TSSs. We found 79% homology between genomic sequence −136 to −28 of the mouse GATA5 and −182 to −74 of the human GATA5 genes. This region contains a conserved E-box sequence (5′-CACGTG-3′, boxed in Fig. 2A) located at −118 to −113 in the mouse and −164 to −159 in the human GATA5 promoters.
Figure 2.
Electrophoretic mobility shift assay of E-box binding. A: The GATA5 promoter region in both mouse and human contains an E-box motif (5′-CACGTG-3′, boxed); the bases mutated in promoter-reporter constructs or oligonucleotides are underlined. B: Nuclear extracts from CMT-93 cells were incubated with labeled mouse probes WT −127 to −100, Met −116, Mut −108/−107, or Mut −115/−114, in the absence or presence of 100 pmole of unlabeled competitor. The retarded bands revealing protein-DNA complex are highlighted by arrow. C: Nuclear extracts from 16HBE14o- cells were incubated with labeled human probes hWT −173 to −146, hMet −162, or hMut −162/−161, in the absence or presence of 100 pmole of unlabeled competitor. Arrow indicates the protein-DNA complex.
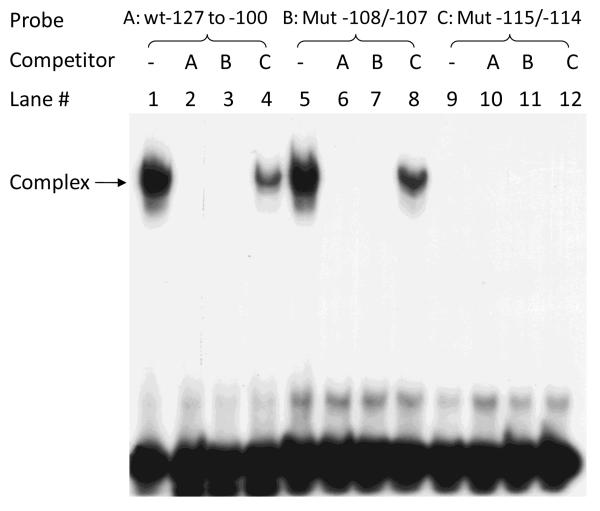
To determine whether this E-box is capable of binding transcription factors, EMSA analysis was performed with wild-type oligonucleotide probe corresponding to −127 to −100 of the mouse GATA5 promoter (WT −127 to −100) and nuclear extracts from mouse CMT-93 cells. A complex of nucleic acid probe bound to protein was observed (Fig. 2B, lane 1) while formation of this protein-DNA complex was specifically competed by addition of 100 pmole of unlabeled probe (Fig. 2B, lane 2). Furthermore, mutation of the E-box from 5′-CACGTG-3′ to 5′-CACACG-3′ within this fragment (Mut −115/−114) abolished the formation of protein-DNA complex (lane 7). On the other hand, mutation at −108/−107 (Mut −108/−107), which left the E-box intact, had no such effect on complex formation (lane 5). Similar findings were observed in EMSA analysis using wild-type probe hWT −173 to −146 or mutated E-box probe hMut −162/−161 and nuclear extracts from human 16HBE14o- cells (Fig. 2C). Additional competition experiments were carried out using mutated oligonucleotide probes with or without intact E-box. As can be seen in Fig. 3, protein-DNA complexes were observed with labeled WT −127 to −100 probe (lane 1) as well as Mut −108/−107 probe (lane 5), but not with labeled Mut −115/−114 probe in which the E-box was disrupted (lane 9). Complex formation was completely prevented by addition of 100 pmole of unlabeled competitor WT −127 to −100 (lane 2, 6) or Mut −108/−107 (lane 3,7), but not by addition of unlabeled Mut −115/−114 (lane 4, 8).
Figure 3.
Competition assay of E-box binding. Nuclear extracts from CMT-93 cells were incubated with the labeled oligonucleotide mouse probe WT −127 to −100 (A), Mut −108/−107 (B), or Mut −115/−114 (C) in the absence or presence of 100 pmole of unlabeled wild-type competitor A (lanes 2, 6, 10) or unlabeled mutated competitor B (lanes 3, 7, 11) or C (lanes 4, 8, 12). Mutation of the two central nucleotides at −115/−114 in the E-box (from CACGTG to CACACG) abolished oligonucleotide binding to nuclear factors, as reflected in the absence of any protein-DNA complex in lanes 9. However, mutation at bp −108/−107 that left the E-box intact did not affect oligonucleotide binding to nuclear proteins, as reflected in the presence of the complex (arrow) in lane 5. Cold competitors A (lane 2, 6) and B (lane 3, 7), but not C (lane 4, 8), abolished protein-DNA complex formation.
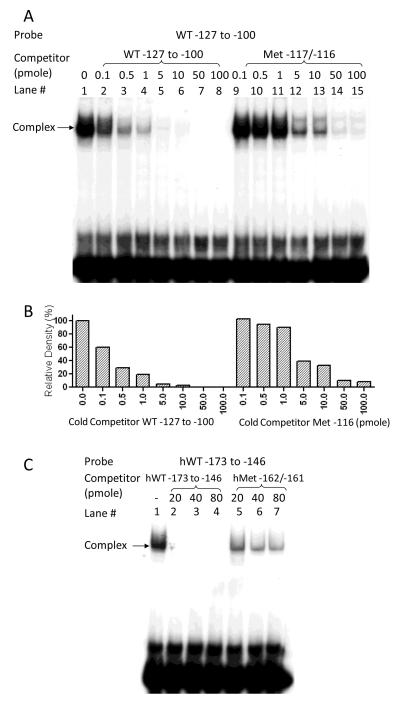
The E-box of GATA5 promoter contains a CpG dinucleotide that potentially might be methylated, and which in turn might alter its ability to bind nuclear factors. We therefore examined the E-box methylation status in mouse intestinal mucosa, which has the highest GATA5 gene expression in all mouse tissues [20], and in HCT116 cells, a human colon cancer cell line without GATA5 gene expression, using bisulfite sequencing analysis. We found that the cytosine of the E-box CpG dinucleotide is unmethylated in mouse intestinal mucosa, but is indeed methylated in HCT116 cells. We therefore tested whether such methylation modifies the ability of this E-box to bind nuclear factors. As seen in Fig. 2B (lane 3), methylation of the E-box in the mouse oligonucleotide probe (Met −117) significantly diminished probe binding of nuclear factors. A competition EMSA analysis also was performed using nuclear extracts from mouse CMT-93 cells and labeled WT −127 to −100 probe and cold WT competitor or cold Met −117 competitor (Fig. 4A). One representative experiment was quantified by densitometry and shown in Fig. 4B. Approximately 80% of protein-DNA complex was efficiently competed by addition of 1 pmole of unlabeled WT −127 to −100 competitor while 1 pmole of unlabeled methylated E-box competitor reduced formation of this complex by only 10%. Analogous results were observed using nuclear extracts from human 16HBE14o- cells and a labeled methylated E-box probe from the human GATA5 promoter (hMet −162) in EMSA analysis (Fig. 2B, lane 3) or in additional competition experiments using a labeled probe (hWT −173 to −146) with a series of dilutions of cold hWT −173 to −146 or cold hMet −162 competitor (Fig. 4C).
Figure 4.
Effect of CpG methylation on mouse GATA5 promoter E-box binding. A: Nuclear extracts from CMT-93 cells were incubated with the labeled mouse oligonucleotide probe WT −127 to −100 in the absence (lane 1) or presence of a series of dilutions of unlabeled wild-type competitor (lanes 2-8) or unlabeled CpG methylation at the E-box competitor Met −116 (lanes 9-15). B: The results from one representative experiment were quantified by densitometry and normalized to the band formed on labeled WT −127 to −100 probe without cold competitor (lane 1). C: A similar competition assay was performed with nuclear extracts from human bronchial epithelial cells using the labeled human oligonucleotide probe hWT −173 to-146 in the absence (lane 1) or presence of a series of dilutions of unlabeled wild-type competitor (lanes 2-4) or E-box methylated competitor hMet-162 (lanes 6-7). CpG methylation at the E-box diminishes its binding to nuclear proteins.
Taken together, these results indicate that the E-box within the GATA5 promoter can bind nuclear factors in vitro, and that methylation prevents such nuclear factor binding.
USF1 specifically binds to E-box of the GATA5 promoter positive regulatory region
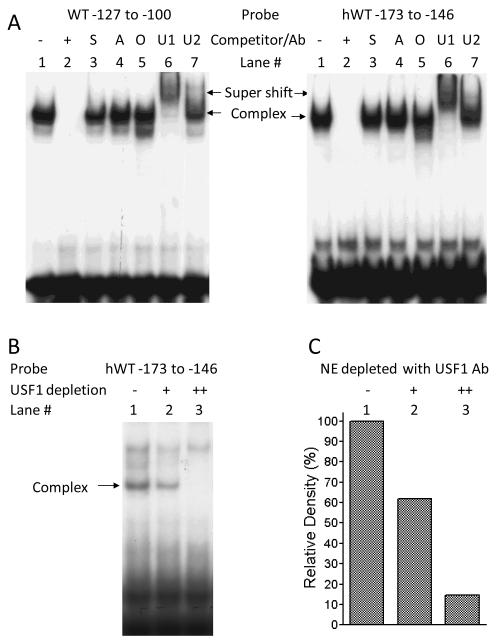
TRANSFAC analysis of the sequence around the E-box suggested possible binding sites for USF, Sp1, AP2, and Oct-1, among other factors. To determine whether any of these was included in the protein-DNA complexes described above, we performed supershift EMSA analysis, using labeled oligonucleotide WT −127 to −100 probe, nuclear extracts from mouse CMT-93 cells, and antibodies directed against the transcription factors named above. As seen in Fig. 5, the protein-DNA complex was specifically supershifted by addition of anti-USF1 antibody, but not by anti-USF2, anti-Sp1, anti-AP2, or anti-Oct-1 antibodies (Fig. 5A, left hand panel). Similar results were found using the human DNA sequence and nuclear proteins from human bronchial epithelial cells (Fig. 5A, right hand panel). To confirm that USF1 specifically binds to the GATA5 promoter E-box, we performed EMSA using USF1-depleted nuclear extracts. 40 μl of 16HBE14o- cell nuclear proteins at 0.5μg/μl was incubated either without antibody or with 1 μl or 2 μl of anti-USF1 antibody (H-86 X, Santa Cruz Biotechnology) overnight at 4°C and then incubated with 20 μl of Salmon Sperm DNA/Protein A Agarose for one hour at 4°C with agitation. 5 μl of the USF1 depleted supernatant fraction and the labeled hWT probe −173 to −146 were used for EMSA analysis as described in Experimental Methods. Depletion of USF1 from HBE cell nuclear extracts using USF1 antibody specifically eliminated the complex formation on the GATA5 promoter E-box (Fig. 5B and 5C). In light of our demonstration that mutation or methylation of the E-box prevents nuclear factor binding (above), the results in Fig. 5 indicate that the GATA5 promoter E-box specifically binds USF1 in vitro.
Figure 5.
Binding of USF1 to the E-box of GATA5 promoter in both mouse and in human. A supershift assay was performed with nuclear extracts from CMT-93 cells (Fig. 5A, left panel) or from 16HBE14o- cells (Fig. 5A, right panel) using the labeled wild-type probes WT −127 to −100 or hWT −173 to-146, respectively, in the absence of antibody (lane 1) or in the presence of 100 pmole of unlabeled competitor (lane 2), anti-Sp1 (lane 3, “S”), anti-AP2 (lane 4, “A”), anti-Oct-1 (lane 5, “O”), anti-USF1 (lane 6, “U1”), or anti-USF-2 (lane 7, “U2”) antibody. A supershifted band was observed in the presence of anti-USF1, but not other antibodies. Depletion of USF1 from HBE cell nuclear extracts using USF1 antibody specifically eliminated the complex formation of GATA5 promoter E-box and nuclear proteins (Fig. 5B ). The results from Fig. 5B were quantified by densitometry and shown in Fig. 5C.
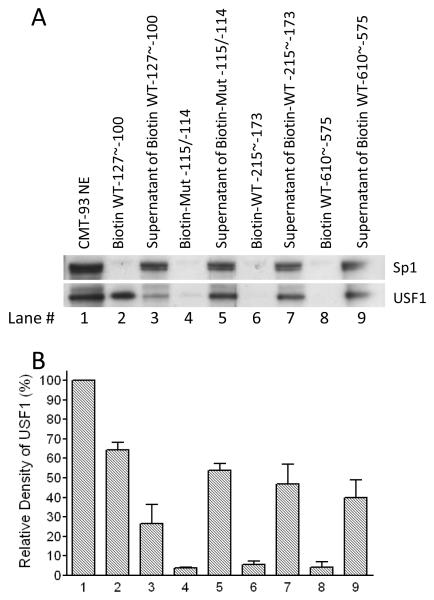
To provide additional evidence that the E-box binds USF1, we performed affinity purification analysis. Double stranded oligonucleotides corresponding to WT −127 to −100 or containing the E-box mutation (Mut −115/−114), or two other sequences corresponding to bp −215 to −173 or bp −610 to −575 in the mouse GATA5 promoter, were labeled with biotin for affinity purification of CMT-93 nuclear extracts. As seen in Fig. 6, USF1 could be recovered from with streptavidin-coated beads bound to biotin-labeled wild-type bp −127 to −100 after incubation with nuclear extracts. However, double-point mutation at −115/−114 to disrupt the E-box eliminated binding to USF1, so that almost all the USF1 remained in the supernatant. Similarly, the genomic sequences −215 to −173 or −610 to −575, which lack E-boxes, also failed to bind USF1. This experiment provides further evidence that USF1 specifically binds to the E-box of the GATA5 promoter region in vitro.
Figure 6.
Biotin affinity purification and western blot analysis. Dynabeads MyOne Streptavidin C1 beads were coated with double-stranded, 5′-biotinylated oligonucleotides and incubated with nuclear extracts from CMT-93 cells. After magnetic separation, the supernatants and the protein/biotinylated oligonucleotides/streptavidin complexes were analyzed by western blotting. USF1, but not Sp1 (included as a negative control) was captured on beads coated with the biotinylated oligonucleotide WT −127 to −100 (lane 2), largely clearing the supernatant (lane 3). Oligonucleotide Mut −115/−114 eliminated binding to USF1 (lane 4), so that almost all USF1 protein remained in the supernatant (lane 5). Similar results were observed using two other 5′-biotinylated oligonucleotides WT −215 to −173 and WT −610 to −575 that do not include any E-box motif (lane 6-9). Densitometry analysis was performed in four experiments and quantitative data are presented in Figure 6B.
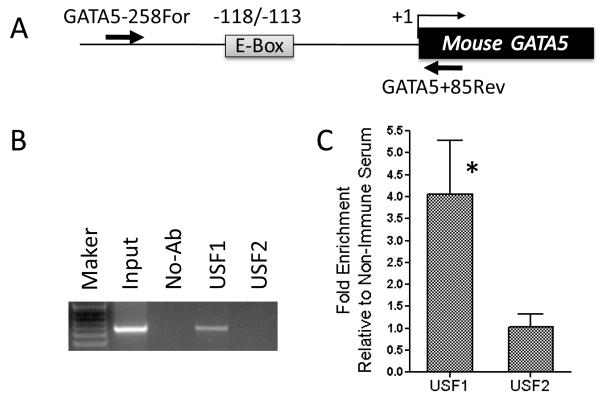
Next, we performed chromatin immunoprecipitation analysis to determine whether USF1 physiologically interacts with the GATA5 promoter positive regulatory region. As shown in Fig. 7, the E-box containing sequence of the GATA5 promoter was recovered after immunoprecipitation of sheared genomic DNA from mouse intestinal mucosa with anti-USF1 antibody, but not after immunoprecipitation with anti-USF2 antibody. Quantitative PCR confirmed that USF1 is present at the GATA5 E-box promoter region and the corresponding genomic DNA was enriched with USF1 antibody, but not USF2 antibody (P < 0.05, Fig. 7C). These data provide additional evidence for the occupancy by USF1, but not USF2, of the GATA5 promoter E-box in vivo.
Figure 7.
Physiological interaction of USF1 with the E-box of GATA5 promoter in mouse intestinal mucosa. The lysate of cross-linked intestinal mucosa was immunoprecipitated with either anti-USF1 or anti-USF2 antibody or without any antibody. Cross-links were reversed and the protein was digested with proteinase K. The recovered DNA was PCR-amplified using the primers listed in Supplementary Table 1 and products were analyzed by agarose gel electrophoresis. A: The location of the primers (arrows) relative to the E-box at −118 to −113 is shown. B: The E-box binding site was PCR-amplified with –258For/+85Rev primers and confirmed by sequencing. C: Quantitative PCR analysis was performed in duplicate for each CHIP experiment. The fold enrichment of USF1 and USF2 antibodies at E-Box promoter region over a non-promoter region of GATA5 gene was calculated. All qChIP data were normalized to non-immune serum control. Significant difference of enrichment between USF1 and USF2 antibodies was observed (P < 0.05).
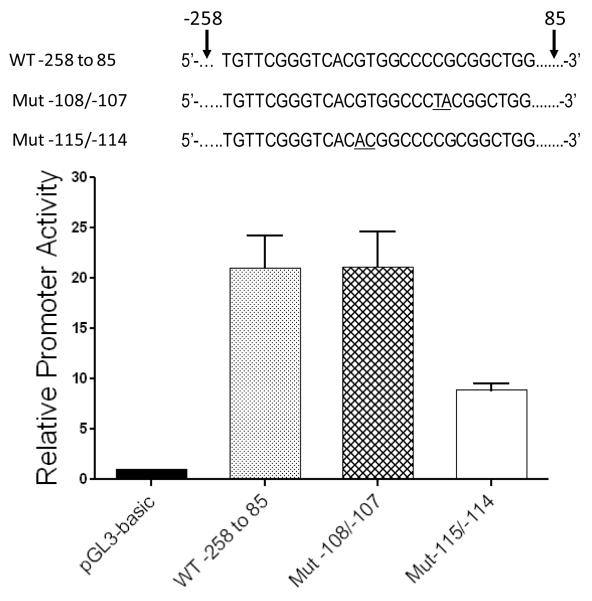
The GATA5 promoter E-box regulates GATA5 expression
To assess the functional role of the E-box in the GATA5 promoter, the wild-type GATA5 promoter fragment bp −258 to +85, or its mutated counterparts harboring the E-box mutation at −115/−114 or the non-E-box mutation at −108/−107, were cloned upstream of the luciferase cDNA in pGL3-basic and transient transfection was performed in HEK 293 cells (Fig. 8). There was significant promoter activity for the wild-type fragment and its mutated counterpart with intact E-box (vs empty pGL3basic luciferase reporter; P=0.0035, P=0.0043, respectively). However, promoter activity was significantly reduced when the E-box was mutated from 5′-CACGTG-3′ to 5′-CACACG-3′ (P=0.021 vs wild-type; P=0.025 vs Mut −108/−107). This indicates that the E-box at bp −118 to −113 is functionally crucial for full GATA5 gene promoter activity.
Figure 8.
Mutation of the E-box reduces GATA5 promoter activity. Genomic fragments corresponding to GATA5 bp −258 to +85 and counterparts harboring Mut −115/−114 or Mut −108/−107 were cloned upstream from the luciferase cDNA and transient co-transfection with Renilla luciferase plasmid was performed in HEK 293 cells. Mutation of the E-box from 5′-CACGTG-3′ to 5′-CACACG-3′ significantly reduced promoter activity compared with the other two promoter fragments in which the E-box remained intact (P=0.021 vs wt, P=0.025 vs Mut −108/−107, respectively). Mean±SD shown; n=3 experiments in each of which each datum is the average of results from 3 wells.
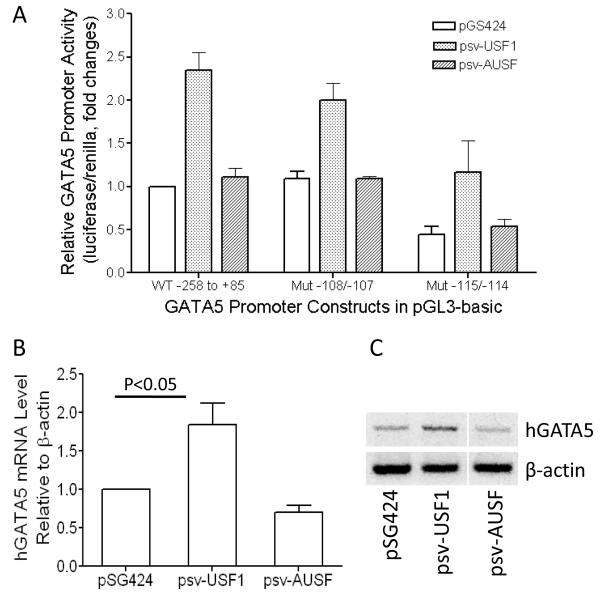
Next, we evaluated whether overexpression of USF further activates transcription from the GATA5 promoter. We co-transfected pGL3-basic-derived plasmids containing the GATA5 promoter region from bp −258 to +85, or its E-box-mutated counterparts, together with expression vectors pGS424, psv-USF1, or psv-AUSF into NIH 3T3 cells, which have no endogenous GATA5 expression [25]. As shown in Fig. 9A, USF1 significantly activated the wild-type GATA5 promoter fragment (as compared with empty pSG424 vector or dominant-negative USF vector; each P<0.05). Mutation of this promoter fragment at −108/−107 (which leaves the E-box intact) did not reduce GATA5 promoter activity during transactivation by USF1 overexpression. However, mutation of the E-box at bp −115/−114 significantly reduced promoter activity even during overexpression of USF1 (P<0.05). These results demonstrate that USF1 transactivates the GATA5 promoter by binding to its E-box.
Figure 9.
Transactivation of GATA5 promoter activity by USF1 overexpression. A: Co-transfection was performed with plasmids containing GATA5 promoter region from bp −258 to +85 or mutated counterparts, together with expression vector pGS424, psv-USF1, or psv-AUSF in NIH 3T3 cells. Overexpression of USF1 significantly activated the wild-type or Mut −108/−107 GATA5 promoter fragments in which the E-box remains intact, but mutation of the E-box at −115/−114 significantly reduced promoter activity both in the absence (pGS424 or psv-AUSF) or presence (psv-USF1) of USF1 overexpression (P<0.05). B: USF1 activates endogenous GATA5 gene expression. Human bronchial epithelial cells were transfected with expression vector pGS424, psv-USF1, or psv-AUSF. GATA5 mRNA expression level from the transfected cells was quantified by real-time PCR and normalized by μ-actin expression level. C: Reverse transcription (RT)-PCR was performed and the products were visualized on 2% agarose gels.
Lastly, we tested whether USF1 increases endogenous GATA5 expression. Human bronchial epithelial cells were transfected with either USF1 expression vector, control vector pSG424 or plasmid expressing dominant negative AUSF. GATA5 mRNA in transfected cells was quantified using real-time PCR. As shown in Fig. 9B, USF1 overexpression increased GATA5 gene expression in human bronchial epithelial cells, as compared with cells transfected with empty pSG424 vector or dominant-negative USF vector. These quantitative results were confirmed qualitatively by RT-PCR (Fig. 9C).
Together, these results indicate that USF1 transactivates GATA5 expression by binding to the E-box in its promoter.
DISCUSSION
Despite its importance in cell differentiation and tissue development [26, 27], little is known about the transcriptional regulation of GATA5 expression. In this study, we identified a positive regulatory region within bp −150 to −71 of the mouse GATA5 5′ flanking sequence (Fig. 1), that contains an E-box consensus sequence (5′-CACGTG-3′) (Fig. 2A). Using EMSA, biotinylated DNA purification, and chromatin immunoprecipitation, we confirmed that this E-box specifically binds USF1 in vitro and in vivo. Such E-box-USF1 binding is functionally important in activating GATA5 expression, because: 1) mutation of the E-box to prevent USF1 binding reduces GATA5 promoter activity (Fig. 3); 2) USF1 overexpression activates E-box-dependent GATA5 promoter activity (Fig. 8); and 3) USF1 overexpression increases endogenous GATA5 mRNA expression (Fig. 9). Taken together, these data demonstrate conclusively that USF1 activates GATA5 transcription.
USFs are members of the basic helix-loop-helix transcription factor family that bind the E-box in a promoter to activate transcription of the target gene. A wide variety of genes are direct targets of USF, including genes involved in the immune response [28], glucid and lipid pathways [29], cell cycle [30, 31], cell proliferation [32], and carcinogenesis [33-36]. In this study, we found that GATA5 is another target gene of USF1 and that GATA5 gene expression is activated through binding of USF1 to the canonical E-box sequence in the GATA5 promoter region, as discussed above. In other genes, it had previously been shown that the E-box modulation by methylation at the core CpG in the USF1 recognition site (5′-CACpGTG-3′) [37-39] significantly hinders transcription factor-DNA interaction and so negatively regulates gene expression. Others have reported that GATA5 gene expression occurs in association with GATA5 promoter hypermethylation in lung, esophagus, colon and gastric cancers [7, 12-14, 17]. Because we demonstrated that the E-box in the GATA5 promoter plays an important role in GATA5 expression, we hypothesized that methylation of CpG dinucleotides in the E-box [37] suppresses USF1 binding, and thereby down-regulates the expression of GATA5. Indeed, our experiments demonstrate that experimental methylation of this E-box does inhibit its binding of USF1 (Fig. 4). Furthermore, we found that the GATA5 promoter E-box is not methylated in mouse intestinal mucosa, which exhibits the greatest endogenous GATA5 mRNA expression [20], whereas the corresponding E-box in human HCT116 colon cancer cells (which lack GATA5 expression) is highly methylated. Finally, Ting et al [40] and McGarvey et al [41] reported that treatment of HCT116 cells with 5-azacytidine to demethylate genomic DNA restored GATA5 expression. Together, these results strongly imply that methylation of the GATA5 promoter E-box in certain cancers is one mechanism by which they lose GATA5 expression.
Besides E-box-USF1 binding, other transcriptional regulatory mechanisms likely also influence GATA5 expression. Using TRANSFAC analysis, we found that the GATA5 gene promoter lacks a TATA box. The 5′ flanking sequence at bp −48 to −38 contains the recently identified X core promoter element [42], which might initiate GATA5 transcription by RNA polymerase II, as well as a potential binding site for stimulating protein 1 (Sp1), which might enhance gene transcription. Furthermore, our 5′ deletion analysis suggests the presence of a negative regulatory region within bp −3053 to −150 (Fig. 1). Additional research is needed to delineate these other transcriptional regulatory mechanisms more completely.
Finally, we recently reported that an N-terminally truncated but still functional isoform of mouse GATA5 is transcribed in intestinal and gastric epithelium from an alternative promoter within intron 1 [20]. Sequence analysis fails to disclose a consensus E-box within 1000 bp of the minor transcription start site (which is 82 bp upstream of exon 2). As such, this alternative GATA5 promoter must be differentially regulated from the principal GATA5 promoter (upstream of exon 1) studied here.
Supplementary Material
Supplementary Table 1 - Oligonucleotide sequences used in plasmid constructs, biotin-affinity purification, EMSAs, methylation analysis and PCR
ACKNOWLEDGEMENTS
We thank Dr. Nanyue Chen (Houston, TX, USA) who kindly provided psv-USF1, psv-AUSF, and pSG424 plasmids.
FUNDING This work was supported in part by NIH grants R01 HL 079398, R01 HL 097805, and K12 HL 090003, and a grant from the Cancer Research Foundation.
Abbreviations used are
- AP2
activating protein 2
- bp
base pair(s)
- ChIP
chromatin immunoprecipitation
- DMEM
Dulbecco’s Modified Essential Medium
- E-box
enhancer box
- EMSA
electrophoretic mobility shift assay
- Oct-1
octamer transcription factor-1
- RT
Reverse-transcription
- Sp1
stimulating protein 1
- TFF
trefoil factor
- USF
upstream stimulatory factor
- AUSF
a dominant-negative USF
- TSS
transcription start site
Footnotes
AUTHORS’ CONTRIBUTIONS B. Chen conceived, designed, and carried out the experiments and drafted the manuscript. R. Hsu, Z. Li, P.C. Kogut, Q. Du, and K. Rouser conducted experiments. B. Camoretti-Mercado contributed to experiments and data interpretation. J. Solway designed the experiments, interpreted data, and revised the manuscript. All authors read and approved the final manuscript.
REFERENCES
- 1.Al-azzeh E, Dittrich O, Vervoorts J, Blin N, Gott P, Luscher B. Gastroprotective peptide trefoil factor family 2 gene is activated by upstream stimulating factor but not by c-Myc in gastrointestinal cancer cells. Gut. 2002;51:685–690. doi: 10.1136/gut.51.5.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bossenmeyer-Pourie C, Kannan R, Ribieras S, Wendling C, Stoll I, Thim L, Tomasetto C, Rio MC. The trefoil factor 1 participates in gastrointestinal cell differentiation by delaying G1-S phase transition and reducing apoptosis. J. Cell Biol. 2002;157:761–770. doi: 10.1083/jcb200108056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao X, Sedgwick T, Shi YB, Evans T. Distinct functions are implicated for the GATA-4, -5, and -6 transcription factors in the regulation of intestine epithelial cell differentiation. Mol. Cell Biol. 1998;18:2901–2911. doi: 10.1128/mcb.18.5.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wright NA, Hoffmann W, Otto WR, Rio MC, Thim L. Rolling in the clover: trefoil factor family (TFF)-domain peptides, cell migration and cancer. FEBS Lett. 1997;408:121–123. doi: 10.1016/s0014-5793(97)00424-9. [DOI] [PubMed] [Google Scholar]
- 5.De Jong WK, Verpooten GF, Kramer H, Louwagie J, Groen HJ. Promoter methylation primarily occurs in tumor cells of patients with non-small cell lung cancer. Anticancer Res. 2009;29:363–369. [PubMed] [Google Scholar]
- 6.Belinsky SA, Grimes MJ, Casas E, Stidley CA, Franklin WA, Bocklage TJ, Johnson DH, Schiller JH. Predicting gene promoter methylation in non-small-cell lung cancer by evaluating sputum and serum. Br. J. Cancer. 2007;96:1278–1283. doi: 10.1038/sj.bjc.6603721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo M, Akiyama Y, House MG, Hooker CM, Heath E, Gabrielson E, Yang SC, Han Y, Baylin SB, Herman JG, Brock MV. Hypermethylation of the GATA genes in lung cancer. Clin. Cancer Res. 2004;10:7917–7924. doi: 10.1158/1078-0432.CCR-04-1140. [DOI] [PubMed] [Google Scholar]
- 8.Lyon CM, Klinge DM, Liechty KC, Gentry FD, March TH, Kang T, Gilliland FD, Adamova G, Rusinova G, Telnov V, Belinsky SA. Radiation-induced lung adenocarcinoma is associated with increased frequency of genes inactivated by promoter hypermethylation. Radiat. Res. 2007;168:409–414. doi: 10.1667/RR0825.1. [DOI] [PubMed] [Google Scholar]
- 9.Fu B, Guo M, Wang S, Campagna D, Luo M, Herman JG, Iacobuzio-Donahue CA. Evaluation of GATA-4 and GATA-5 methylation profiles in human pancreatic cancers indicate promoter methylation patterns distinct from other human tumor types. Cancer Biol. Ther. 2007;6:1546–1552. doi: 10.4161/cbt.6.10.4708. [DOI] [PubMed] [Google Scholar]
- 10.Wakana K, Akiyama Y, Aso T, Yuasa Y. Involvement of GATA-4/-5 transcription factors in ovarian carcinogenesis. Cancer Lett. 2006;241:281–288. doi: 10.1016/j.canlet.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 11.Huggins GS, Wong JY, Hankinson SE, De Vivo I. GATA5 activation of the progesterone receptor gene promoter in breast cancer cells is influenced by the +331G/A polymorphism. Cancer Res. 2006;66:1384–1390. doi: 10.1158/0008-5472.CAN-05-2715. [DOI] [PubMed] [Google Scholar]
- 12.Guo M, House MG, Akiyama Y, Qi Y, Capagna D, Harmon J, Baylin SB, Brock MV, Herman JG. Hypermethylation of the GATA gene family in esophageal cancer. Int. J. Cancer. 2006;119:2078–2083. doi: 10.1002/ijc.22092. [DOI] [PubMed] [Google Scholar]
- 13.Akiyama Y, Watkins N, Suzuki H, Jair KW, van Engeland M, Esteller M, Sakai H, Ren CY, Yuasa Y, Herman JG, Baylin SB. GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer. Mol. Cell Biol. 2003;23:8429–8439. doi: 10.1128/MCB.23.23.8429-8439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wen XZ, Akiyama Y, Pan KF, Liu ZJ, Lu ZM, Zhou J, Gu LK, Dong CX, Zhu BD, Ji JF, You WC, Deng DJ. Methylation of GATA-4 and GATA-5 and development of sporadic gastric carcinomas. World J. Gastroenterol. 16:1201–1208. doi: 10.3748/wjg.v16.i10.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuasa Y, Nagasaki H, Akiyama Y, Hashimoto Y, Takizawa T, Kojima K, Kawano T, Sugihara K, Imai K, Nakachi K. DNA methylation status is inversely correlated with green tea intake and physical activity in gastric cancer patients. Int. J. Cancer. 2009;124:2677–2682. doi: 10.1002/ijc.24231. [DOI] [PubMed] [Google Scholar]
- 16.Derks S, Postma C, Moerkerk PT, van den Bosch SM, Carvalho B, Hermsen MA, Giaretti W, Herman JG, Weijenberg MP, de Bruine AP, Meijer GA, van Engeland M. Promoter methylation precedes chromosomal alterations in colorectal cancer development. Cell Oncol. 2006;28:247–257. doi: 10.1155/2006/846251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hellebrekers DM, Lentjes MH, van den Bosch SM, Melotte V, Wouters KA, Daenen KL, Smits KM, Akiyama Y, Yuasa Y, Sanduleanu S, Khalid-de Bakker CA, Jonkers D, Weijenberg MP, Louwagie J, van Criekinge W, Carvalho B, Meijer GA, Baylin SB, Herman JG, de Bruine AP, van Engeland M. GATA4 and GATA5 are potential tumor suppressors and biomarkers in colorectal cancer. Clin. Cancer Res. 2009;15:3990–3997. doi: 10.1158/1078-0432.CCR-09-0055. [DOI] [PubMed] [Google Scholar]
- 18.McGarvey KM, Van Neste L, Cope L, Ohm JE, Herman JG, Van Criekinge W, Schuebel KE, Baylin SB. Defining a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells. Cancer Res. 2008;68:5753–5759. doi: 10.1158/0008-5472.CAN-08-0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corre S, Galibert MD. Upstream stimulating factors: highly versatile stress-responsive transcription factors. Pigment Cell Res. 2005;18:337–348. doi: 10.1111/j.1600-0749.2005.00262.x. [DOI] [PubMed] [Google Scholar]
- 20.Chen B, Yates E, Huang Y, Kogut P, Ma L, Turner JR, Tao Y, Camoretti-Mercado B, Lang D, Svensson EC, Garcia JG, Gruber PJ, Morrisey EE, Solway J. Alternative promoter and GATA5 transcripts in mouse. Am. J. Physiol. Gastrointest Liver Physiol. 2009;297:G1214–1222. doi: 10.1152/ajpgi.00165.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ismail PM, Lu T, Sawadogo M. Loss of USF transcriptional activity in breast cancer cell lines. Oncogene. 1999;18:5582–5591. doi: 10.1038/sj.onc.1202932. [DOI] [PubMed] [Google Scholar]
- 22.Strobl LJ, Zimber-Strobl U. Magnetic DNA affinity purification of a cellular transcription factor. Methods Mol. Biol. 2001;174:271–277. doi: 10.1385/1-59259-227-9:271. [DOI] [PubMed] [Google Scholar]
- 23.Mukhopadhyay A, Deplancke B, Walhout AJ, Tissenbaum HA. Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat. Protoc. 2008;3:698–709. doi: 10.1038/nprot.2008.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 25.Kakita T, Hasegawa K, Morimoto T, Kaburagi S, Wada H, Sasayama S. p300 protein as a coactivator of GATA-5 in the transcription of cardiac-restricted atrial natriuretic factor gene. J. Biol. Chem. 1999;274:34096–34102. doi: 10.1074/jbc.274.48.34096. [DOI] [PubMed] [Google Scholar]
- 26.Singh MK, Li Y, Li S, Cobb RM, Zhou D, Lu MM, Epstein JA, Morrisey EE, Gruber PJ. Gata4 and Gata5 cooperatively regulate cardiac myocyte proliferation in mice. J. Biol. Chem. 2010;285:1765–1772. doi: 10.1074/jbc.M109.038539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molkentin JD, Tymitz KM, Richardson JA, Olson EN. Abnormalities of the genitourinary tract in female mice lacking GATA5. Mol. Cell Biol. 2000;20:5256–5260. doi: 10.1128/mcb.20.14.5256-5260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galibert MD, Boucontet L, Goding CR, Meo T. Recognition of the E-C4 element from the C4 complement gene promoter by the upstream stimulatory factor-1 transcription factor. J. Immunol. 1997;159:6176–6183. [PubMed] [Google Scholar]
- 29.Roth U, Jungermann K, Kietzmann T. Modulation of glucokinase expression by hypoxia-inducible factor 1 and upstream stimulatory factor 2 in primary rat hepatocytes. Biol. Chem. 2004;385:239–247. doi: 10.1515/BC.2004.018. [DOI] [PubMed] [Google Scholar]
- 30.North S, Espanel X, Bantignies F, Viollet B, Vallet V, Jalinot P, Brun G, Gillet G. Regulation of cdc2 gene expression by the upstream stimulatory factors (USFs) Oncogene. 1999;18:1945–1955. doi: 10.1038/sj.onc.1202506. [DOI] [PubMed] [Google Scholar]
- 31.Pawar SA, Szentirmay MN, Hermeking H, Sawadogo M. Evidence for a cancer-specific switch at the CDK4 promoter with loss of control by both USF and c-Myc. Oncogene. 2004;23:6125–6135. doi: 10.1038/sj.onc.1207806. [DOI] [PubMed] [Google Scholar]
- 32.Goueli BS, Janknecht R. Regulation of telomerase reverse transcriptase gene activity by upstream stimulatory factor. Oncogene. 2003;22:8042–8047. doi: 10.1038/sj.onc.1206847. [DOI] [PubMed] [Google Scholar]
- 33.Hale TK, Braithwaite AW. Identification of an upstream region of the mouse p53 promoter critical for transcriptional expression. Nucleic Acids Res. 1995;23:663–669. doi: 10.1093/nar/23.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reisman D, Rotter V. The helix-loop-helix containing transcription factor USF binds to and transactivates the promoter of the p53 tumor suppressor gene. Nucleic Acids Res. 1993;21:345–350. doi: 10.1093/nar/21.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis PL, Miron A, Andersen LM, Iglehart JD, Marks JR. Isolation and initial characterization of the BRCA2 promoter. Oncogene. 1999;18:6000–6012. doi: 10.1038/sj.onc.1202990. [DOI] [PubMed] [Google Scholar]
- 36.Jaiswal AS, Narayan S. Upstream stimulating factor-1 (USF1) and USF2 bind to and activate the promoter of the adenomatous polyposis coli (APC) tumor suppressor gene. J. Cell Biochem. 2001;81:262–277. doi: 10.1002/1097-4644(20010501)81:2<262::aid-jcb1041>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 37.Prendergast GC, Ziff EB. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991;251:186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- 38.Watt F, Molloy PL. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988;2:1136–1143. doi: 10.1101/gad.2.9.1136. [DOI] [PubMed] [Google Scholar]
- 39.Belanger AS, Tojcic J, Harvey M, Guillemette C. Regulation of UGT1A1 and HNF1 transcription factor gene expression by DNA methylation in colon cancer cells. BMC Mol. Biol. 11:9. doi: 10.1186/1471-2199-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ting AH, Jair KW, Suzuki H, Yen RW, Baylin SB, Schuebel KE. CpG island hypermethylation is maintained in human colorectal cancer cells after RNAi-mediated depletion of DNMT1. Nat. Genet. 2004;36:582–584. doi: 10.1038/ng1365. [DOI] [PubMed] [Google Scholar]
- 41.McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66:3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- 42.Tokusumi Y, Ma Y, Song X, Jacobson RH, Takada S. The new core promoter element XCPE1 (X Core Promoter Element 1) directs activator-, mediator-, and TATA-binding protein-dependent but TFIID-independent RNA polymerase II transcription from TATA-less promoters. Mol. Cell Biol. 2007;27:1844–1858. doi: 10.1128/MCB.01363-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 - Oligonucleotide sequences used in plasmid constructs, biotin-affinity purification, EMSAs, methylation analysis and PCR