Abstract
We performed a genome-wide association study in non-Hispanic white subjects with fibrotic idiopathic interstitial pneumonias (N=1616) and controls (N=4683); replication was assessed in 876 cases and 1890 controls. We confirmed association with TERT and MUC5B on chromosomes 5p15 and 11p15, respectively, the chromosome 3q26 region near TERC, and identified 7 novel loci (PMeta = 2.4×10−8 to PMeta = 1.1×10−19). The novel loci include FAM13A (4q22), DSP (6p24), OBFC1 (10q24), ATP11A (13q34), DPP9 (19p13), and chromosomal regions 7q22 and 15q14-15. Our results demonstrate that genes involved in host defense, cell-cell adhesion, and DNA repair contribute to the risk of fibrotic IIP.
The idiopathic interstitial pneumonias (IIPs) represent a group of lung diseases commonly characterized by pulmonary fibrosis or progressive scarring of the alveolar interstitium which can lead to significant morbidity and mortality due to hypoxemic respiratory insufficiency 1. While some forms of pulmonary fibrosis are associated with known environmental exposures (e.g. asbestos), drug toxicity 2, radiation exposure, or collagen vascular diseases (e.g. scleroderma), the IIPs have no known etiology. The most common and severe IIP is idiopathic pulmonary fibrosis (IPF) 1 which has a median survival of 2–3 years after diagnosis. There are no IPF pharmacologic therapies approved for use in the United States, and lung transplantation is the only intervention known to prolong life 3. Although all IIPs have a variable clinical course, they often progress to end-stage lung disease and death. While it appears that risk of IIP is likely determined by multiple genetic variants and environmental toxins, the specific causes of IIP are only beginning to emerge.
The evidence for a genetic component to the risk of IIP is substantial and includes familial clustering of disease, the occurrence of pulmonary fibrosis as part of systemic genetic syndromes, considerable variability in the risk of pulmonary fibrosis among those with similar exposures to known environmental agents such as asbestos, and identification of genetic risk loci in IIP. Rare mutations in the TERT, TERC, SFTPC, and SFTPA2 genes have been associated with familial interstitial pneumonia (FIP; defined as 2 or more family members with IIP) and IPF 4–9, and a common polymorphism in TERT has been associated with IPF 10. Recently, we have identified a promoter variant in the MUC5B gene (rs35705950) that is present in approximately 50–60% of individuals with FIP or IPF and is estimated to increase risk 6-fold for heterozygotes and 20-fold for homozygotes 11. The identification of MUC5B as a common risk factor has altered our view of the pathogenesis of pulmonary fibrosis from focusing primarily on alveolar epithelial cells and lung matrix to inclusion of mucus-producing cells in the distal airways of the lung 11,12. However, the MUC5B variant is observed in ~19% of unaffected individuals and approximately one third of individuals with IIP do not have any identifiable genetic risk factors for this disease, suggesting that other genetic variants contribute to disease risk alone or in combination with the MUC5B variant.
With the goal of identifying additional genetic risk factors that collectively further our understanding of IIP, we have completed a case-control genome-wide association study (GWAS; 1616 cases and 4683 controls) and replication study (876 cases and 1890 controls) of IIP. We included all types of fibrotic IIP in our case group since: a) distinguishing among the IIP diagnoses is often problematic due to substantial clinical, pathological, and radiological overlap; and b) there is strong evidence for shared genetic susceptibility; over 40% of families with FIP have more than one type of IIP among the affected family members 13. We also included both familial and sporadic IIPs since the MUC5B, TERT, TERC, and SFTPC variants provide suggestive evidence that sporadic IIP is genetically similar to the familial form of this disease. We hypothesized that IIPs are caused by multiple genetic variants, acting independently or in combination, and that the same genetic variants can lead to different histologic types of IIP.
Results
Genome-wide Discovery
We genotyped 1914 self-reported non-Hispanic white fibrotic IIP cases on the Illumina 660 Quad beadchip. Of those, 298 were excluded based on being a genetic outlier (N=14), evidence for being a first degree relative of another case (N=126), high heterozygosity (N=8), or missing >2% of genotypes across all SNPs (N=150, see Statistical Methods); 1616 cases were included in analyses (Supplementary Tables 1–3). Among 15,352 out-of study controls without phenotypic information also genotyped on the Illumina 660 Quad beadchip in the same laboratory, we selected 4,683 controls most genetically similar to our cases based on genome-wide identity-by-state comparisons who met the same quality control criteria as cases (see Online Methods and Supplementary Table 1).
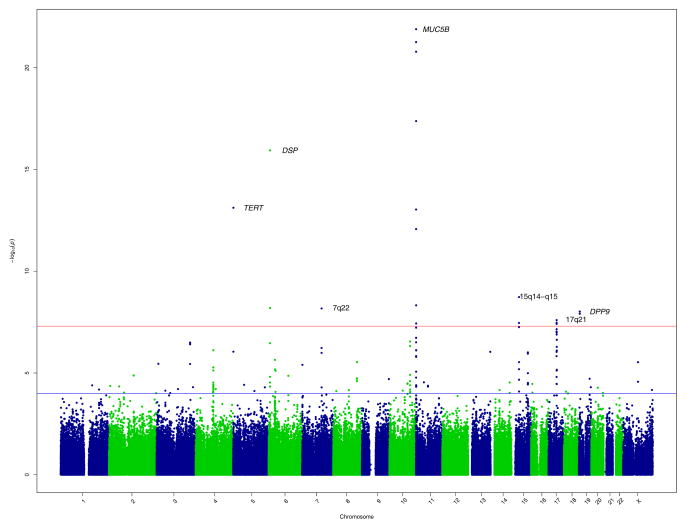
We compared the cases of IIP and controls at 439,828 SNPs with 1) MAF > .05, 2) HWE P-value > 0.0001 in cases and controls evaluated separately, and 3) P-value for differential missingness between cases and controls > 0.001 if less than 2% missing and > 0.05 if between 2% and 5% missing. Neither the QQ-plot of P-values (Supplementary Figure 1) nor the estimated genomic inflation factor (λ) of 0.99 suggested any systematic biases, such as those related to population stratification. Under an additive model for the minor allele at each SNP, we identified 19 SNPs, representing 7 chromosomal locations, with genome-wide significant (P < 5×10−8) associations (Figure 1, Table 1 and Supplementary Table 4). In secondary analyses, we identified another genome-wide significant SNP (rs1379326) representing a unique locus, under a recessive model (Supplementary Table 4).
Figure 1.
GWAS results at 439,828 SNPs with 1616 cases and 4683 controls under additive model. SNPs above red line were genome-wide significant at P < 5×10−8. These SNPs and SNPs between red and blue lines, corresponding to 5×10−8 < P-value <.0001, were selected for follow-up in 876 cases and 1890 controls.
Table 1.
Genome-wide Significant Loci in Discovery GWAS and Meta-Analysis (P-value < 5×10−8)
| Discovery GWAS | Replication | Meta-Analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Positiona | Geneb | Annotation | Minor Allele | MAF Case | OR (95% CI) | P-valuec | MAF Case | OR (95% CI) | P-valuec | P-valuec | |
| Chr. 5p15 | |||||||||||
| rs2736100 | 1339516 | TERT | intronic | C | 0.43 | 0.73 (0.67,0.79) | 7.60e-14 | 0.43 | 0.74 (0.65,0.83) | 4.05e-07 | 1.71e-19 |
| Chr. 6p24 | |||||||||||
| rs2076295 | 7508231 | DSP | intronic | T | 0.54 | 1.43 (1.32,1.55) | 1.14e-16 | 0.52 | 1.26 (1.13,1.42) | 6.28e-05 | 1.08e-19 |
| Chr. 7q22 | |||||||||||
| rs4727443 | 99431282 | intergenic | A | 0.46 | 1.30 (1.20,1.41) | 6.72e-09 | 0.42 | 1.11 (0.98,1.24) | 0.093 | 1.17e-08 | |
| Chr. 11p15 | |||||||||||
| rs7934606 | 1083945 | MUC2 | intronic | C | 0.52 | 1.52 (1.40,1.65) | 5.46e-22 | 0.51 | 1.56 (1.39,1.76) | 1.49e-13 | 6.87e-34 |
| Chr. 15q14-15 | |||||||||||
| rs2034650 | 38504594 | intronic | G | 0.42 | 0.77 (0.71,0.84) | 1.86e-09 | 0.42 | 0.82 (0.74,0.93) | 0.00098 | 9.76e-12 | |
| Chr. 17q21 | |||||||||||
| rs1981997 | 41412603 | MAPT | intronic | A | 0.17 | 0.71 (0.64,0.78) | 2.52e-08 | 0.16 | 0.67 (0.58,0.79) | 4.74e-07 | 8.87e-14 |
| Chr. 19p13 | |||||||||||
| rs12610495 | 4668672 | DPP9 | intronic | G | 0.34 | 1.29 (1.18,1.41) | 9.57e-09 | 0.34 | 1.30 (1.15,1.47) | 3.94e-05 | 1.68e-12 |
Based on NCBI Build 36,
Name of gene if SNP falls in body of gene,
Adjusted for sex
MAF: Minor allele frequency; minor allele defined as minor allele in combined case and control group; OR: Odds ratio for the minor allele; CI: Confidence Interval
Replication and Meta-Analysis
We selected the 20 genome-wide significant SNPs and an additional 178 SNPs with 5×10−8 < P-value <.0001 (143 under an additive model shown between red and blue lines in Figure 1; see Supplementary Tables 5 and 6 for SNP location, genotype and HWE information and Supplementary Table 4 for association information for all 198 SNPs) for genotyping in a replication cohort of 1027 cases of IIP and 2138 controls (Supplementary Tables 1, 3, and 7). After genotype quality control, we included 876 cases and 1890 controls (Supplementary Tables 1–3) successfully genotyped on 181 of the SNPs. Six of the 8 genome-wide significant loci (13 of 20 SNPs) were associated with IIP in the replication cohort at P <0.0025, corresponding to conservative Bonferroni correction for 20 tests (Table 1 middle columns, Supplementary Table 4). Seven of the 8 loci (18 of 20 SNPs, Figure 2) were genome-wide significant in the meta-analysis (Table 1 last column, Supplementary Table 4). Four additional loci (Table 2, Figure 3) were represented among 25 additional SNPs (Supplementary Table 4) that were genome-wide significant under an additive model in the meta-analysis but not in the GWAS discovery.
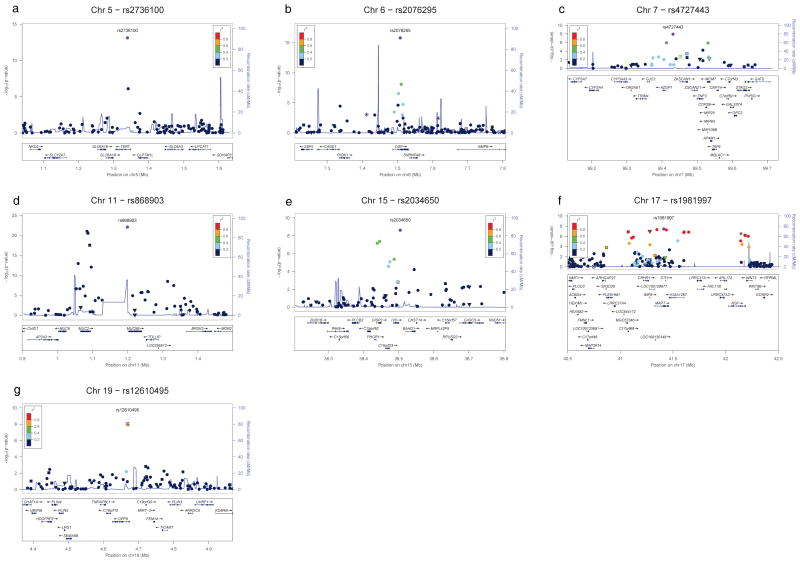
Figure 2.
Locus-specific plots corresponding to discovery GWAS results for all loci reaching genome-wide significance in the GWAS discovery analysis and meta-analysis of the discovery and replication results (a–g). For each plot, the –log10 P values (y axis) of the SNPs are shown according to their chromosomal positions (x axis). The significant loci are on chromosomes 5p15 (a), 6p24 (b), 7q22 (c), 11p15 (d), 15q14-15 (e), 17q21 (f), and 19p13 (g). The estimated recombination rates (cM/Mb) from the HapMap Project (NCBI Build 36) are shown as light blue lines, and the genomic locations of genes within the regions of interest in the NCBI Build 36 human assembly are shown as arrows. SNPs shown in red, orange, green, light blue and blue have r2 ≥ 0.8, r2 ≥ 0.6, r2 ≥ 0.4, r2 ≥ 0.2 and r2 < 0.2 with the most highly-associated SNP, respectively. SNP annotation key: Circles, squares, triangles, star (*), and squares with an x represent no annotation, synonymous or 3′ UTR, nonsynonymous, TFBScons and MCS44 placental, respectively. Genotyped SNPs shown; analogous plots with imputed SNP genotypes are shown in Supplementary Figure 2.
Table 2.
Genome-wide Significant Loci from Meta-analysis (GWAS 5×10−8 < P-value <.0001 and Meta-analysis P-value < 5×10−8)
| Discovery GWAS | Replication | Meta-Analysis | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Positiona | Geneb | Annotation | Minor Allele | MAF Case | OR (95% CI) | P-valuec | MAF Case | OR (95% CI) | P-valuec | P-valuec | |
| Chr. 3q26 | |||||||||||
| rs6793295 | 171001149 | LRRC34 | missense | C | 0.32 | 1.30 (1.19, 1.42) | 3.20e-07 | 0.33 | 1.39 (1.23,1.58) | 2.37e-07 | 8.33e-13 |
| Chr. 4q22 | |||||||||||
| rs2609255 | 90030218 | FAM13A | intronic | G | 0.26 | 1.29 (1.18, 1.42) | 5.27e-06 | 0.28 | 1.43 (1.25,1.64) | 2.56e-07 | 2.20e-11 |
| Chr. 10q24 | |||||||||||
| rs11191865 | 105662832 | OBFC1 | intronic | G | 0.45 | 0.80 (0.74,0.87) | 2.82e-07 | 0.46 | 0.87 (0.77,0.97) | 0.017 | 2.44e-08 |
| Chr. 13q34 | |||||||||||
| rs1278769 | 112584628 | ATP11A | 3′ UTR | A | 0.20 | 0.79 (0.72, 0.88) | 9.11e-07 | 0.20 | 0.80 (0.70, 0.92) | 0.002 | 6.72e-09 |
Based on NCBI Build 36,
Name of gene if SNP falls in body of gene,
Adjusted for sex
MAF: Minor allele frequency; minor allele defined as minor allele in combined case and control group; OR: Odds ratio for the minor allele; CI: Confidence Interval
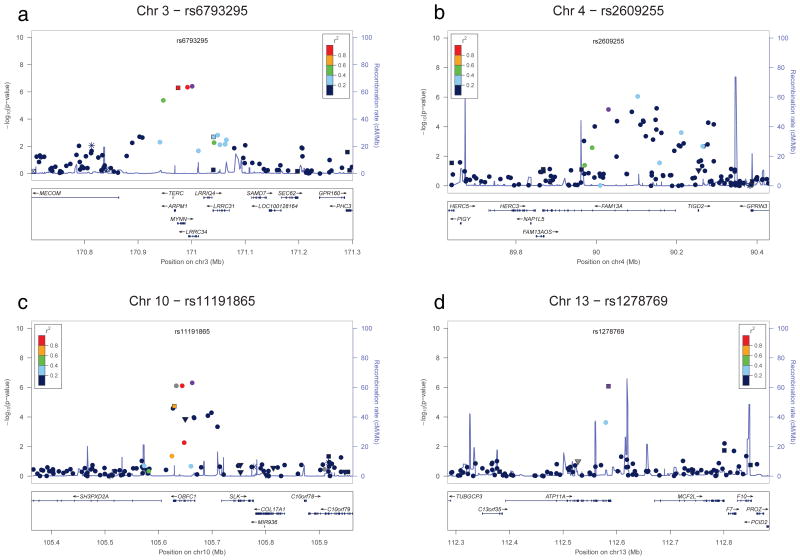
Figure 3.
Locus-specific plots corresponding to discovery GWAS results for four additional loci reaching genome-wide significance after the meta-analysis of the discovery and replication results (a–d). For each plot, the –log10 P values (y axis) of the SNPs are shown according to their chromosomal positions (x axis). The significant loci are on chromosomes 3q26 (a), 4q22 (b), 10q24 (c), and 13q34 (d). The estimated recombination rates (cM/Mb) from the HapMap Project (NCBI Build 36) are shown as light blue lines, and the genomic locations of genes within the regions of interest in the NCBI Build 36 human assembly are shown as arrows. SNPs shown in red, orange, green, light blue and blue have r2 ≥ 0.8, r2 ≥ 0.6, r2 ≥ 0.4, r2 ≥ 0.2 and r2 < 0.2 with the most highly-associated SNP, respectively. SNP annotation key: Circles, squares, triangles, star (*), and squares with an x represent no annotation, synonymous or 3′ UTR, nonsynonymous, TFBScons and MCS44 placental, respectively. Genotyped SNPs shown; analogous plots with imputed SNP genotypes are shown in Supplementary Figure 3.
The most highly associated SNP in the GWAS discovery, rs868903 (PGWAS = 1.3×10−22; PMeta = 9.2×10−26), is in the promoter of the MUC5B gene at chromosome 11p15, which we have previously reported to be associated with IPF and FIP 11 and has been confirmed in other studies 14,15. Ten additional SNPs in the MUC5B region, including SNPs in the MUC2 and TOLLIP genes were also genome-wide significant in the joint analysis and not in strong LD with rs868903 (Figure 2d). The SNPs rs2736100 (PMeta = 1.7×10−19) and rs2853676 (PMeta = 3.3×10−8) at chromosome 5p15 are in the TERT gene (Figure 2a) and rs1881984 (PMeta = 4.5×10−8) is near the TERC gene (Figure 3a); rare mutations in TERT and TERC have been reported to be associated with FIP and IPF 4,5, and rs2736100 in the TERT gene has previously been reported10.
The remaining 8 genome-wide significant loci are novel IIP loci. Five of the association signals on chromosomes 4q22, 6p24, 10q24, 13q34, and 19p13 appear localized to single genes (Figures 2 and 3). SNP rs2609255 (PMeta = 2.2×10−11) is in the FAM13A gene (family with sequence similarity 13, member A) at chromosome 4q22 (Figure 3b). SNPs rs10484326 (PMeta = 5.5×10−9) and rs2076295 (PMeta = 1.1×10−19) are in the DSP gene (desmoplakin) at chromosome 6p24 (Figure 2b). SNPs rs10748858 (PMeta = 2.7×10− −8), rs2067832 (PMeta = 3.7×10−8), and rs11191865 (PMeta = 2.4×10−8) are in the OBFC1 gene (oligonucleotide-binding fold containing 1) at chromosome 10q24 (Figure 3c). SNP rs1278769 (PMeta = 6.7×10−9) is in the ATP11A gene (ATPase, class VI, type 11A) at chromosome 13q34 (Figure 3d). SNPs rs12610495 (PMeta = 1.7×10−12) and rs2109069 (PMeta = 2.4×10−11) are in the DPP9 gene (dipeptidyl-peptidase 9) at chromosome 19p13 (Figure 2g). The other three chromosomal regions (7q22, 15q14-15, and 17q21) have either no significant SNP in any gene or SNPs with significant associations in multiple genes (Tables 1 and 2 and Figure 2c, 2e, 2f, respectively). The estimated odds ratios for all of the genome-wide significant SNPs range from ~1.1 to ~1.6 (Tables 1 and 2; ORs for MAF that are less than 1 correspond to ORs for major allele in this same range).
Investigation of local ancestry on chromosome 17q21
Several SNPs in the 17q21 locus were found to be significantly associated with IIP. However, chromosome 17q21 contains a common inversion polymorphism 16,17 and the haplotypes (collectively referred to as H2) that contain the inversion show marked frequency differences across European populations16,17 . We stratified our GWAS discovery samples by carriage of the H2 haplotypes and tested for association genome-wide among those without the H2 haplotypes to assess the potential for confounding by local ancestry in the chromosome 17q21 region. We similarly stratified our replication sample for association testing at each of the replication SNPs. The association signal at chromosome 17q21 was completely confounded by carriage of an H2 haplotype in both the discovery and replication cohorts. Among those with no H2 haplotypes, most of the 17q21 SNPs had too little variation to allow robust tests of association; all P-values for SNPs that could be tested across the region increased dramatically and none were significant (all nominal P > 0.01; Supplementary Tables 8 [GWAS] and 9 [Replication]).
However, after the stratification by carriage of the H2 haplotypes, the other genome-wide significant associations were either essentially unchanged or reduced in statistical significance consistent with the smaller sample size in the non-H2 carrier group (Supplementary Table 8). Importantly, in both the discovery and replication cohorts, the ORs for each of the other loci were nearly identical in the full and stratified analyses, providing further evidence that the ancestry differences at chromosome 17q21 were not driving any of the other associations we identified.
Imputation across genome-wide significant loci
We imputed genotypes for HapMap3 SNPs using Impute 18 across the significantly-associated regions to better understand the range over which the association signal extended and to identify additional SNPs potentially associated with IIP (see Supplementary Figures 2 and 3 and Supplementary Table 10). In general, the imputation results were entirely consistent with the genotyped SNP results. However, the imputation results implicate the TERC gene more strongly than the real genotype data (Supplementary Figure 3a) and appear to better localize the association signal on chromosome 7q22 to the ZKSCAN1 gene (Supplementary Figure 2c).
Investigation of adjusted models for genome-wide significant SNPs
To adjust for the previously discovered MUC5B promoter SNP (rs35705950; not on the Illumina 660 Quad beadchip), we genotyped a subset of the GWAS discovery cases on the same platform and at the same time as the replication cases for the replication SNPs (those listed in Supplementary Table 4). We combined the raw genotypes from these cases (n=859) with the replication cases and controls for joint analyses.
To assess the evidence for multiple independent association signals within each region, we tested for association with each SNP in a given region after adjusting for the most significant SNP in that region based on the meta-analysis. For the chromosome 11p15 region, we adjusted for rs35705950 given our prior findings and the strength of the association we observed between rs35705950 and IIP in our current study population (OR [95%CI]: 4.51 [3.91, 5.21], PJoint = 7.21×10−95). After adjustment for rs35705950, only one of the SNPs at 11p15 (rs4077759) remained nominally associated with IIP (P=.03; Table 3) while rs35705950 remained highly significant in all models (all P < 1.81×10−80), suggesting that the associations we observed with other SNPs were due to weak LD with rs35705950 (Table 3; see Supplementary Figure 4 for LD among all the SNPs). The reductions in significance of SNPs in the other regions after adjustment for the top SNP were consistent with the LD among the SNPs (Supplementary Table 11) and do not provide evidence for multiple association signals. Interestingly, SNP rs1881984 near the TERC gene was no longer significant after adjustment for SNP rs6793295 in the LRRC34 gene.
Table 3.
Chromosome 11p15 genome-wide significant SNPs adjusted for rs35705950 in the MUC5B promoter
| Joint Analysisa | Joint Analysis Adjusted for rs35705950b | LD with rs35705950 | |||
|---|---|---|---|---|---|
| SNP | OR (95% CI) | P-value | OR (95% CI) | P-value | Pair-wise r2 |
| rs35705950c | 4.51 (3.91,5.21) | 7.21e-95 | N/A | N/A | N/A |
| rs2301160 | 1.17 (1.06,1.29) | 1.50e-03 | 1.02 (0.92,1.14) | 0.68 | 0.01 |
| rs7942850 | 1.15 (1.04,1.27) | 5.63e-03 | 0.94 (0.85,1.05) | 0.31 | 0.02 |
| rs7934606 | 1.61 (1.46,1.78) | 3.47e-21 | 1.06 (0.94,1.18) | 0.34 | 0.15 |
| rs6421972 | 1.62 (1.46,1.78) | 1.85e-21 | 1.06 (0.94,1.18) | 0.34 | 0.15 |
| rs7480563 | 0.82 (0.75,0.91) | 7.10e-05 | 1.10 (0.99,1.23) | 0.08 | 0.07 |
| rs4077759 | 0.87 (0.78,0.96) | 4.86e-03 | 1.13 (1.02,1.27) | 0.03 | 0.04 |
| rs868903 | 0.74 (0.67,0.81) | 5.74e-10 | 1.04 (0.93,1.16) | 0.46 | 0.07 |
| rs2857476 | 0.82 (0.75,0.91) | 7.90e-05 | 1.10 (0.99,1.23) | 0.07 | 0.07 |
| rs3829223 | 0.78 (0.71,0.86) | 7.23e-06 | 1.03 (0.93,1.15) | 0.56 | 0.06 |
| rs2334659 | 0.72 (0.63,0.83) | 3.99e-06 | 0.89 (0.77,1.03) | 0.13 | 0.02 |
| rs7122936 | 0.79 (0.72,0.88) | 7.23e-06 | 1.01 (0.91,1.13) | 0.85 | 0.05 |
Based on joint analysis of a subset of GWAS and all replication cases compared to replication controls to allow for adjustment for rs35705950, which is not on GWAS panel; a subset of GWAS cases were re-genotyped for Supplementary Table 4 SNPs and rs35705950 using same platform and at same time as replication cases and controls. All SNP associations adjusted for sex.
Each SNP was tested for association in a logistic regression model that also included rs35705950 in addition to sex.
P-values for rs35705950 were all < 1.81×10−80 after adjustment for each SNP and sex in individual logistic regression models.
Given the sex differences in IIP risk, we tested for an interaction between each of the GWAS-significant SNPs and sex. We found no strong evidence for differential effects of the SNPs based on sex after correction for the 43 tests (all Pinteraction > 0.01). Finally, we adjusted for age in addition to sex for all of the genome-wide significant SNPs; with the exception of rs7942850 on chromosome 11 (Page-adjusted = 0.06), all SNPs remained nominally significant after adjustment (Supplementary Table 11).
Expression of key genes in lung tissue
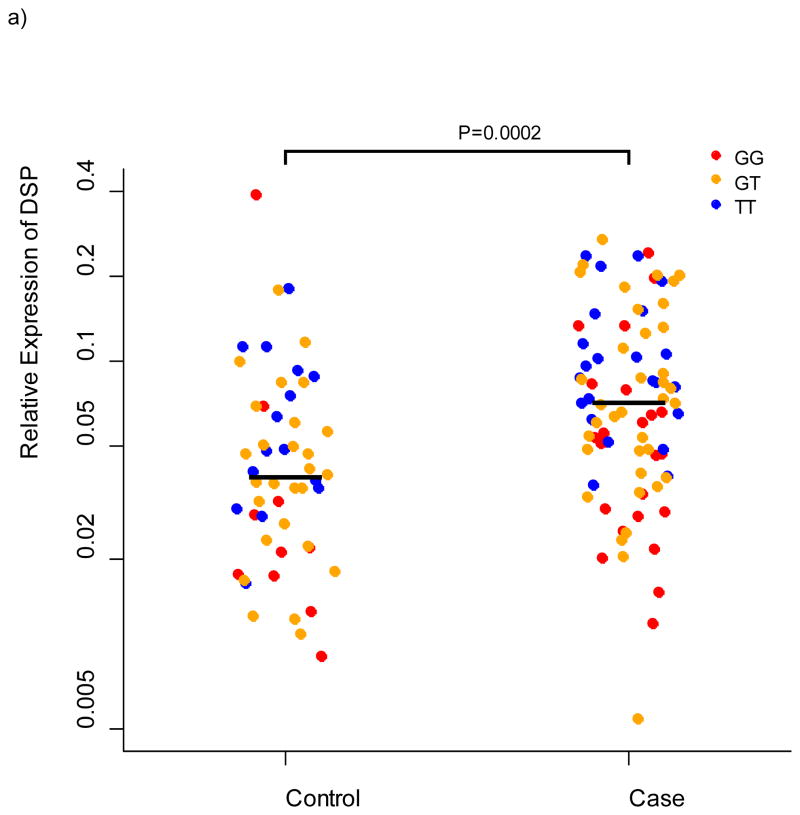
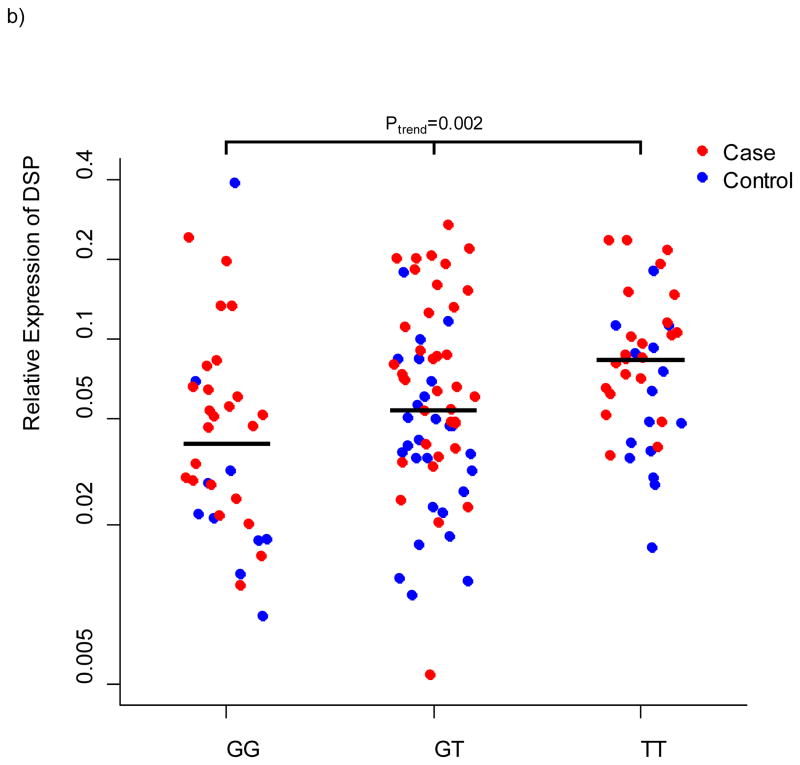
We selected 11 genes for lung tissue expression studies based on localized evidence for novel IIP association (DPP9, DSP, FAM13A, IVD, DISP2, OBFC1, ATP11A, and MUC2) and/or close proximity to an association signal coupled with a priori evidence for expression differences of the gene family in IIP compared to controls (MUC5B, MUC2, WNT3, and WNT9B). We measured expression of these genes in lung tissue from 100 cases of IPF and 94 controls using quantitative PCR and validated Taqman Genotyping Assays (Applied Biosystems, Foster, City, CA) to test for differences between cases and controls and to test for association between the genotypes at the most-highly associated SNPs in each gene with expression of that gene. We confirmed our results from a smaller study 11 that MUC5B is more highly expressed in lung tissue of cases compared to controls (P = 5.6×10−11) but consistent with our previous findings for rs35705950 among cases of IPF, rs868903 was not associated with expression of MUC5B. DSP was more highly expressed in cases compared to controls (P = 0.0002), and expression differed by genotype at rs2076295 (P = 0.002); relative expression of DSP increased with the number of copies of the putative risk allele (Figure 4). There are two isoforms of desmoplakin generated by alternative splicing. rs2076295 is contained in a binding site for transcription factor PU.1, which has been implicated in alternative splicing of target genes 19; however, we saw no evidence for a differential effect of rs2076295 genotype on expression of the primary isoform compared to the alternative isoform (data not shown). There was nominal evidence for higher expression of DPP9 in cases compared to controls (P = 0.03), but neither rs12610495 (P = 0.46) nor rs2109069 (P =0.72) were associated with DPP9 expression. Neither FAM13A, IVD, OBFC1, nor ATP11A differed in expression between cases and controls or by genotype (all P > 0.12); MUC2, DISP2, WNT3, and WNT9B showed little or no expression in these lung samples.
Figure 4.
Relative expression of DSP in lung tissue from 100 cases and 94 controls. a) relative expression by case/control status b) relative expression by genotype at rs2076295 in DSP.
Percent variation in disease risk explained by GWAS SNPs
We estimated the percent of disease risk explained by all the 439,828 GWAS SNPs tested for association using a variance components model 20 across a range of prevalence estimates for IIP (50 per 100,000 to 100 per 100,000). We found that the GWAS SNPs could account for an estimated 28% (s.e. 2%) to 31% (s.e. 3%) of the risk of IIP. Since we did not include the MUC5B promoter SNP (rs35705950) in this analysis (it was not genotyped in the 4,683 out-of study control population), this may be a conservative estimate of the contribution of common SNPs to risk of IIP.
Discussion
These findings provide convincing evidence that common genetic variation is an important contributor to risk of IIP. We have identified 7 novel genetic risk loci (4q22, 6p24, 7q22, 10q24, 13q34, 15q14-15, and 19p13), and confirmed the role of risk variants in three previously reported genes/loci (TERC [3q26], TERT [5p15], and MUC5B [11p15]) for IIP. Prior to our report, the only consistently IIP-associated common variant was MUC5B (rs35705950). In aggregate, the common risk variants associated with IIP suggest that this disease is primarily initiated by defects in host defense, cell-cell adhesion, and DNA repair. Moreover, our findings can be used to guide intervention trials for this complex disease.
Secreted mucins (MUC5B) in the distal airways appear to play a role in the development of IIP. Our data do not suggest any effects of SNPs in other genes (MUC2 or TOLLIP) in the 11p15 region after accounting for the effect of the MUC5B promoter SNP rs35705950, previously identified as a key risk factor for IIP 11. SNP rs868903 in the promoter of the MUC5B gene was one of the most strongly associated SNPs in the GWAS, replication, and meta-analysis, is not in strong LD (r2=0.13) with rs35705950, and is closer to the transcription start site for MUC5B than rs35705950 (1.5 kb vs. 3 kb, respectively). Although lung tissue from patients with IIP has higher concentrations of MUC5B than controls, neither of these MUC5B promoter variants appear to be entirely responsible for the increased expression of MUC5B in patients with IIP, suggesting that other gene variants or environmental toxins are likely to play a role in this disease. We speculate that dysregulated lung mucins initiate or exacerbate lung fibrosis through one of the following mechanisms: 1) altered mucosal defense 21; 2) interference with alveolar repair 22,23; or 3) direct cell toxicity (endoplasmic reticulum stress or apoptosis 24) stimulating a fibroproliferative response initiated by unfolded intracellular MUC5B.
Genes that maintain the length of telomeres appear to play a role in the development of IIP. Prior to this report, the associations between pulmonary fibrosis and TERT and TERC involved rare variants of TERT and TERC 4,5 and potentially one common variant of TERT 10. Mutations in these genes are associated with shortened telomeres in alveolar epithelial cells 25, suggesting that these gene variants may increase the risk of pulmonary fibrosis through disruption of intracellular homeostatic mechanisms. Moreover, dyskeratosis congenita, a congenital disorder that resembles premature aging and frequently involves pulmonary fibrosis, has been attributed to mutations in TERT and TERC 4. Our GWAS identified common variants in TERT and near TERC, and in another gene that influences telomere length, OBFC1. A common variant in OBFC1 has been associated with telomere length in two GWAS studies of human leukocyte telomere length in the general population 26,27. Whether the common variants identified here represent common risk variation or are markers of a collection of rare variants in these genes needs to be established 28,29. However, it appears that risk associated with these genes is not limited to rare variants. In aggregate, these findings underscore the importance of telomerase activity, telomere length, and possibly early cell senescence in the pathogenesis of pulmonary fibrosis.
Our results implicate alterations in cell-cell adhesion in risk of developing IIP. Variants in the DSP gene were strongly associated with IIP and the expression of DSP in the lung tissue of patients with IIP. DSP encodes the protein desmoplakin, a component of the desmosome, an adhesive intercellular molecule that tightly links adjacent cells and forms a dynamic structure with other proteins (plakogobin and plakophilins) that tether the cytoskeleton to the cell membrane 30. Desmosomes are particularly important for maintaining the integrity of tissues that experience mechanical stress (such as the peripheral portions of the lung), and there is strong evidence that perturbation of the desmosome disrupts epithelial homeostasis 30. Mutations in DSP have been associated with arrhythmogenic right ventricular dysplasia 31, keratodermas 32,33, and alopecia 34,35, directly implicating desmoplakin in diseases with loss of tissue integrity. More specifically, mutations in DSP have been associated with cardiac interstitial fibrosis based on over-expression in mouse cardiac tissue 36. An additional potential mechanism for the involvement of DSP is through alterations in the wnt/β-catenin signaling pathway which have been observed in pulmonary fibrosis 37,38. Desmoplakin has been shown to influence the wnt/β-catenin signaling pathway through regulation of another component of the desmosome, γ-catenin 39. These studies and our finding that over-expression of DSP in IIP is associated with the variant allele of rs2076295 provide strong biomechanical or biologic rationales for a role of genetic variation in DSP underlying pulmonary fibrosis.
Our results also implicate other cell adhesion molecules in the risk of IIP development. The DPP9 gene is a member of the same protein family as fibroblast activation protein, which has been shown to be expressed in fibroblastic foci but not in adjacent healthy lung in IPF 40. DPP9 is expressed in epithelia and has been shown to alter cell adhesion in human embryonic kidney cells 41. In addition, the catenin cadherin-associated protein alpha 3 (CTNNA3) gene was nearly significant in the meta-analysis (Pmeta = 9.8×10−07), is located at 10q22, and is a cell adhesion molecule that physically interacts with β-catenin 42 and mediates cell adhesion 43. In aggregate, these findings suggest that pulmonary fibrosis may be caused by defects in cell-cell adhesion or the cytoskeleton that are unable to accommodate the stress associated with mechanical stretch of the lung.
FAM13A is a signal transduction gene that is responsive to hypoxia and a SNP (rs7671167) in this gene has recently been found to be protective in chronic obstructive lung disease 44. The ATP11A gene encodes one of the ATP-binding cassette (ABC) transporter (ABCA1), a gene thought to produce a transmembrane protein with general transport function 45. Another ABC transporter (ABCA3) is expressed by type II alveolar cells, and mutations in ABCA3 have been shown to interfere with lamellar body formation and surfactant protein function, cause surfactant protein deficiency in newborns 46, and have been associated with desquamative interstitial pneumonitis 47 and usual interstitial pneumonia 48 in children.
The other genome-wide significant loci are not as well localized to a single gene, although there are interesting candidates. On chromosome 7q22, an imputed SNP (rs6963345) is the most strongly-associated SNP and is in an intron of the ZKSCAN1 gene. The ZKSCAN1 gene is in the same family as ZKSCAN3, variation in which has been associated with FEV1/FVC in a large meta-analysis of pulmonary function 49. The dispatched homolog 2 (DISP2) gene on 15q14-15 encodes a multi-transmembrane protein involved in hedgehog signaling, which is integral to embryogenesis, tissue regeneration and carcinogenesis 50. The strongest associations on chromosome 15q14-15, however, are in or immediately upstream from the isovaleryl-CoA dehydrogenase (IVD) gene; IVD is a mitochondrial matrix enzyme involved in leucine catabolism. The association signal on chromosome 17q21 was completely confounded with local ancestry at that genomic region, marked by carriage of a common inversion polymorphism, in both the discovery and replication cohorts. As such, determining whether the haplotypes that carry the inversion contain protective variants for IIP will require investigation beyond statistical analysis, such as examination of gene expression differences between cases and controls. An obvious candidate among the genes in the region is the WNT3 gene because alterations in wnt signaling have been observed in IIP 37; however, we found no evidence for WNT3 expression in the lung.
While it has been proposed that pulmonary fibrosis results from activation of developmental pathways 51 or aberrant lung repair 52, our findings suggest that these mechanisms are secondary to a primary defect in host defense or cell-cell adhesion. Since we discovered that several genes involved in the integrity of lung epithelia (DSP, DPP9, and CTNNA3) and lung mucins (MUC5B) are IIP risk variants, we hypothesize that defects in these mechanisms primarily contribute to the development of pulmonary fibrosis. Given the importance of environmental exposures (e.g., exposure to cigarette smoke, asbestos, and silica) in the development of interstitial lung disease, it is logical to speculate that common inhaled particles might, over years, cause exaggerated interstitial injury in persons who have defects in lung host defense or cell-cell adhesion. Our view is that shortened telomeres and consequent changes in cell survival and persistent tissue injury may primarily alter host defense or may enhance the ‘host defense challenge’ to the lung through endogenous mechanisms. Thus, excessive lung injury either through enhanced environmental exposure, endogenous defects in critical homeostatic mechanisms, or subtle defects in host defense may, over years, lead to pulmonary fibrosis. We believe that more attention should be directed to host defense and cell-cell adhesion when considering drugable targets for this complex disease.
Our findings should substantially influence future genetic, diagnostic, and pharmacologic studies of IIP. We estimated that the cumulative GWAS SNPs (excluding rs35705950) reported in this manuscript explain approximately one-third of the variability in risk of developing IIP, suggesting that further examination of common variation with larger cohorts is warranted in addition to studies of rare variation, epigenetic features, and gene-environment interactions. While the clinical manifestations of these diseases have been well defined 1, it is becoming increasingly clear that each type of IIP is caused by multiple gene variants that likely have distinct prognoses which may respond differently to pharmacologic intervention. Consequently, genotyping IIP subjects in therapeutic trials may inform drug development by identifying agents that are effective in selected groups of patients. In fact, the lack of attention to pharmacogenetic approaches in IIP trials may explain why few agents have been found to alter the course of these diseases. Moreover, the genetic heterogeneity of IIP suggests that genetic variants may prove helpful in redefining the types of IIP and may provide more accurate prognostic information for our patients and their families.
Online methods
Study populations
Case definition
We used standard criteria established by the American Thoracic Society/European Respiratory Society 1 to determine diagnostic classification of all patients in the discovery and replication phases (Supplementary Tables 1–3 and 7). We excluded cases with known explanations for development of fibrotic IIP including infections, systemic disorders, or relevant exposures (e.g. asbestos). To maximize power and minimize potential confounding by ancestry, we included only non-Hispanic white (NHW) participants in the GWAS and replication studies. All subjects gave written informed consent as part of IRB-approved protocols for their recruitment and the GWAS study was approved by the National Jewish Health IRB and Colorado Combined Institutional Review Board (COMIRB).
GWAS Discovery
We genotyped 1914 patients with IIP from 6 cohorts (familial interstitial pneumonia [n=566], National Jewish Health IIP population [n=238], InterMune IPF trials [n=720], UCSF [n=66], Vanderbilt University IIP population [n=105], and the National Heart Lung and Blood Institute Lung Tissue Research Consortium [n=219]) and compared them to genotypes from 4683 out-of-study controls (Supplementary Tables 1–3). After genotype quality control, we included 1616 cases in analyses.
A family with familial interstitial pneumonia (FIP) is defined by the presence of at least 2 cases of definite or probable IIP in individuals who are 3rd degree relatives or closer. Recruitment of families based at three major referral centers (Vanderbilt University, Duke University and National Jewish Health) has been ongoing since 1999. We included only 1 IIP case among first degree relatives. The National Jewish Health IIP cohort consists of patients with sporadic IIP who were clinically evaluated and enrolled at National Jewish Health as part of ongoing research protocols associated with clinical care. Details of the recruitment criteria for the cases from the Intermune IPF γ-Interferon Intervention Trial have been described in detail 53. Briefly, eligible patients had IPF, were 40 to 79 years old with clinical symptoms for at least 3 months and evidence of disease progression within the previous 12 months. We included all available cases regardless of treatment assignment. The National Heart Lung and Blood Institute Lung Tissue Research Consortium (NHLBI LTRC) was established to provide lung tissue and DNA for the research community. We included DNA from those subjects with a diagnosis of IIP.
We used de-identified control genotypes generated at Centre d’Étude du Polymorphisme Humain (CEPH) as part of other studies. Potential controls were those who self-reported NHW, had been genotyped on the same platform as our cases, and were appropriately approved for use as controls in other studies. We selected a subset of controls, corresponding to approximately 3 controls for 1 case, based on genetic similarity to the cases which passed our genotyping quality control thresholds (see Statistical Analyses below for details).
Replication
We genotyped a total of 1027 NHW IIP cases (See Supplementary Table 7 for breakdown by replication sample) and 2138 NHW controls for replication of the top SNPs from the GWAS. The replication controls were a subset (n=2000) of the controls from the Chronic Obstructive Pulmonary Disease (COPD) Gene Study 54 and 138 controls from the University of Pittsburgh. We selected controls to be frequency matched to the replication cases based on age and gender. After quality control, we included 876 cases and 1890 controls in any analyses that included replication samples.
Expression
We measured gene expression on a subset of Lung Tissue Research Consortium and National Jewish Health IIP cases from the GWAS (n=100) and National Jewish Health controls (n=94). Whole-lung samples were obtained from International Institute for the Advancement of Medicine (Edison, NJ). Eligible cases and controls had sufficient RNA from lung tissue biopsy available for assay; cases with IPF were preferentially chosen over other IIP diagnoses. National Jewish Health controls also had genome-wide SNP data available.
DNA preparation, storage, and quality control
Genomic DNA was isolated from both whole blood and biopsied lung tissue on either the Autopure LS (Qiagen) or Qiacube (Qiagen) automation platform, respectively. Prior to extraction on the Qiacube using the DNAeasy kit, fibrotic lung tissues were first homogenized using Lysing Matrix D tubes and a FastPrep-24 benchtop homogenizer (MPBiomedicals). Following isolation, all DNA was assayed for concentration and purity on the NanoDrop ND-1000 Spectrophotometer. Samples were excluded if DNA was < 50ng/ul or had an A260/A280 ratio outside of the 1.7–2.0 range.
For GWAS genotyping, prior to submission to the Centre National de Genotypage at CEPH, all samples were re-quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen), normalized with 1xTE, and aliquotted into individually barcoded screw-cap tubes. Due to volume limitations with liquid handling robots, an absolute minimum quantity for submission to the CNG was 30ul at 50ng/ul. If samples did not meet this minimum quantity, an alternate extraction was performed or the sample was withheld from the study.
For replication genotyping, upon receipt, samples were transferred into 96-well robotics compatible plates, quantified with PicoGreen, and normalized with 1xTE. 400ng of DNA was submitted for each member of the GWAS and the replication cohorts sent for replication genotyping. In an effort to minimize confounding by batch effects, samples were aliquotted into 96-well plates in a randomized fashion across all cohorts with two duplicates per plate using the Tecan Evo200 liquid handling robot.
Genome-wide genotyping
Genome-wide genotyping was carried out at CEPH using the Illumina 660 Quad beadchip. Barcoded DNA samples were received in standard tubes together with sample information, and were subjected to stringent quality control (QC). Concentration, fragmentation and response to PCR were determined. Samples from cases and controls were randomly distributed on 96-well plates. Processing was carried out under full LIMS control in a fully automated Illumina BeadLab equipped with 8 Tecan liquid handling robots, 6 Illumina BeadArray readers and 2 Illumina iScans.
Replication genotyping
Replication genotyping was carried out at the Biomedical Genomics Center at the University of Minnesota. We genotyped 198 SNPs with P-values less than 0.0001 (see Statistical Analyses) in 1027 independent cases and 2000 COPDgene controls. We also genotyped the MUC5B promoter SNP rs35705950, which is not on the Illumina 660 Quad beadchip, to allow adjustment of other SNPs on chromosome 11p15 for rs35705950. In addition, to allow follow-up joint statistical tests (using raw genotypes from both GWAS cases and replication cases and controls) with adjustment for covariates that were not available on the out-of-study controls, we also genotyped a subset of GWAS cases. Details of the validation assays are described below. After genotyping quality control, we included 876 cases and 1890 controls in the replication, meta- and joint analyses and 859 of the GWAS cases in the joint analyses.
Prior to genotyping, all samples were quality controlled by real-time Q-PCR quantitation (“QC1”) and uniplex genotyping using Taqman (“QC2”). Samples that failed QC1 or QC2, although carried forward through genotyping, were later removed from analysis.
Validation genotyping was accomplished with a combination of multiplexed (Sequenom iPLEX) and uniplex (Taqman) assays. First, assay design for multiplexed Sequenom iPLEX genotyping was performed on an input set of 198 SNPs (Supplementary Table 4), using a combination of web-based (AssayDesigner Suite) and desktop (AssayDesigner) software tools (Sequenom, San Diego). Of 198 input SNPs, 193 were efficiently placed into a set of 6 assays of the following plexities: 35, 35, 35, 35, 31, and 22 SNPs. Sequenom iPLEX genotyping is based on multiplexed locus-specific PCR amplification, multiplexed single-based extension (SBE) from locus-specific amplicons, and multiplexed resolution of SBE products base calling using matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOD) mass spectrometry.
Primers for the Sequenom assay were purchased from IDT (Coralville, Iowa), and all steps of the iPLEX procedure were carried out using reagents and methods from Sequenom (San Diego, CA) according to the manufacturer’s instructions. Reactions were carried out in 384-well plates and analyzed using the Sequenom MassARRAY Analyzer 4 system with iPLEX Gold reagents and SpectroCHIP arrays. Results were analyzed using a combination of commercial software (Typer 4, Sequenom) and custom tools for data management. Of 193 assays in 6 multiplexes, 176 were successful in generating usable genotyping data.
The remaining 5 SNPs that were not successfully included in the original Sequenom iPLEX designs (rs2736100, rs35705950, rs13225346, rs10822856, rs10139381, rs10751635), as well as a sixth SNP (rs35705950) published in earlier studies, were genotyped using commercial Taqman assays (Life Technologies, San Diego, CA). Reactions were carried out in 384-well plates and fluorescence read out using an Applied Biosystems ABI 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA).
Gene Expression
Total RNA was isolated from approximately 30 mg of snap-frozen or RNA-later preserved lung tissue using the Ambion mirVana kit (Life Technologies). RNA concentration was determined by Nanodrop ND-1000 (Thermo Scientific) and RNA integrity was determined using the 2100 Bioanalyzer (Agilent). cDNA single strand conversions were performed using the Superscript III First-Strand Synthesis System (Invitrogen) and expression analysis was performed using pre-designed Taqman assays run on the Viia7 Real-Time PCR instrument (Life Technologies). (DPP9: Hs00373589; DSP: Hs00189422 and the DSP variant 1 assay is Hs00950584; FAM13A: Hs00208453; IVD: Hs01064832; MUC5B: Hs00861588; MUC2: Hs00149374; OBFC1: Hs00998588; WNT3: Hs00902257; WNT9B: Hs00916642; GAPDH: 4333764F) ATP11A: Hs00392589. All assays were run in triplicate with GAPDH used as the endogenous control. As an additional control, one sample per plate was run in duplicate from the cDNA conversion step.
Statistical analyses
Selection of out-of-study controls for GWAS discovery
We obtained controls based on genetic matching to cases from a large database of anonymous genotypes from Europeans who had been genotyped at CEPH on the Illumina 660 Quad beadchip. An ancestry analysis was carried out using the EIGENSTRAT3.0 software 55. HapMap data on 618 individuals (CEU, YRI, JPT and CHB) and samples of reference Europeans were used as representatives of European, West African and East Asian populations to infer ancestry-informative principal components (PCs) which were projected onto the case and control samples 56. Putative non-European samples were flagged as outliers and eliminated from all subsequent analyses. We selected a subset of the available controls to obtain three matching controls per case by first jointly clustering the cases and controls into subpopulations based on the top 10 PCs using a support vector machine approach 57 (R package “e1071” with radial basis function). We then applied an optimal paired matching algorithm within each cluster to choose the best three controls for each case based on a distance matrix defined by the top 10 PCs (“pairmatch” function in R package “optmatch” 58).
Removal of first degree relatives among cases and controls
We included only one individual among first degree relatives based on an estimated kinship coefficient ≥ 0.45. For estimation of the percent variation in disease risk explained by the GWAS SNPs which is particularly sensitive to cryptic relatedness, we kept only one individual among those with estimated kinship coefficient > 0.025.
Exclusion of individuals and prioritization of SNPs for discovery GWAS
In addition to individuals excluded by the laboratory, we excluded cases and controls with 1) evidence for being a genetic outlier based on a pairwise identity-by-state (IBS) estimate with the 5th closest neighbor that was > 4 standard deviations from the mean pairwise IBS estimate across all pairs, 2) unresolved sex mix-match between clinical and genomic data, 3) heterozygosity across the SNPs greater or less than 4 standard deviations from the mean heterozygosity across all individuals, and 4) genotype calls at less than 98% of SNPs that pass laboratory quality control. Based on this quality control, we excluded 298 cases and 145 controls. In addition to the laboratory quality control measures, we prioritized SNPs for follow-up based on other criteria. We tested for differential missingness via a chi-squared test of proportions of missingness between cases and controls and for departures from HWE via a 1-df goodness of fit test. We prioritized SNPs with 1) MAF > .05, 2) HWE P-value > 0.0001 in cases and controls evaluated separately, 3) p-value for differential missingness between cases and controls > 0.001 if less than 2% missing and > 0.05 if between 2% and 5% missing 59.
GWAS association testing
We tested for association between each SNP and IIP using an exact mixed model approach to account for both subtle relatedness and population stratification among our cases and controls implemented in the genome-wide efficient mixed-model association (GEMMA) software package 60. We tested for association under an additive model for our primary analysis and in a secondary analysis took the minimum of the recessive and dominant model p-values if there was significant lack of fit to the additive model (p<.05) from a linear regression that assumed independence among the samples (such a test is not currently implementable in the GEMMA software). We adjusted for sex in all models. We compared the distribution of p-values obtained under the additive model to that expected under the null hypothesis of no association across the genome and report the quantile-quantile (Q-Q plot) and genomic inflation factor (λ) to verify the absence of systematic biases due to experimental or other confounding factors such as population stratification. We selected all SNPs with a P-value <0.0001 for follow-up in the replication populations. We visually inspected genotype spectra for all 198 selected SNPs to assure genotype call quality. We calculated odds ratios and 95% confidence intervals (CIs) from a logistic regression model adjusted for sex assuming independence among the cases and controls since the linear model in GEMMA uses the identity link rather than the log-odds link function. As such, the CIs may be slightly narrower than those based on the full mixed models.
Replication association
We tested for association between each replication SNP and IIP in the replication cases and controls using the freely available SNPGWA software (see URLs). We tested for association under the genetic model from the GWAS that gave the minimum p-value (143 under an additive model, 24 under a dominant model and 31 under a recessive model). A p-value <.0025 was considered statistically significant replication for the 20 genome-wide significant GWAS SNPs. The p-values for the other 178 SNPs were used in the meta-analysis of the GWAS and replication cohorts.
Meta-analysis
To obtain an over-all measure of association between each of the 181 successfully genotyped SNPs in the replication set and IIP, we performed a meta-analysis of the GWAS and replication results. We used the weighted inverse normal method. Let Zi (i=GWAS or replication) be the test statistic from the test of association in the ith study and let vi (i= GWAS or replication) be the corresponding weight. Here we took the weight to be the square root of the total sample size in the ith study since effect estimates from the GWAS and replication were not on the same scale. Note that this method explicitly accounts for the directionality of the association. Thus, highly significant associations with conflicting directions do not exhibit strong statistical association in this meta-analysis. We used METAL (61) to perform our meta-analysis. SNPs with PMeta<5×10−8 were considered genome-wide statistically significant. We created locus-specific plots 62 of the discovery GWAS results for all loci that were genome-wide significant in the meta-analysis.
Stratified analyses by H1 and H2 haplotypes on Chromosome 17
We stratified our discovery and replication cohorts using rs17563986, which completely determines the H1 or H2 haplotypes 17. We tested for association among those carrying two H1 haplotypes given the small sample sizes of H2 carriers. There were 1127 cases and 2832 controls included in the stratified GWAS analysis and 617 cases and 1138 controls in the stratified replication analysis.
Imputation
We imputed genotypes using the combined case and control discovery samples for all HapMap SNPs across a 5Mb region that covered the association signal for each genome-wide significant locus. We used the multi-population reference panel data from HapMap3 for pre-phasing using Shapeit with appropriate default parameters 63,64. We performed imputation using Impute 65,66 and tested for association at only those SNPs with imputation information as measured by .info > 0.5 using Snptest (v2; 67) with multiple Newton-Raphson interations to estimate parameters.
Multi-SNP models
To assess the independence of effects of the meta-analysis genome-wide significant SNPs, we used logistic regression models within each locus using a combined case group (subset of GWAS and all replication) and the replication controls using SAS (v. 9.2). Specifically, within each locus with a genome-wide significant SNP, we tested for association between IIP and each of the other validation panel SNPs within that locus after adjusting for the most significantly associated SNP in that locus (on chromosome 11p15, we adjusted for rs35705950). To assess the robustness of each SNP association to age effects in addition to sex, we tested for association between IIP and each SNP adjusted for both age and sex. Finally, to test for effect modification of SNP associations by sex, we tested for association between IIP and each SNP by sex interaction.
Expression analyses
We tested for differential gene expression in the lung between 100 cases and 94 controls using a two-sample t-test. We also tested for differential expression by genotype using the combined case and control group via a test for trend across the three genotype groups unless there < 5 individuals in a genotype group; we grouped the rare homozygote and heterozygote groups in that case. A P-value < .05 was considered statistically significant.
Supplementary Material
Acknowledgments
We gratefully acknowledge the individuals who participated in the studies that contributed to this work. This research was supported by the National Heart, Lung and Blood Institute (R01-HL095393, R01-HL097163, P01-HL092870, RC2-HL101715, U01-HL089897, U01-HL089856, U01-HL108642, and P50-HL0894932), the Veterans Administration (1I01BX001534), the Dorothy P. and Richard P. Simmons Center, and InterMune, Inc.
Footnotes
Author contributions: TEF and DAS designed the study; KKB, MPS, JEL, GPC, DL, SG, HRC, PLW, RMD, CKG, MSD, GG, HJI, NK, YZ, KFG, LHL, WRM, TMM, PLM, AUW, JDC, BJM, EAR, and MIS performed clinical, radiological, and pathological phenotyping of study subjects; WXB, KK, and SDS provided data and samples from the InterMune subjects; JT, RNK, CRM coordinated the clinical evaluations; EM supervised and coordinated the laboratory work; EM, JDC, DSW, JJD, DZ, KS performed the laboratory work; DM organized the database; KBB supervised the replication genotyping; ML supervised the genome wide genotyping; MFM, MS, AP, DSK, and MIS provided advice on the design and relevance to pulmonary fibrosis; TEF, WZ, ALP, BSP, and YK analyzed the data; TEF, ML, and DAS developed the conceptual approaches to data analysis; TEF and DAS wrote the manuscript.
References Cited
- 1.American Thoracic Society/European Respiratory Society International Multidisciplinary Consensus Classification of the Idiopathic Interstitial Pneumonias. This joint statement of the American Thoracic Society (ATS), and the European Respiratory Society (ERS) was adopted by the ATS board of directors, June 2001 and by the ERS Executive Committee. Am J Respir Crit Care Med. 2001 Jun;165:277–304. doi: 10.1164/ajrccm.165.2.ats01. 2002. [DOI] [PubMed] [Google Scholar]
- 2.Hubbard R, et al. Exposure to commonly prescribed drugs and the etiology of cryptogenic fibrosing alveolitis: a case-control study. Am J Respir Crit Care Med. 1998;157:743–7. doi: 10.1164/ajrccm.157.3.9701093. [DOI] [PubMed] [Google Scholar]
- 3.Raghu G. Idiopathic pulmonary fibrosis: new evidence and an improved standard of care in 2012. Lancet. 2012;380:699–701. doi: 10.1016/S0140-6736(12)61256-2. [DOI] [PubMed] [Google Scholar]
- 4.Armanios MY, et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N Engl J Med. 2007;356:1317–26. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 5.Tsakiri KD, et al. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc Natl Acad Sci U S A. 2007;104:7552–7. doi: 10.1073/pnas.0701009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas AQ, et al. Heterozygosity for a surfactant protein C gene mutation associated with usual interstitial pneumonitis and cellular nonspecific interstitial pneumonitis in one kindred. Am J Respir Crit Care Med. 2002;165:1322–8. doi: 10.1164/rccm.200112-123OC. [DOI] [PubMed] [Google Scholar]
- 7.Lawson WE, et al. Genetic mutations in surfactant protein C are a rare cause of sporadic cases of IPF. Thorax. 2004;59:977–80. doi: 10.1136/thx.2004.026336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, et al. Genetic defects in surfactant protein A2 are associated with pulmonary fibrosis and lung cancer. Am J Hum Genet. 2009;84:52–9. doi: 10.1016/j.ajhg.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Moorsel CH, et al. Surfactant protein C mutations are the basis of a significant portion of adult familial pulmonary fibrosis in a dutch cohort. Am J Respir Crit Care Med. 2010;182:1419–25. doi: 10.1164/rccm.200906-0953OC. [DOI] [PubMed] [Google Scholar]
- 10.Mushiroda T, et al. A genome-wide association study identifies an association of a common variant in TERT with susceptibility to idiopathic pulmonary fibrosis. J Med Genet. 2008;45:654–6. doi: 10.1136/jmg.2008.057356. [DOI] [PubMed] [Google Scholar]
- 11.Seibold MA, et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med. 2011;364:1503–12. doi: 10.1056/NEJMoa1013660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boucher RC. Idiopathic pulmonary fibrosis--a sticky business. N Engl J Med. 2011;364:1560–1. doi: 10.1056/NEJMe1014191. [DOI] [PubMed] [Google Scholar]
- 13.Steele MP, et al. Clinical and pathologic features of familial interstitial pneumonia. Am J Respir Crit Care Med. 2005;172:1146–52. doi: 10.1164/rccm.200408-1104OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Noth I, Garcia JG, Kaminski N. A variant in the promoter of MUC5B and idiopathic pulmonary fibrosis. N Engl J Med. 2011;364:1576–7. doi: 10.1056/NEJMc1013504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stock CJ, et al. Mucin 5B promoter polymorphism is associated with idiopathic pulmonary fibrosis but not with development of lung fibrosis in systemic sclerosis or sarcoidosis. Thorax. 2013 doi: 10.1136/thoraxjnl-2012-201786. [DOI] [PubMed] [Google Scholar]
- 16.Boettger LM, Handsaker RE, Zody MC, McCarroll SA. Structural haplotypes and recent evolution of the human 17q21.31 region. Nat Genet. 2012;44:881–5. doi: 10.1038/ng.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steinberg KM, et al. Structural diversity and African origin of the 17q21.31 inversion polymorphism. Nat Genet. 2012;44:872–80. doi: 10.1038/ng.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 19.Guillouf C, Gallais I, Moreau-Gachelin F. Spi-1/PU.1 oncoprotein affects splicing decisions in a promoter binding-dependent manner. J Biol Chem. 2006;281:19145–55. doi: 10.1074/jbc.M512049200. [DOI] [PubMed] [Google Scholar]
- 20.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rose MC, Voynow JA. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol Rev. 2006;86:245–78. doi: 10.1152/physrev.00010.2005. [DOI] [PubMed] [Google Scholar]
- 22.Crouch E. Pathobiology of pulmonary fibrosis. Am J Physiol (Lung Cell Mol Physiol 4) 1990;259:L159–L184. doi: 10.1152/ajplung.1990.259.4.L159. [DOI] [PubMed] [Google Scholar]
- 23.Kuhn IC, et al. An immunohistochemical study of architectural remodeling and connective tissue synthesis in pulmonary fibrosis. Am Rev Respir Dis. 1989;140:1693–1703. doi: 10.1164/ajrccm/140.6.1693. [DOI] [PubMed] [Google Scholar]
- 24.Korfei M, et al. Epithelial endoplasmic reticulum stress and apoptosis in sporadic idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2008;178:838–46. doi: 10.1164/rccm.200802-313OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alder JK, et al. Short telomeres are a risk factor for idiopathic pulmonary fibrosis. Proc Natl Acad Sci U S A. 2008;105:13051–6. doi: 10.1073/pnas.0804280105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levy D, et al. Genome-wide association identifies OBFC1 as a locus involved in human leukocyte telomere biology. Proc Natl Acad Sci U S A. 2010;107:9293–8. doi: 10.1073/pnas.0911494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mangino M, et al. Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orozco G, Barrett JC, Zeggini E. Synthetic associations in the context of genome-wide association scan signals. Hum Mol Genet. 2010;19:R137–44. doi: 10.1093/hmg/ddq368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delva E, Tucker DK, Kowalczyk AP. The desmosome. Cold Spring Harb Perspect Biol. 2009;1:a002543. doi: 10.1101/cshperspect.a002543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Awad MM, Calkins H, Judge DP. Mechanisms of disease: molecular genetics of arrhythmogenic right ventricular dysplasia/cardiomyopathy. Nat Clin Pract Cardiovasc Med. 2008;5:258–67. doi: 10.1038/ncpcardio1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armstrong DK, et al. Haploinsufficiency of desmoplakin causes a striate subtype of palmoplantar keratoderma. Hum Mol Genet. 1999;8:143–8. doi: 10.1093/hmg/8.1.143. [DOI] [PubMed] [Google Scholar]
- 33.Whittock NV, et al. Striate palmoplantar keratoderma resulting from desmoplakin haploinsufficiency. J Invest Dermatol. 1999;113:940–6. doi: 10.1046/j.1523-1747.1999.00783.x. [DOI] [PubMed] [Google Scholar]
- 34.Whittock NV, et al. Compound heterozygosity for non-sense and mis-sense mutations in desmoplakin underlies skin fragility/woolly hair syndrome. J Invest Dermatol. 2002;118:232–8. doi: 10.1046/j.0022-202x.2001.01664.x. [DOI] [PubMed] [Google Scholar]
- 35.Al-Owain M, et al. Novel homozygous mutation in DSP causing skin fragility-woolly hair syndrome: report of a large family and review of the desmoplakin-related phenotypes. Clin Genet. 2011;80:50–8. doi: 10.1111/j.1399-0004.2010.01518.x. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z, et al. Desmosomal dysfunction due to mutations in desmoplakin causes arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circ Res. 2006;99:646–55. doi: 10.1161/01.RES.0000241482.19382.c6. [DOI] [PubMed] [Google Scholar]
- 37.Chilosi M, et al. Aberrant Wnt/beta-catenin pathway activation in idiopathic pulmonary fibrosis. Am J Pathol. 2003;162:1495–502. doi: 10.1016/s0002-9440(10)64282-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henderson WR, Jr, et al. Inhibition of Wnt/beta-catenin/CREB binding protein (CBP) signaling reverses pulmonary fibrosis. Proc Natl Acad Sci U S A. 2010;107:14309–14. doi: 10.1073/pnas.1001520107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang L, et al. Desmoplakin acts as a tumor suppressor by inhibition of the Wnt/beta-catenin signaling pathway in human lung cancer. Carcinogenesis. 2012;33:1863–70. doi: 10.1093/carcin/bgs226. [DOI] [PubMed] [Google Scholar]
- 40.Acharya PS, Zukas A, Chandan V, Katzenstein AL, Pure E. Fibroblast activation protein: a serine protease expressed at the remodeling interface in idiopathic pulmonary fibrosis. Hum Pathol. 2006;37:352–60. doi: 10.1016/j.humpath.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 41.Yu DM, Wang XM, Ajami K, McCaughan GW, Gorrell MD. DP8 and DP9 have extra-enzymatic roles in cell adhesion, migration and apoptosis. Adv Exp Med Biol. 2006;575:63–72. doi: 10.1007/0-387-32824-6_7. [DOI] [PubMed] [Google Scholar]
- 42.Janssens B, et al. alphaT-catenin: a novel tissue-specific beta-catenin-binding protein mediating strong cell-cell adhesion. J Cell Sci. 2001;114:3177–88. doi: 10.1242/jcs.114.17.3177. [DOI] [PubMed] [Google Scholar]
- 43.Lauren J, Airaksinen MS, Saarma M, Timmusk T. A novel gene family encoding leucine-rich repeat transmembrane proteins differentially expressed in the nervous system. Genomics. 2003;81:411–21. doi: 10.1016/s0888-7543(03)00030-2. [DOI] [PubMed] [Google Scholar]
- 44.Cho MH, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat Genet. 2010;42:200–2. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang B, Groffen J, Heisterkamp N. Resistance to farnesyltransferase inhibitors in Bcr/Abl-positive lymphoblastic leukemia by increased expression of a novel ABC transporter homolog ATP11a. Blood. 2005;106:1355–61. doi: 10.1182/blood-2004-09-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shulenin S, et al. ABCA3 gene mutations in newborns with fatal surfactant deficiency. N Engl J Med. 2004;350:1296–303. doi: 10.1056/NEJMoa032178. [DOI] [PubMed] [Google Scholar]
- 47.Bullard JE, Wert SE, Whitsett JA, Dean M, Nogee LM. ABCA3 mutations associated with pediatric interstitial lung disease. Am J Respir Crit Care Med. 2005;172:1026–31. doi: 10.1164/rccm.200503-504OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Young LR, et al. Usual interstitial pneumonia in an adolescent with ABCA3 mutations. Chest. 2008;134:192–5. doi: 10.1378/chest.07-2652. [DOI] [PubMed] [Google Scholar]
- 49.Soler Artigas M, et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011;43:1082–90. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Katoh Y, Katoh M. Hedgehog signaling pathway and gastric cancer. Cancer Biol Ther. 2005;4:1050–4. doi: 10.4161/cbt.4.10.2184. [DOI] [PubMed] [Google Scholar]
- 51.Selman M, Pardo A, Kaminski N. Idiopathic pulmonary fibrosis: aberrant recapitulation of developmental programs? PLoS Med. 2008;5:e62. doi: 10.1371/journal.pmed.0050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gross TJ, Hunninghake GW. Idiopathic pulmonary fibrosis. N Engl J Med. 2001;345:517–25. doi: 10.1056/NEJMra003200. [DOI] [PubMed] [Google Scholar]
- 53.King TE, Jr, et al. Effect of interferon gamma-1b on survival in patients with idiopathic pulmonary fibrosis (INSPIRE): a multicentre, randomised, placebo-controlled trial. Lancet. 2009;374:222–8. doi: 10.1016/S0140-6736(09)60551-1. [DOI] [PubMed] [Google Scholar]
- 54.Regan EA, et al. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heath SC, et al. Investigation of the fine structure of European populations with applications to disease association studies. Eur J Hum Genet. 2008;16:1413–29. doi: 10.1038/ejhg.2008.210. [DOI] [PubMed] [Google Scholar]
- 57.Cortes C, Vapnik V. Support-vector Networks. Machine Learning. 1995;20:273–297. [Google Scholar]
- 58.Hansen BB, Klopfer SO. Optimal Full Matching and Related Designs Via Network Flows. Journal of Computational and Graphical Statistics. 2006;15:1–19. [Google Scholar]
- 59.Harley JB, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–10. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821–4. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pruim RJ, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 64.Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–81. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- 65.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–70. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.