Abstract
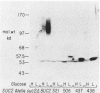
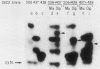
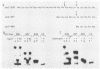
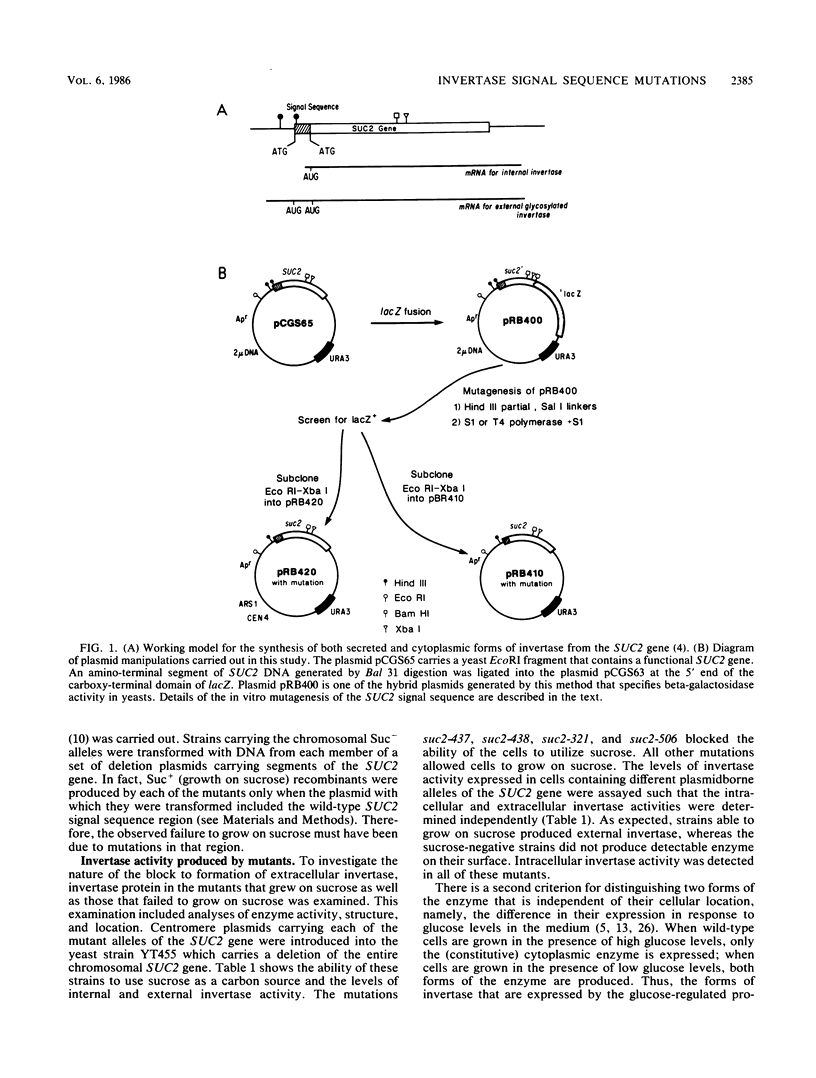
Nine mutations in the signal sequence region of the gene specifying the secreted Saccharomyces cerevisiae enzyme invertase were constructed in vitro. The consequences of these mutations were studied after returning the mutated genes to yeast cells. Short deletions and two extensive substitution mutations allowed normal expression and secretion of invertase. Other substitution mutations and longer deletions blocked the formation of extracellular invertase. Yeast cells carrying this second class of mutant gene expressed novel active internal forms of invertase that exhibited the following properties. The new internal proteins had the mobilities in denaturing gels expected of invertase polypeptides that had retained a defective signal sequence and were otherwise unmodified. The large increase in molecular weight characteristic of glycosylation was not seen. On nondenaturing gels the mutant enzymes were found as heterodimers with a normal form of invertase that is known to be cytoplasmic, showing that the mutant forms of the enzyme are assembled in the same compartment as the cytoplasmic enzyme. All of the mutant enzymes were soluble and not associated with the membrane components after fractionation of crude cell extracts on sucrose gradients. Therefore, these signal sequence mutations result in the production of active internal invertase that has lost the ability to enter the secretory pathway. This demonstrates that the signal sequence is required for the earliest steps in membrane translocation.
Full text
PDF



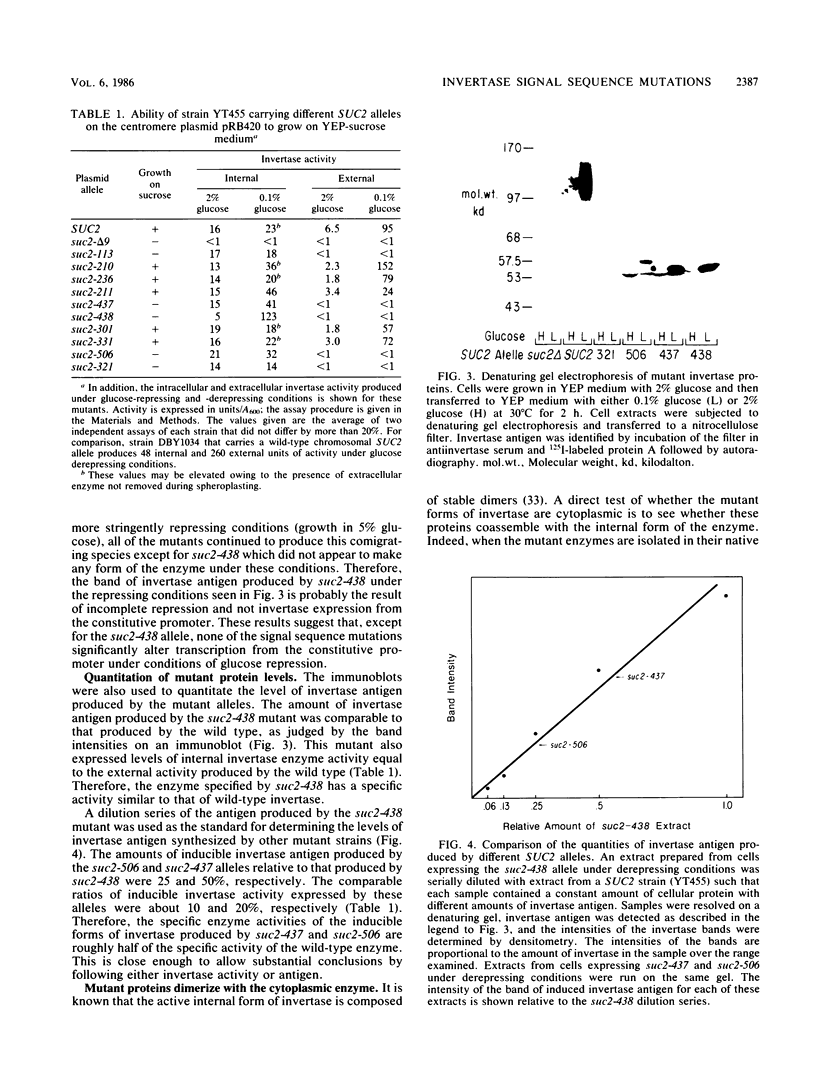

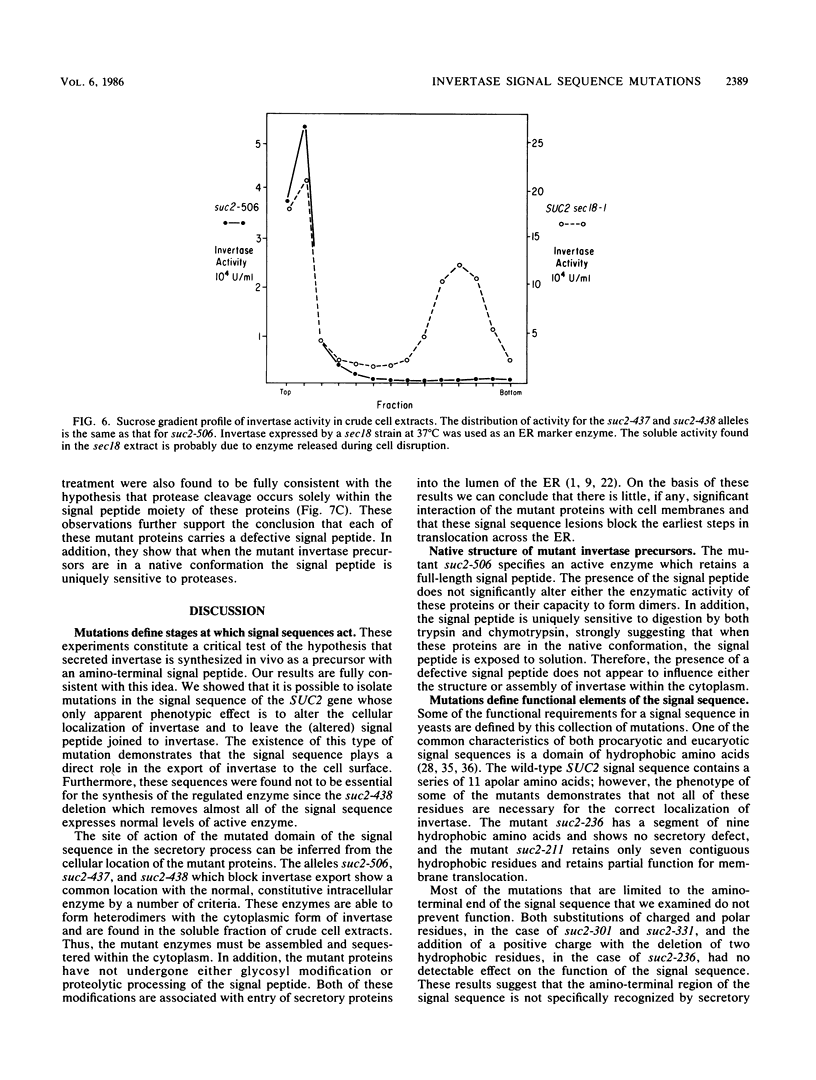


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blobel G., Dobberstein B. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J Cell Biol. 1975 Dec;67(3):835–851. doi: 10.1083/jcb.67.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Brown P. A., Halvorson H. O., Raney P., Perlman D. Conformational alterations in the proximal portion of the yeast invertase signal peptide do not block secretion. Mol Gen Genet. 1984;197(3):351–357. doi: 10.1007/BF00329928. [DOI] [PubMed] [Google Scholar]
- Carlson M., Botstein D. Two differentially regulated mRNAs with different 5' ends encode secreted with intracellular forms of yeast invertase. Cell. 1982 Jan;28(1):145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Carlson M., Osmond B. C., Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981 May;98(1):25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M., Taussig R., Kustu S., Botstein D. The secreted form of invertase in Saccharomyces cerevisiae is synthesized from mRNA encoding a signal sequence. Mol Cell Biol. 1983 Mar;3(3):439–447. doi: 10.1128/mcb.3.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE LA FUENTE G., SOLS A. Transport of sugars in yeasts. II. Mechanisms of utilization of disaccharides and related glycosides. Biochim Biophys Acta. 1962 Jan 1;56:49–62. doi: 10.1016/0006-3002(62)90526-7. [DOI] [PubMed] [Google Scholar]
- Emr S. D., Schauer I., Hansen W., Esmon P., Schekman R. Invertase beta-galactosidase hybrid proteins fail to be transported from the endoplasmic reticulum in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Nov;4(11):2347–2355. doi: 10.1128/mcb.4.11.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmon B., Novick P., Schekman R. Compartmentalized assembly of oligosaccharides on exported glycoproteins in yeast. Cell. 1981 Aug;25(2):451–460. doi: 10.1016/0092-8674(81)90063-5. [DOI] [PubMed] [Google Scholar]
- Falco S. C., Rose M., Botstein D. Homologous Recombination between Episomal Plasmids and Chromosomes in Yeast. Genetics. 1983 Dec;105(4):843–856. doi: 10.1093/genetics/105.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro-Novick S., Hansen W., Schauer I., Schekman R. Genes required for completion of import of proteins into the endoplasmic reticulum in yeast. J Cell Biol. 1984 Jan;98(1):44–53. doi: 10.1083/jcb.98.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel O., Wang S. F. Determination of enzymatic activity in polyacrylamide gels. I. Enzymes catalyzing the conversion of nonreducing substrates to reducing products. Anal Biochem. 1969 Mar;27(3):545–554. doi: 10.1016/0003-2697(69)90068-2. [DOI] [PubMed] [Google Scholar]
- Gascón S., Lampen J. O. Purification of the internal invertase of yeast. J Biol Chem. 1968 Apr 10;243(7):1567–1572. [PubMed] [Google Scholar]
- Gascón S., Neumann N. P., Lampen J. O. Comparative study of the properties of the purified internal and external invertases from yeast. J Biol Chem. 1968 Apr 10;243(7):1573–1577. [PubMed] [Google Scholar]
- Goldstein A., Lampen J. O. Beta-D-fructofuranoside fructohydrolase from yeast. Methods Enzymol. 1975;42:504–511. doi: 10.1016/0076-6879(75)42159-0. [DOI] [PubMed] [Google Scholar]
- Grossmann M. K., Zimmermann F. K. The structural genes of internal invertases in Saccharomyces cerevisiae. Mol Gen Genet. 1979 Sep;175(2):223–229. doi: 10.1007/BF00425540. [DOI] [PubMed] [Google Scholar]
- Hahn S., Hoar E. T., Guarente L. Each of three "TATA elements" specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall M. N., Gabay J., Schwartz M. Evidence for a coupling of synthesis and export of an outer membrane protein in Escherichia coli. EMBO J. 1983;2(1):15–19. doi: 10.1002/j.1460-2075.1983.tb01373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Milstein C., Brownlee G. G., Harrison T. M., Mathews M. B. A possible precursor of immunoglobulin light chains. Nat New Biol. 1972 Sep 27;239(91):117–120. doi: 10.1038/newbio239117a0. [DOI] [PubMed] [Google Scholar]
- Nagawa F., Fink G. R. The relationship between the "TATA" sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P., Ferro S., Schekman R. Order of events in the yeast secretory pathway. Cell. 1981 Aug;25(2):461–469. doi: 10.1016/0092-8674(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Novick P., Field C., Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980 Aug;21(1):205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Ottolenghi P. Some properties of five non-allelic -D-fructofuranosidases (invertases) of Saccharomyces. C R Trav Lab Carlsberg. 1971;38(13):213–221. [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983 Jun 25;167(2):391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O., Cannon L. E. Presecretory and cytoplasmic invertase polypeptides encoded by distinct mRNAs derived from the same structural gene differ by a signal sequence. Proc Natl Acad Sci U S A. 1982 Feb;79(3):781–785. doi: 10.1073/pnas.79.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. Distinct repressible mRNAs for cytoplasmic and secreted yeast invertase are encoded by a single gene. Cell. 1981 Aug;25(2):525–536. doi: 10.1016/0092-8674(81)90071-4. [DOI] [PubMed] [Google Scholar]
- Rodriguez L., Lampen J. O., MacKay V. L. SUC1 gene of Saccharomyces: a structural gene for the large (glycoprotein) and small (carbohydrate-free) forms of invertase. Mol Cell Biol. 1981 May;1(5):469–474. doi: 10.1128/mcb.1.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S., Davis R. W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimble R. B., Maley F. Subunit structure of external invertase from Saccharomyces cerevisiae. J Biol Chem. 1977 Jun 25;252(12):4409–4412. [PubMed] [Google Scholar]
- Vlasuk G. P., Inouye S., Ito H., Itakura K., Inouye M. Effects of the complete removal of basic amino acid residues from the signal peptide on secretion of lipoprotein in Escherichia coli. J Biol Chem. 1983 Jun 10;258(11):7141–7148. [PubMed] [Google Scholar]
- Walter P., Blobel G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol. 1981 Nov;91(2 Pt 1):557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. On the hydrophobic nature of signal sequences. Eur J Biochem. 1981 May 15;116(2):419–422. doi: 10.1111/j.1432-1033.1981.tb05351.x. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985 Jul 5;184(1):99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]